Abstract
Telomeres are nucleoprotein structures at the ends of eukaryotic chromosomes that protect them from degradation, end-to-end fusions, and fragility. In mammals, telomeres are composed of TTAGGG tandem repeats bound by a protein complex called shelterin, which has fundamental roles in the regulation of telomere protection and length. The telomeric repeat binding factor 1 (TERF1 or TRF1) is one of the components of shelterin and has been shown to be essential for telomere protection. Telomeric repeats can also be found throughout the genome, as Internal or Interstitial Telomeric Sequences (ITSs). Some of the components of shelterin have been described to bind to ITSs as well as other extra-telomeric regions, which in the case of RAP1 exert a key role in transcriptional regulation. Here, we set to address whether TRF1 can be found at extra-telomeric sites both under normal conditions and upon induction of telomere shortening. In particular, we performed a ChIP-sequencing technique to map TRF1 binding sites in MEFs wild-type and deficient for the telomerase RNA component (Terc−/−), with increasingly short telomeres. Our findings indicate that TRF1 is exclusively located at telomeres both under normal conditions, as well as under extreme telomere shortening. These results indicate that in mice not all members of shelterin have extra-telomeric roles as it was described for RAP1.
Keywords: ChIP-sequencing, interstitial telomeric sequences, shelterin, telomeres, telomere shortening, TRF1
Abbreviations
- ChIP
chromatin Immunoprecipitation
- FDR
false discovery rate
- ITS
interstitial telomeric sequence
- MEF
mouse embryonic fibroblast
- Terc
telomerase RNA component
- TRF1
telomeric repeat binding factor 1
Introduction
Telomeres are nucleoprotein structures at the end of the chromosomes that protect them from being detected as double-strand breaks, preventing fusions, recombination and degradation.1,2 Mammalian telomeres consist of tandem repeats of the TTAGGG sequence bound by a complex of proteins called telosome or shelterin, which has essential roles in the protection of chromosome ends and the regulation of telomere length. Telomeric repeats can be also found at internal chromosomal sites, forming the so-called Interstitial Telomeric Sequences (ITSs).3 Two classes of ITSs exist in mammals: long blocks of mainly pericentromeric, heterochromatic ITSs (het-ITSs), present only in some species, for instance the Chinese Hamster, and short ITSs, present at chromosome arms in ostensibly all vertebrates genomes. Some of these short ITSs are part of the subtelomeric (S) regions, the 3 Mbp regions adjacent to telomeres. In humans and mice, 83 and 244 non-subtelomeric short ITSs have been described, respectively,4 however, no het-ITSs have been found in these 2 species. ITSs in rodents are longer than in primates.4,5 The origin of ITSs seems to be related with chromosome fusions or telomeric hexamers insertions in the genomes during the repair of DNA double-strand breaks.6 However, their biological role still remains unknown.
The shelterin complex is composed of 6 core proteins: TRF1, TRF2, POT1, TPP1, TIN2 and RAP1.2 Only 3 of these proteins bind directly to DNA: TRF1 and TRF2, which bind to double-stranded DNA, and POT1, which binds to the single-stranded 3′ overhangs. TPP1 interacts with POT1 and TIN2. TIN2 is recruited to the telomere through binding to TRF1 and TRF2 via independent domains and is able to recruit the TPP1-POT1 complex, constituting the bridge among the different shelterin components.7-9 RAP1 is not able to directly bind DNA and its recruitment to telomeres is dependent on its interaction with TRF2.10,11
Aside from their fundamental role in telomere biology, recent evidence suggests a role of some shelterins independent of their binding to telomeric repeats. In particular, the ortholog of mammalian RAP1 in budding yeast, scRap1, binds not only telomeric but also extra-telomeric DNA, acting as a transcription factor.12,13 By analysis of in vivo RAP1 binding to chromatin using chromatin immunoprecipitation linked to sequencing (ChiP-seq), we recently described that this extra-telomeric function of RAP1 is conserved in mammals. In particular, mouse RAP1 is able to bind to inter-genic and intra-genic extra-telomeric sites, and also shows enrichment at subtelomeric regions, where RAP1 binding induces gene silencing.14 RAP1 and TRF2 also bind to ITSs outside subtelomeres.14 Similarly, a recent study in humans described that TRF2 and RAP1 proteins occupy a limited number of interstitial regions throughout the genome and also regulate gene expression.15 Interestingly, in yeast telomeric alterations can lead to delocalization from telomeres of Rap1-associated heterochromatin factors that are able to operate at interstitial genomic sites.16,17 It is also known that gradual reduction in the telomere length associated to aging is linked to global deregulation of the transcriptome and loss of maintenance of epigenetic silencing mechanisms.18
In line with extra-telomeric roles for shelterin proteins, Taz1, the fission yeast ortholog of mammalian TRF1 and TRF2, is also involved in telomere protection and recruitment of Rap1 to telomeres19,20 and was recently described to bind to the internal telomeric repeats and play an essential role in the “replication timing control” (RTC), with about half of late origins and almost all origins in subtelomeric regions being regulated by the Taz1-mediated mechanism.21 In agreement with this, a recent study reported restricted abilities of TRF1 and TRF2 proteins in binding extra-telomeric sites of the genome in human tumor cell lines, most of them ITSs. In particular, 38% of the TRF1 and TRF2 common binding sites were located in S regions.22 In addition, human TRF1 can stabilize common fragile sites.23 Furthermore, in Chinese hamster cells, which contain long blocks of het-ITSs that represent about the 5% of the genome,24 TRF1 is involved in protection of these internal non-telomeric (TTAGGG)n repeats from double-strand breaks and chromosome rearrangements.25,26 Together, these data suggest that TRF1 could bind extra-telomeric sites and have important roles in genome stability.
Here, we set to address in vivo binding of mouse TRF1 to extra-telomeric sites by using genome-wide Chip-seq analysis of mouse embryonic fibroblasts (MEF) wild-type or deficient for Trf1 as negative control. In addition, we hypothesized that upon telomere shortening, TRF1 could delocalize from telomeres to other regions of the genome, having additional functions independent from telomere biology and contributing to the gene expression changes associated with aging. To address this, we have generated MEFs deficient for the telomerase RNA component or Terc for successive mouse generations.27
Our results indicate that TRF1 does not show any significant binding to regions outside telomeric repeats, showing only binding to telomeres both in conditions of normal telomere length or when telomeres are critically short.
Results
Identification of TRF1 binding sites in MEFs by ChIP- sequencing analysis
To address the in vivo TRF1 binding to telomeric and extra-telomeric regions we performed a whole-genome ChIP sequencing analysis. To this end we used Trf1+/+ p53 −/−-Cre MEFs as well as Trf1-deficient Trf1Δ/Δ p53−/−-Cre MEFs as a negative control for TRF1 peak specificity.28 Trf1 excision in Trf1Δ/Δ p53−/−-Cre MEFs was confirmed both by PCR (Fig. 1A) and Western blot (Fig. 1B). We next tested our home-made anti-TRF1 polyclonal antibody for its ability to pull down TRF1 protein in MEFs by using dot-blot analysis. Shown in Fig. 1C, the antibody was able to specifically immunoprecipitate TRF1, which was bound to telomeric DNA but not centromeric DNA. Upon chromatin immunoprecipitation with the TRF1 antibody and before alignment of the immunoprecipitated sequences with the mouse genome, we found a strong over-representation of raw 40-base-pair (bp) sequences containing the telomeric (TTAGGG)5 or the complementary (CCCTAA)5 repeats in Trf1+/+ p53 −/−-Cre MEFs ChIP-seq compared with that in Trf1-null control (Trf1Δ/Δ p53−/−-Cre MEFs) and the input DNA (input pool 1), which corresponds to the sheared chromatin before incubation with the anti-TRF1 antibody (Fig. 2A). The enrichment in reads containing 2, 3, 4 and 6 repeats of the TTAGGG or CCCTAA sequences was also noticeable in the Trf1+/+ p53 −/−-Cre sample (Table S1). These results demonstrate in vivo binding of TRF1 to telomeric repeats.
Figure 1.
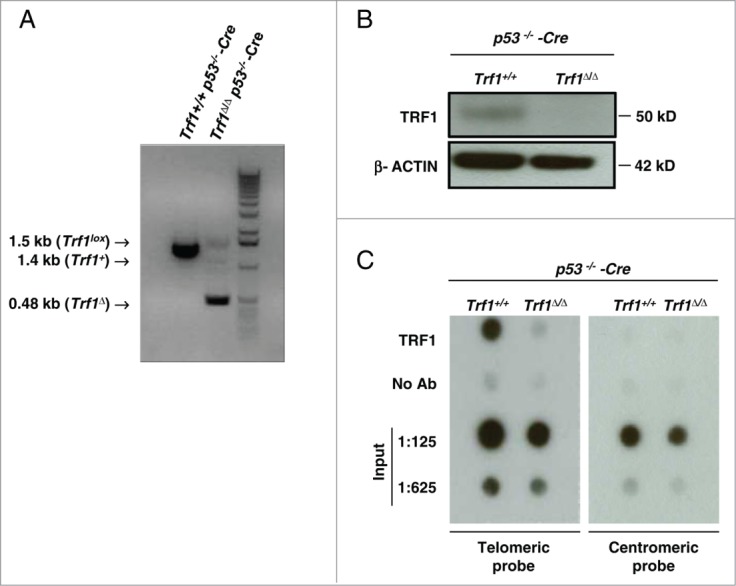
(A) Trf1 deletion in Trf1Δ/Δ p53−/− Cre-infected MEFs was confirmed by PCR. (B) Western blot analysis demonstrating the decrease in TRF1 upon infection with retroviral Cre recombinase. (C) ChIP experiment with TRF1 antibody immunoprecipitation. The co-precipitated DNA was analyzed by dot-blotting with a telomere probe to detect telomeric sequences. The same blot was hybridized with a probe against centromeric DNA (major satellite) to test the ChIP specificity, as TRF1 is bound to telomeres but not centromeres. “No ab” corresponds to the same cross-linked DNA incubated with the pre-immune TRF1 antibody serum. “Input” corresponds to the chromatin fraction before incubation with the antibody.
Figure 2.
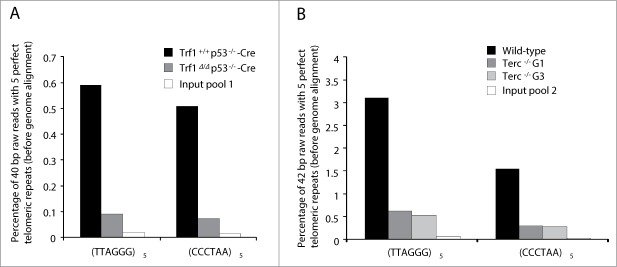
Enrichment of telomere sequences bound to TRF1 in ChIP-seq experiments. The percentage of 40 or 42-bp raw reads before alignment with mouse genome containing perfect (TTAGGG)5 or (CCCTAA)5 repeats is shown for the different samples. In the first experiment, input pool 1 is a combination of equal amounts of input DNA (total DNA before immunoprecipitation) from Trf1+/+ p53−/−-Cre and Trf1Δ/Δ p53−/−-Cre samples (A). In the second experiment, input pool 2 is a combination of input DNA from wild-type, Terc−/− G1 and Terc−/− G3 samples (B).
Our ChIP-seq experiment yielded >11 × 106 uniquely mapped short reads for Trf1+/+ p53−/−-Cre, >32 × 106 for Trf1Δ/Δ p53−/−-Cre and almost 20 × 106 for input pool 1 (Table 1). Significantly read-enriched peaks were detected using the MACS v1.4 software with a P value cut-off of 1 × 10−5. To define extra-telomeric TRF1 binding sites, we retained only peaks that were present in Trf1+/+ p53−/−-Cre sample but not in Trf1-null sample (Trf1+/+ p53−/−-Cre vs Trf1Δ/Δ p53−/−-Cre). In the comparisons with the controls, the statistical significance of enriched sites is measured by empirical FDR, which is the expected proportion of incorrectly identified sites among those found to be significant. A False Discovery Rate (FDR) of 10 was applied for this software. 1,165 peaks were found in the comparison Trf1+/+ p53−/−-Cre vs Trf1Δ/Δ p53−/−-Cre. We also did the comparison Trf1+/+ p53−/−-Cre vs input pool 1, in this case obtaining 1,288 peaks. Out of the 1,165 peaks from the comparison with the Trf1-null control, 290 were discarded in the comparison with the input. Unfortunately, all the 875 remaining peaks showed a FDR of 100%, meaning that TRF1 specific peaks did not have enough statistical support in the comparison with Trf1-null sample.
Table 1.
Summary of ChIP-seq experiments. Overall reads and length obtained in ChIP-seq experiments. % of alignment (PF) is the percentage of filtered reads that were uniquely aligned to the reference. In the first experiment, read length of 40 bp, in the second, 42 bp
| Sample ID | Raw sequenced reads | Size of raw sequenced reads (bp) | % of alignment (PF) | Uniquely mapped reads | Size of uniquely mapped reads (bp) |
|---|---|---|---|---|---|
| Trf1+/+ p53−/−-Cre | 21,493,914 | 859,756,560 | 58.36 | 11,170,552 | 446,822,080 |
| Trf1Δ/Δ p53−/−-Cre | 52,426,641 | 2,097,065,640 | 91.51 | 32,673,672 | 1,306,946,880 |
| Input Pool 1 | 37,408,510 | 1,496,340,400 | 71.53 | 19,932,267 | 797,290,680 |
| Wildtype | 29,609,663 | 1,243,605,846 | 58.54 | 14,498,028 | 608,917,176 |
| Terc−/−G1 | 31,473,982 | 1,321,907,244 | 63.43 | 16,542,830 | 694,798,860 |
| Terc−/−G3 | 30,741,283 | 1,291,133,886 | 59.88 | 15,476,230 | 650,001,660 |
| Input Pool 2 | 35,229,238 | 1,479,627,996 | 73.78 | 21,381,218 | 898,011,156 |
Contrary to what was reported for mouse RAP114 and human RAP1, TRF2 and TRF1,15,22 mouse TRF1 peaks were not enriched in subtelomeric regions (Fig. S1), as we only found 29 peaks (2.36% from the total of peaks) in S regions. If the same analysis was done randomly, the expected enrichment would be that 2.27% of the peaks would be subtelomeres, so there is no enrichment of peaks in subtelomeric regions. In chromosomes 4 and 8 there is a slight enrichment of 6% and 5.4% respectively. Besides, most of them were not associated with genes, as mapped in regions at more than 10 kb from genes transcription start sites (TSS), with just 13% of the peaks (117 peaks out of 875) at less than 10 kb from genes TSS (Fig. S2A). From the peaks that mapped inside a gene, 90% of them were intronic (Fig. S2B). Taking all these results together, we conclude that mouse TRF1 does not bind to chromosomal regions other than the telomeres in wild-type MEFs.
Identification of TRF1 binding sites upon telomere shortening by ChIP-sequencing
In order to elucidate whether telomere shortening could result in redistribution of TRF1 binding from telomeres to chromosome arms, we performed a second ChIP-seq experiment in Trf1+/+ MEFs Terc+/+ (wild-type), and Terc−/−of the first (Terc−/− G1) and third (Terc−/− G3) generation, which display progressively shorter telomeres.
Again, we first confirmed the enrichment in telomeric repeats before sequence alignment with the mouse genome in wild-type, Terc−/− G1 and Terc−/− G3 MEFs compared with their corresponding input DNA (input pool 2). In agreement with shorter telomeres owing to telomerase deficiency, Terc−/− G1 and G3 samples showed gradual decrease in the percentage of reads containing perfect telomeric repeats (Fig. 2B). A similar enrichment was seen for 2, 3, 4 and 6 repeats of the TTAGGG or CCCTAA sequences (Table S2).
This ChIP-seq experiment yielded >14 × 106 uniquely mapped short reads for wild-type, >16 × 106 for Terc−/− G1, >15 × 106 for Terc−/− G3 and >21 × 106 for input pool 2 (Table 1). Peak detection and statistical analysis was done as described above. We compared Terc−/− G3 vs wild-type, Terc−/− G1 vs wild-type and Terc−/− G3 vs Terc−/− G1, so as to obtain new TRF1 binding sites to the genome while telomeres get shorter. We obtained 2,179 peaks in the comparison Terc−/− G3 vs wild-type, 2,207 peaks in the comparison Terc−/− G1 vs wild-type and 2,524 peaks in the comparison Terc−/− G3 vs Terc−/− G1. However, none of these peaks were of statistical significance and were discarded from the analysis. Once again, we did not see any enrichment in S regions in the comparison Terc−/− G3 vs wild-type (1.93% compared to 2.27% of the random average) (Fig. S3), suggesting that telomeric shortening and the subsequently decreased binding of TRF1 to the telomere does not induce TRF1 interaction with other regions of the genome.
Overlapping with mouse RAP1 ChIP-seq
To further analyze whether TRF1 binding to chromatin is restricted to telomeres in mice, we compared our TRF1 ChIP-seq data with that previously obtained by us for RAP114 and sought peaks that overlapped in both experiments. Just a few TRF1 regions (14 sites, 1.6% of the 875 peaks from the comparison Trf1+/+ p53−/−-Cre vs Trf1Δ/Δ p53−/−-Cre) were found to overlap with RAP1 binding sites and only 5 of them contained telomeric (TTAGGG)n≥2 tracks (Table 2). While dozens of peaks associated to ITSs were found in RAP1 ChIP-seq, in TRF1 ChIP-seq we only detected a few peaks in ITSs and with no statitistical significance. In this comparison, we also noticed that peaks obtained in TRF1 ChIP-seq are much longer than RAP1 peaks (RAP1 peaks had an average width of 75 bp, while TRF1 peaks are 576 bp on average). This distribution in broad peaks suggests that peaks obtained in this experiment are peaks associated with background instead of peaks showing real binding of TRF1. Lastly, RAP1 ChIP-seq showed a much higher number of RAP1 binding peaks (30,000 peaks) compared to only 1,165 TRF1 binding peaks.
Table 2.
Peaks that overlap with RAP1 ChIP-seq in the Trf1+/+ p53−/−-Cre vs Trf1Δ/Δ p53−/−-Cre comparison and that contain tracks with (TTAGGG)n ≥ 2 repeats
| Chr | RAP1 Peak number | Start | End | TRF1 Peak number | Start | End | FDR (%) |
|---|---|---|---|---|---|---|---|
| 2 | 4 | 28,040,118 | 28,040,149 | 603 | 28,039,717 | 28,040,535 | 100 |
| 2 | 1 | 57,482,074 | 57,482,124 | 620 | 57,481,892 | 57,482,277 | 100 |
| 6 | 2027 | 4,873,918 | 4,873,946 | 874 | 4,873,501 | 4,874,305 | 100 |
| 8 | 2 | 73,957,000 | 73,957,029 | 980 | 73,956,797 | 73,957,467 | 100 |
| 9 | 7 | 95,326,334 | 95,326,363 | 1073 | 95,326,167 | 95,326,779 | 100 |
Chip-seq validation by qRT-PCR
Even though none of the TRF1 peaks obtained by ChiP-seq passed the statistical significance threshold, we decided to further study those TRF1 peaks that overlapped with the RAP1 ChIP-seq peaks and that corresponded to ITS or subtelomeric regions to be certain that TRF1 does not show significant binding outside of telomeres. We tested the 5 peaks associated to TTAGGG repeats that overlapped with RAP1: peak 603 (chr2: 28,039,717- 28,040,535), peak 620 (chr2: 57,481,892-57,482,277), peak 874 (chr6: 52,133,777-52,134,592), peak 980 (chr8: 73,956,797-73,957,467) and peak 1073 (chr9: 95,326,167- 95,326,779) (Table 2). In addition, we also selected for validation peaks in the subtelomeric regions of chromosome 4 and 8, which were the chromosomes that showed a slight enrichment in peaks in the S regions: peak 804 (chr4: 153,217,553-153,218,190), peak 805 (chr4: 154,765,741-154,766,224), peak 806 (chr4: 155,161,073-155,161,670), peak 807 (chr4: 155,334,959-155,335,528), peak 1014 (chr8: 128,849,051-128,849,629), peak 1015 (chr8: 129,108,238-129,108,707) and peak 1016 (chr8: 129,423,259-129, 423,839) (Table 3).
Table 3.
Peaks from subtelomeres of chromosomes 4 and 8, which are the chromosomes that showed higher enrichment of peaks in S regions in the comparison Trf1+/+ p53−/−-Cre vs Trf1Δ/Δ p53−/−-Cre
| Chr | Peak number | Start | End | FDR (%) |
|---|---|---|---|---|
| 4 | 804 | 153,217,553 | 153,218,190 | 100 |
| 4 | 805 | 154,765,741 | 154,766,224 | 100 |
| 4 | 806 | 155,161,073 | 155,161,670 | 100 |
| 4 | 807 | 155,334,959 | 155,335,528 | 100 |
| 8 | 1014 | 128,849,051 | 128,849,629 | 100 |
| 8 | 1015 | 129,108,238 | 129,108,707 | 100 |
| 8 | 1016 | 129,423,259 | 129,423,839 | 100 |
| 8 | 1017 | 131,239,943 | 131,240,518 | 100 |
To study TRF1 binding to these peaks, we carried out independent ChIP experiments with the TRF1 antibody in 2 independent MEFs per genotype: Trf1+/+ p53−/−-Cre MEFs and Trf1Δ/Δ p53−/−-Cre MEFs deficient for Trf1 as negative control for peak specificity, followed by qPCR analysis with primers for the extra-telomeric regions mentioned above (see primer sequences in Table S3). We failed to detect TRF1 binding to the indicated ITSs (Fig. 3A) or to subtelomeric regions (Fig. 3B), thus confirming that the peaks obtained in the ChIP-seq were false positives in agreement with the FDR indicator.
Figure 3.
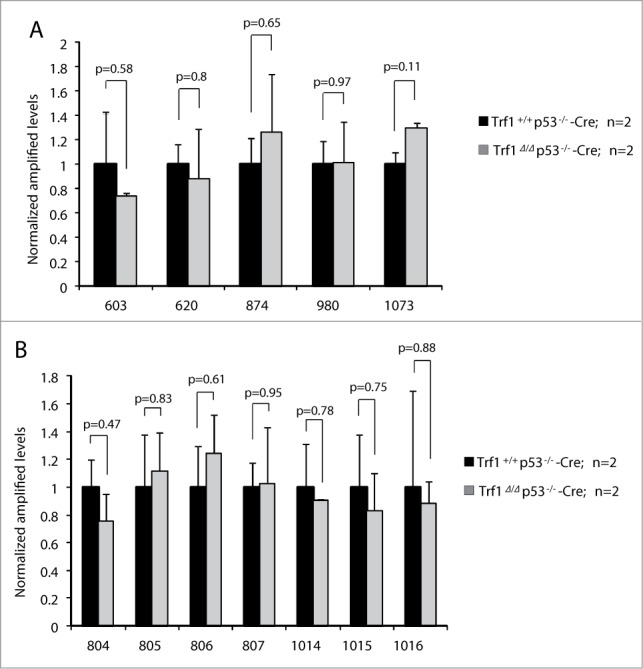
Validation of TRF1-binding peaks by ChIP with anti-TRF1 antibody followed by qPCR. The peak ranks of the regions tested are indicated on the x axis. (A) Validation of peaks that overlapped with RAP1 ChIP-seq and contain (TTAGGG)n≥2 repeats. (B) Validation of peaks from subtelomeric regions of chromosomes 4 and 8. The results were normalized to the input and relativized to wild-type levels. No decrease in TRF1 binding was detected in Trf1-null MEFs. n = number of independent MEFs analyzed. Error bars: s.e.m. P values were calculated by Student's t test.
Discussion
The aim of this study was to establish the genome-wide DNA binding patterns of the telomeric protein TRF1 in MEFs and find out whether telomere shortening could induce delocalization of TRF1 from telomeres and binding to other regions in the genome. When we analyzed TRF1 specific binding sites by ChIP sequencing, however, none of them had enough statistical significance and were statistically considered false positives. In agreement with this, we could not validate the TRF1 peaks by ChIP-qPCR, further indicating that mouse TRF1 binding to chromatin is restricted to telomeres, at least in MEFs. This is in apparent conflict with what was previously observed in a work done in humans, where they identified a limited number of extra-telomeric TRF1 binding sites that largely comprised ITSs in human tumor cell lines.22 Nevertheless, we have to take into consideration that not only protein binding profiles but also ITSs structure and length differ between humans and mice, which may explain the differences in TRF1 binding observed in both species. The fact that the human studies were performed in transformed cell lines, while we studied TRF1 binding to chromatin in normal primary cells, may also account for the different results, as the specific cellular context may have an effect on TRF1 levels and its binding to chromatin. It has also been reported that TRF1 binds to ITSs in immortalized Chinese hamster cells, where it is involved in the stability of these repeated sequences.25,26 Unlike mouse cells, however, the Chinese hamster cells contain large blocks of cytologically detectable het-ITSs that correspond to the 5% of their genome, which probably have completely different stabilization and protection mechanisms, and this may also explain the difference with our study.
Finally, we found very few TRF1 peaks associated with ITSs in MEFs when compared with an analogous RAP1 ChIP-seq also in MEFs and we could not validate them by ChIP-RT-PCR, suggesting that indeed those putative TRF1 peaks may be artifacts of the sequencing technique owing to erroneous alignment of telomeres to ITSs rather than bona fide TRF1 binding sites. When studying genome-wide DNA binding profiles, protein concentration can be critical in the identification of statistically significant peaks. In fact, in a ChIP-seq experiment done with anti-TRF2 and anti-RAP1 antibodies in humans, they demonstrated that TRF2 concentration could considerably influence this protein's binding to ITSs.15 Even so, we previously published a TRF1 ChIP-seq done in induced Pluripotent Stem (iPS) cells, which display high TRF1 levels, showing no significant TRF1 binding to putative regulatory regions nor intragenic binding of this protein outside telomeric regions.29
Given our negative findings with TRF1, it would be interesting to study the genome-wide DNA binding profiles for TRF2, other shelterin that binds to double-stranded DNA and recruits RAP1 to telomeres. In fact, we previously demonstrated that TRF2 binds to TTAGGG-rich extra-telomeric RAP1 binding sites.14
Telomere shortening is accompanied by a change in the architecture of telomeric and subtelomeric chromatin leading to a more “open” chromatin state. However, this loss of telomeric repeats in G2 and G5 Terc−/− MEFs has no effect on TRF1 density at telomeres, inferring that no alterations of TRF1 binding to telomeres are associated with telomere shortening.30 Besides, it is known that TRF1 protein levels are significantly reduced in late generation Terc−/− mice.31 These 2 data together with our results suggest that TRF1 expression and degradation are highly regulated in mice and that telomeric shortening induces a decrease in TRF1 levels rather than a relocalization of this protein to other regions in the genome. Further studies of the extra-telomeric binding of other shelterins in a context of telomere shortening and other pathological conditions are needed. Besides, a deeper knowledge of the structure, function and stability of these ITSs in the different species will help us to understand the dissimilar binding of shelterins to these regions.
Experimental Procedures
Cell culture and retroviral infections
Cells were cultured in complete standard DMEM medium (Gibco) supplemented with 10% of FBS (Gibco) and antibiotic-antimicotic solution (Gibco). MEFs were isolated from Trf1lox/lox p53−/−, Trf1+/+ p53−/−, wild-type, first and third generation of telomerase null (Terc−/− G1 and G3) embryos at day 13.5 as described.27 Retroviral supernatants were produced in HEK293T cells (12.5 × 106 cells per 150 mm diameter dish) transfected with the ecotropic packaging plasmid and pBabe-Cre. Trf1lox/lox p53−/− and Trf1+/+ p53−/− MEFs were seeded the following day (2 × 106 cells per 150 mm diameter dish) and infected after 24 hours with the diluted retroviral supernatants (2/5). Infected cells were selected by addition of puromycin (2 μg/ml) after 48 hours. The removal of exon 1 of Trf1 in Trf1lox/lox MEFs upon retroviral infection with Cre recombinase was confirmed by PCR with E1-Popout (Forward, 5′-ATAGTGATCAAAATGTGGTCCTGGG-3′) and SAR1 (Reverse, 5′-GCTTGCCAAATTGGGTTGG-3′) primers as previously described.28
Western blotting
Nuclear protein extracts prepared as described32,28 were used for Western blot analysis. Protein concentration was determined using the Bradford assay (Sigma). 30 μg of protein per extract were loaded in a NuPAGE 4–12% Bis-Tris gel 1.0 mm (Invitrogen) and electrophoresed in MES SDS Running Buffer (Invitrogen) before Western blotting. The following antibodies were used: for TRF1, Abcam, ab-10579 (1:1000) and for β-actin, Sigma, a2228 (1:10,000).
Chromatin immunoprecipitation assays and telomere dot-blots
ChIP assays were carried out essentially as described previously,33 with some modifications. Briefly, we cross-linked and sonicated each sample for 50 minutes using the Bioruptor sonication system (Diagenode). Chromatin from 4 × 106 cells was used per immunoprecipitation. We pre-cleared each immunoprecipitation with 50 μl of protein A/G Plus agarose beads (Santa Cruz Biotechnology, sc-2003) and incubated with 4 μl of rabbit polyclonal anti-mouse TRF1 serum antibody (generated in our laboratory)33 or preimmune serum. Inputs correspond to the total DNA sample, 1:10 dilution of the amount of lysate used in the immunoprecipitation. The precipitated DNA was eluted and transferred to a Hybond+ membrane by dot-blotting. The membrane was then hybridized with either a telomeric probe recognizing TTAGGG repeats or a probe recognizing major satellite sequences, characteristic of pericentric heterochromatin.
ChIP-sequencing
For each sample we used around 100 × 106 cells. Each sample was independently processed into sequencing libraries with a ChIP-Seq sample preparation kit (Illumina) in accordance with the manufacturer's instructions.34 Inputs samples from Trf1+/+ p53−/−-Cre and Trf1Δ/Δ p53−/−-Cre were pooled in a single library (input pool 1) and inputs from wild-type, Terc−/− G1 and Terc−/− G3 MEFs in another library (input pool 2). 29, 37, 40, 16, 15, 26 and 40 ng of DNA (as quantitated by PicoGreen Fluorometry) were respectively used for Trf1+/+ p53−/−-Cre, Trf1Δ/Δ p53−/−-Cre, input pool 1, wild-type, Terc−/− G1, Terc−/− G3 and input pool 2 samples. Each sample was electrophoresed on agarose gel and a fraction of 100–150 bp was taken. Extracted DNA was processed through subsequent enzymatic treatments of end-repair, dA-tailing, and ligation to adapters as in Illumina's “ChIP Sequencing Sample Prep Guide” (part # 11257047 Rev. A), with the exception that no further gel extraction was performed. Adapter-ligated library was PCR amplified with Illumina PE primers. The resulting purified DNA libraries were applied to an Illumina flow cell for cluster generation and sequenced on a Genome Analyzer IIx (GA2) by following manufacturer's protocol and using 40 (first ChIP-seq experiment) or 42 (second experiment)-base read run.
Data analysis
Image analysis was performed with Illumina Real Time Analysis software (RTA1.8 for Trf1Δ/Δ p53−/−-Cre, Trf1+/+ p53−/−-Cre and input pool 1 samples and RTA1.6 for wild-type, Terc−/− G1, Terc−/− G3 and input pool 2 samples). Sequence alignment to the reference genome (NCBI m37/mm9 mouse assembly, April 2007, strain C57BL/6J) was made with Illumina's ELANDv2 algorithm on its “eland_extended” mode from within CASAVA-1.7 package. ELANDv2 performs multiseed alignment with consecutive read substrings of 16 to 32 bases separately. The seeds are aligned to multiple candidate positions in the reference genome, with a maximum of 2 mismatches allowed per 32 bases seed; then they are extended to the full read using gapped alignment, allowing for any number of mismatches and potential gaps (indels) of up to 20 bases. The best alignment among the multiple candidate positions is chosen based on quality scores. Uniquely aligned 40 or 42-bp-length reads were pooled into 7 datasets: Trf1+/+ p53−/−-Cre, Trf1Δ/Δ p53−/−-Cre, input pool 1, wild-type, Terc−/− G1 and Terc−/− G3 and input pool 2. Peak detection was performed with MACS version 1.4 software (p value = 1 × 10−5; FDR = 10%; 300-bp window).
Validation of ChIP-seq results by real-time qPCR
For ChIP-seq validation, real-time qPCR was performed using an ABI PRISM 7700 thermocycler (Applied Biosystems) and the Power SYBR Green PCR Master mix (Life technologies) according to the manufacturer's protocol. Immunoprecipitated and input DNA from independent ChIP experiments in Trf1+/+ p53−/−-Cre and Trf1Δ/Δ p53−/−-Cre samples were quantified by qPCR with oligos designed to amplify DNA fragments corresponding to the following peaks: peak 603 (chr2: 28,039,717- 28,040,535), peak 620 (chr2: 57,481,892-57,482,277), peak 874 (chr6: 52,133,777- 52,134,592), peak 980 (chr8: 73,956,797- 73,957,467), peak 1073 (chr9: 95,326,167- 95,326,779), peak 804 (chr4: 153,217,553- 153,218,190), peak 805 (chr4: 154,765,741- 154,766,224), peak 806 (chr4: 155,161,073- 155,161,670), peak 807 (chr4: 155,334,959- 155,335,528), peak 1014 (chr8: 128,849,051-128,849,629), peak 1015 (chr8: 129,108,238- 129,108,707) and peak 1016 (chr8: 129,423,259- 129, 423,839). The primers sequences are listed in Supplementary Table S3. The results were normalized to the value obtained in the input DNA, by calculating de ΔCt values between the levels obtained in input DNA and that of the precipitated DNA. The results were relativized to wild-type levels. All values were obtained in triplicates.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
IG is the recipient of a “Carlos III Institute” PhD fellowship. Research in the Blasco lab is funded by the Spanish Ministry of Economy and Competitiveness Projects SAF2008-05384 and CSD2007-00017, the Madrid Regional Government Project S2010/BMD-2303 (ReCaRe), The European Research Council (ERC) Project GA#232854 (TEL STEM CELL), the Preclinical Research Award from Fundación Lilly (Spain), Fundación Botín (Spain), the European Union FP7 projects 2007-A-20088 (MARK-AGE) and 2010-259749 (EuroBATS) and the AXA Research Fund (Life Risks Project).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Chan SW, Blackburn EH. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene 2002; 21:553-63; PMID:11850780; http://dx.doi.org/ 10.1038/sj.onc.1205082 [DOI] [PubMed] [Google Scholar]
- 2. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 2005; 19:2100-10; PMID:16166375; http://dx.doi.org/ 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- 3. Meyne J, Baker RJ, Hobart HH, Hsu TC, Ryder OA, Ward OG, Wiley JE, Wurster-Hill DH, Yates TL, Moyzis RK. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma 1990; 99:3-10; PMID:2340757; http://dx.doi.org/ 10.1007/BF01737283 [DOI] [PubMed] [Google Scholar]
- 4. Nergadze SG, Santagostino MA, Salzano A, Mondello C, Giulotto E. Contribution of telomerase RNA retrotranscription to DNA double-strand break repair during mammalian genome evolution. Genome Biol 2007; 8:R260; PMID:18067655; http://dx.doi.org/ 10.1186/gb-2007-8-12-r260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruiz-Herrera A, Nergadze SG, Santagostino M, Giulotto E. Telomeric repeats far from the ends: mechanisms of origin and role in evolution. Cytogenet Genome Res 2008; 122:219-28; PMID:19188690; http://dx.doi.org/ 10.1159/000167807 [DOI] [PubMed] [Google Scholar]
- 6. Messier W, Li SH, Stewart CB. The birth of microsatellites. Nature 1996; 381:483; PMID:8632820; http://dx.doi.org/ 10.1038/381483a0 [DOI] [PubMed] [Google Scholar]
- 7. Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 2008; 319:1092-6; PMID:18202258; http://dx.doi.org/ 10.1126/science.1151804 [DOI] [PubMed] [Google Scholar]
- 8. Kim SH, Beausejour C, Davalos AR, Kaminker P, Heo SJ, Campisi J. TIN2 mediates functions of TRF2 at human telomeres. J Biol Chem 2004; 279:43799-804; PMID:15292264; http://dx.doi.org/ 10.1074/jbc.M408650200 [DOI] [PubMed] [Google Scholar]
- 9. Ye JZ, Hockemeyer D, Krutchinsky AN, Loayza D, Hooper SM, Chait BT, de Lange T. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2TRF1 complex. Genes Dev 2004; 18:1649-54; PMID:15231715; http://dx.doi.org/ 10.1101/gad.1215404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol 2005; 7:712-8; PMID:15968270; http://dx.doi.org/ 10.1038/ncb1275 [DOI] [PubMed] [Google Scholar]
- 11. Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell 2000; 101:471-83; PMID:10850490; http://dx.doi.org/ 10.1016/S0092-8674(00)80858-2 [DOI] [PubMed] [Google Scholar]
- 12. Buchman AR, Lue NF, Kornberg RD. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol 1988; 8:5086-99; PMID:3072472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Capieaux E, Vignais ML, Sentenac A, Goffeau A. The yeast H+-ATPase gene is controlled by the promoter binding factor TUF. J Biol Chem 1989; 264:7437-46; PMID:2523395 [PubMed] [Google Scholar]
- 14. Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, Dominguez O, Pisano DG, Tarsounas M, Blasco MA. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol 2010; 12:768-80; PMID:20622869; http://dx.doi.org/ 10.1038/ncb2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. Human telomeric proteins occupy selective interstitial sites. Cell Res 2011; 21:1013-27; PMID:21423278; http://dx.doi.org/ 10.1038/cr.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes Dev 1996; 10:1796-811; PMID:8698239; http://dx.doi.org/ 10.1101/gad.10.14.1796 [DOI] [PubMed] [Google Scholar]
- 17. Marcand S, Buck SW, Moretti P, Gilson E, Shore D. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev 1996; 10:1297-309; PMID:8647429; http://dx.doi.org/ 10.1101/gad.10.11.1297 [DOI] [PubMed] [Google Scholar]
- 18. Schoeftner S, Blanco R, Lopez de Silanes I, Munoz P, Gomez-Lopez G, Flores JM, Blasco MA. Telomere shortening relaxes X chromosome inactivation and forces global transcriptome alterations. Proc Natl Acad Sci U S A 2009; 106:19393-8; PMID:19887628; http://dx.doi.org/ 10.1073/pnas.0909265106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cooper JP, Nimmo ER, Allshire RC, Cech TR. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature 1997; 385:744-7; PMID:9034194; http://dx.doi.org/ 10.1038/385744a0 [DOI] [PubMed] [Google Scholar]
- 20. Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 2001; 11:1624-30; PMID:11676925; http://dx.doi.org/ 10.1016/S0960-9822(01)00503-6 [DOI] [PubMed] [Google Scholar]
- 21. Tazumi A, Fukuura M, Nakato R, Kishimoto A, Takenaka T, Ogawa S, Song JH, Takahashi TS, Nakagawa T, Shirahige K, et al. Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes Dev 2012; 26:2050-62; PMID:22987637; http://dx.doi.org/ 10.1101/gad.194282.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simonet T, Zaragosi LE, Philippe C, Lebrigand K, Schouteden C, Augereau A, Bauwens S, Ye J, Santagostino M, Giulotto E, et al. The human TTAGGG repeat factors 1 and 2 bind to a subset of interstitial telomeric sequences and satellite repeats. Cell Res 2011; 21:1028-38; PMID:21423270; http://dx.doi.org/ 10.1038/cr.2011.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bosco N, de Lange T. A TRF1-controlled common fragile site containing interstitial telomeric sequences. Chromosoma 2012; 121:465-74; PMID:22790221; http://dx.doi.org/ 10.1007/s00412-012-0377-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Day JP, Limoli CL, Morgan WF. Recombination involving interstitial telomere repeat-like sequences promotes chromosomal instability in Chinese hamster cells. Carcinogenesis 1998; 19:259-65; PMID:9498274; http://dx.doi.org/ 10.1093/carcin/19.2.259 [DOI] [PubMed] [Google Scholar]
- 25. Krutilina RI, Oei S, Buchlow G, Yau PM, Zalensky AO, Zalenskaya IA, Bradbury EM, Tomilin NV. A negative regulator of telomere-length protein trf1 is associated with interstitial (TTAGGG)n blocks in immortal Chinese hamster ovary cells. Biochem Biophys Res Commun 2001; 280:471-5; PMID:11162541; http://dx.doi.org/ 10.1006/bbrc.2000.4143 [DOI] [PubMed] [Google Scholar]
- 26. Krutilina RI, Smirnova AN, Mudrak OS, Pleskach NM, Svetlova MP, Oei SL, Yau PM, Bradbury EM, Zalensky AO, Tomilin NV. Protection of internal (TTAGGG)n repeats in Chinese hamster cells by telomeric protein TRF1. Oncogene 2003; 22:6690-8; PMID:14555982; http://dx.doi.org/ 10.1038/sj.onc.1206745 [DOI] [PubMed] [Google Scholar]
- 27. Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 1997; 91:25-34; PMID:9335332; http://dx.doi.org/ 10.1016/S0092-8674(01)80006-4 [DOI] [PubMed] [Google Scholar]
- 28. Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernández-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev 2009; 23:2060-75; PMID:19679647; http://dx.doi.org/ 10.1101/gad.543509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schneider RP, Garrobo I, Foronda M, Palacios JA, Marion RM, Flores I, et al. TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nat Commun 2013; 4:1946; PMID:23735977; http://dx.doi.org/ 10.1038/ncomms2946 [DOI] [PubMed] [Google Scholar]
- 30. Benetti R, Garcia-Cao M, Blasco MA. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat Genet 2007; 39:243-50; PMID:17237781; http://dx.doi.org/ 10.1038/ng1952 [DOI] [PubMed] [Google Scholar]
- 31. Franco S, Alsheimer M, Herrera E, Benavente R, Blasco MA. Mammalian meiotic telomeres: composition and ultrastructure in telomerase-deficient mice. Eur J Cell Biol 2002; 81:335-40; PMID:12113474; http://dx.doi.org/ 10.1078/0171-9335-00259 [DOI] [PubMed] [Google Scholar]
- 32. Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol 2000; 20:8602-12; PMID:11046155; http://dx.doi.org/ 10.1128/MCB.20.22.8602-8612.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 2004; 36:94-9; PMID:14702045; http://dx.doi.org/ 10.1038/ng1278 [DOI] [PubMed] [Google Scholar]
- 34. Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat Method 2008; 5:1005-10; PMID:19034268; http://dx.doi.org/ 10.1038/nmeth.1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


