Abstract
Nuf2 plays an important role in kinetochore-microtubule attachment and thus is involved in regulation of the spindle assembly checkpoint in mitosis. In this study, we examined the localization and function of Nuf2 during mouse oocyte meiotic maturation. Myc6-Nuf2 mRNA injection and immunofluorescent staining showed that Nuf2 localized to kinetochores from germinal vesicle breakdown to metaphase I stages, while it disappeared from the kinetochores at the anaphase I stage, but relocated to kinetochores at the MII stage. Overexpression of Nuf2 caused defective spindles, misaligned chromosomes, and activated spindle assembly checkpoint, and thus inhibited chromosome segregation and metaphase-anaphase transition in oocyte meiosis. Conversely, precocious polar body extrusion was observed in the presence of misaligned chromosomes and abnormal spindle formation in Nuf2 knock-down oocytes, causing aneuploidy. Our data suggest that Nuf2 is a critical regulator of meiotic cell cycle progression in mammalian oocytes.
Keywords: kinetochore-microtubule attachments, meiosis, Nuf2, oocyte, spindle assembly checkpoint
Introduction
Genomic stability of a cell significantly depends on accurate chromosome segregation, which requires dynamic associations between microtubules and the kinetochores. A large protein structure established in the centromere region of each chromosome serves as critical link between microtubules and kinetochores.1,2 Kinetochores have a remarkable ability to track microtubule ends, thereby contributing to the mechanisms that pull the chromosome to the poles. Furthermore, kinetochores can also detect whether the attached microtubules are generating tension and thus to ensure that microtubules are bound correctly to the kinetochores. If this is not the case, the spindle assembly checkpoint (SAC) will exert its key role in ensuring the fidelity of this process.3,4 SAC surveys the interactions between chromosomes and microtubules, and will delay cell cycle timing to allow additional time for corrections.5-8 The Ndc80 complex has been identified as a bridge of kinetochore-microtubule attachment and a necessary factor for SAC regulation.9-14
Nuf2 is the core component of the Ndc80 complex,15,16 and the complex is a heterotetramer of Ndc80 (also called Hec1 in humans15), Nuf2, Spc24 and Spc25.17,18 The Ndc80 and Nuf2 proteins compose a dimer that associates with Spc24 and Spc25.19,20 The head of Ndc80 and Nuf2 form a microtubule binding domain,21 while the Spc24 and Spc25 form a centromere-binding domain.22,23 In HeLa cells, when Nuf2 is depleted by RNA interference, spindle formation occurs normally, but kinetochores fail to form attachments to spindle microtubules and fail to activate the spindle assembly checkpoint.10,11,24 In S. cerevisiae and X. laevis, experiments revealed that siRNA depletion of Nuf2 or Ndc80 inactivated the spindle assembly checkpoint while multiple outer kinetochore proteins failed to properly function.13,25 Another RNAi study in HeLa cells demonstrated the necessity of HsNuf2 in stabilizing microtubule-kinetochore attachments, maintaining tension, and silencing the spindle checkpoint.26 In addition, perturbation of Ndc80 and Spc25 in mouse oocytes disrupted chromosome alignment and proper spindle checkpoint signaling.27,28 Therefore, we hypothesized that Nuf2 may be a critical player in the regulation of kinetochore-microtubule attachment and spindle assembly checkpoint function in female germ cell meiosis.
In the present study, Myc6-Nuf2 mRNA was injected into mouse oocytes to overexpress Nuf2 and siRNA was employed to suppress Nuf2 function in order to clarify the role of Nuf2 during meiosis. We determined that Nuf2 is required for the regulation of meiotic progression and for ensuring high fidelity segregation of chromosomes in mouse oocytes.
Results
Expression and subcellular localization of Nuf2 during oocyte meiotic maturation
To investigate the role of Nuf2 during meiosis, the expression level and subcellular localization of this protein was first examined. A total of 100 oocytes were collected after oocytes had been cultured for 0, 4, 8, 9.5 and 12 h, corresponding to GV, GVBD, MI, ATI and MII stages, respectively. As shown in Figure 1A, the mRNA level was moderate at GV and GVBD stages, and reached the highest level at MI and MII stages. At ATI, Nuf2 was reduced to a similar level as seen during the GV stage. Compared to the GV oocytes, the mRNA levels of Nuf2 in MI and MII oocytes were significantly increased (288.23 ± 93.71%, 727.82 ± 187.24% vs. 100%, P < 0.05). Because of the limited availability of workable antibody for immunofluorescent staining to determine the subcellular localization of Nuf2, a low concentration of exogenous Myc6-Nuf2 mRNA in nuclease-free water was injected into oocytes at the GV stage; for controls, only nuclease-free water was injected at the same amount. Anti-Myc6 antibody was used to detect the subcellular localization of Myc6-Nuf2. In the GV oocytes, no evident signal of Myc6-Nuf2 was detected (Fig. 1Ba). Shortly after GVBD (2–3 h in maturation culture medium), clear staining was observed at the kinetochores (Fig. 1Bb). At the pro-MI and MI stage, the signal of Myc6-Nuf2 was obvious at the kinetochores (Fig. 1Bc.d). However, at the AI/TI stage, the signal for Myc6-Nuf2 disappeared at the kinetochores (Fig. 1Be). When oocytes reached the MII stage, the Myc6-Nuf2 staining was concentrated again at the kinetochores of chromosomes, similar to the MI stage (Fig. 1Bf).
Figure 1.
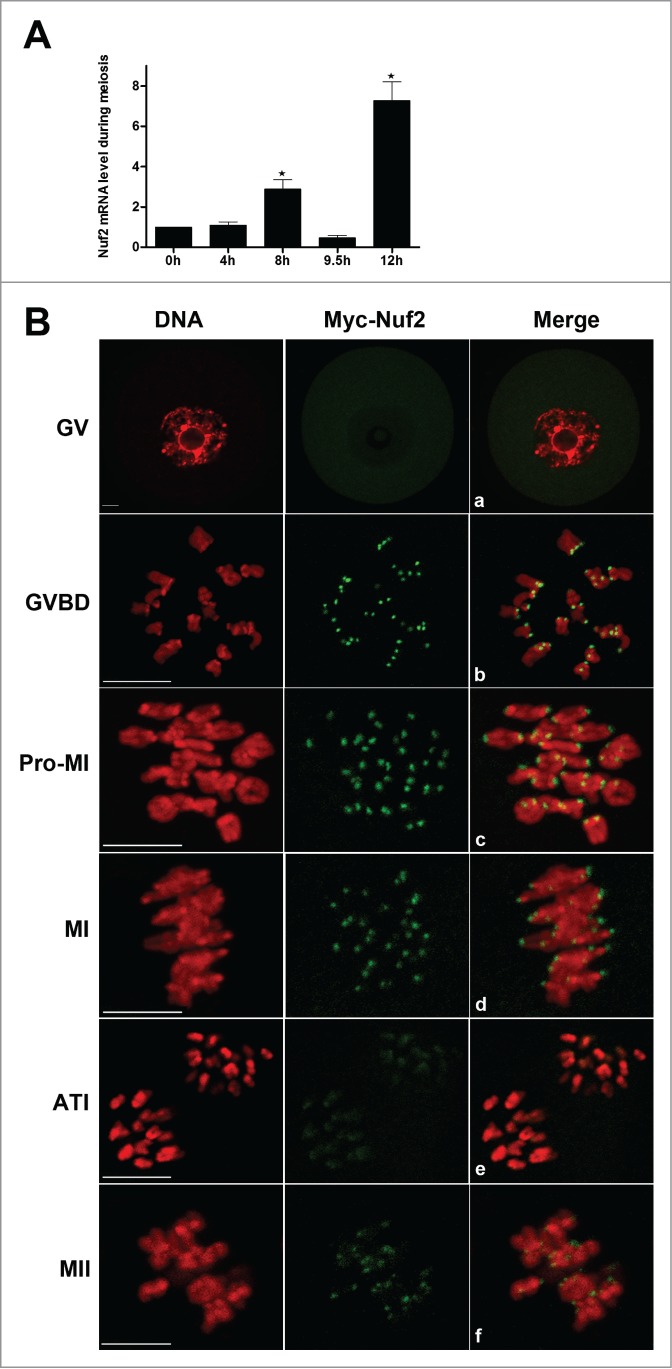
Expression and subcellular localization of Nuf2 during mouse oocyte meiotic maturation. (A) Expression of Nuf2 mRNA as revealed by real time RT-PCR analysis. Samples of 100 oocytes were collected after culture for 0, 4, 8, 9.5 or 12 h, the time points when most oocytes had reached the GV, GVBD, MI, ATI and MII stages, respectively. (B) Confocal microcopy showing the subcellular localization of myc6-Nuf2 (green) in mouse oocytes at GV, GVBD, pro-MI, MI, ATI and MII stages. DNA (red) was counterstained with Hoechst 33342. Scale bars: 10 µm.
To further define the kinetochore localization of Nuf2, Bub3 (one main component of SAC) and Myc6-Nuf2 were double stained. By detection in chromosome spreads, Myc6-Nuf2 localized in the outer domain of Bub3 at the MI and MII stages (Fig. 2). The results indicate that Nuf2 is a component of the kinetochore outer plate, and may contribute to the kinetochore-microtubule attachment in mammalian meiosis.
Figure 2.
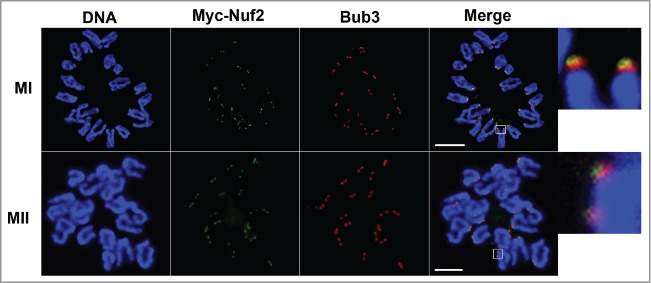
The localization of Bub3 and Myc6-Nuf2 on chromosome spreads at MI and MII stages. Oocytes were injected with a low concentration of Myc6-Nuf2 mRNA and cultured to MI and MII stages, then chromosomes were spread and stained with anti-Myc6 antibody (green), anti-Bub3 antibody (red) and Hoechst 33342 (blue). Magnifications of the boxed regions are shown on the right of each main panel. Scale bars: 10 µm.
Overexpressed Nuf2 causes pro-MI/MI arrest, reduces PB1 extrusion and activation of the SAC
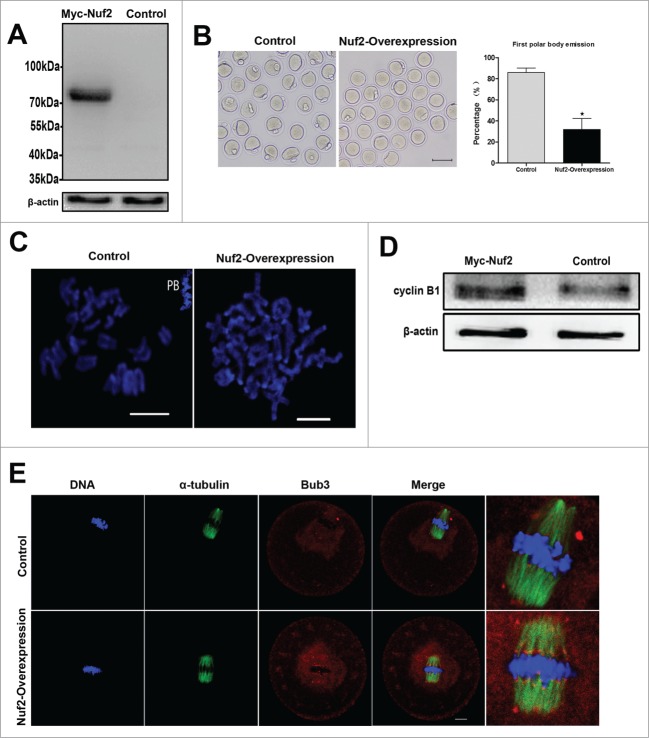
The localization implied that Nuf2 may function in regulating meiotic cell cycle progression. Nuf2 overexpression was employed to investigate its roles in oocyte meiotic maturation. GV oocytes were injected with Myc6-Nuf2 or control mRNA, and cultured for 5h with 2.5 µM milrinone. First, Western blot was conducted to confirm the Myc6-Nuf2 fusion protein expression after exogenous mRNA injection. As presented in Figure 3A, in the overexpression group, a clear Myc6-Nuf2 blot was observed (54 (Nuf2) kDa+ approximate 20 (Myc6) kDa = approximate 74 kDa), however, in the control group, no specific blot was detected.
Figure 3.
Overexpression of Nuf2 inhibits the MI-ATI transition in meiosis I. (A) Samples from control and overexpression groups were collected to test the expression of Myc6-Nuf2 mRNA. Control, 200 oocytes injected with the same amount of Myc6-cotrol mRNA; Myc6-Nuf2, 200 oocytes injected with 2.5 mg/ml Myc6-Nuf2 mRNA solution. (B) Rates of fist polar body extrusion when Myc6-Nuf2 mRNA-injected oocytes were cultured for 14h after release from milrinone. *Significantly different (P < 0.05). (C) Chromosome spreads in Myc6-Nuf2 mRNA injected and control oocytes after 14 h of culture. Scale bars: 10 µm. (D) protein gel blotting showing expression levels of cyclin B1 10 h of oocyte culture. Each sample contained 200 oocytes. (E) Confocal microcopy showing the detection of Bub3 at 10 hours of culture. Bub3 (red); α-tubulin (green); DNA (blue). Magnifications of the boxed regions are shown on the right of each main panel. Scale bars: 10 µm.
The effect of Nuf2 overexpression was examined by quantifying polar body extrusion after Myc6-Nuf2 mRNA injection. As shown in Figure 3B, after culture for 14 h, the PB1 extrusion rate in the overexpression group (31.9 ± 10.4%, n = 191) was considerably lower than that in the control group (85.8 ± 4.4%, n = 198, P < 0.05). Chromosome spreading indicated that most oocytes contained homologous chromosomes, while a large proportion of oocytes in the control group had entered meiosis II (Fig. 3C).
To further confirm the oocyte arrest at the Pro-MI/MI stage by overexpressed Nuf2, the level of cyclin B1 was examined after culture for 10h. As presented in Figure 3D, negligible signal of cyclin B1 in control oocytes was observed by western blot, while the cyclin B1 level was significantly higher in the overexpression oocytes. Next, localization of Bub3, one of primary component of SAC, was analyzed in oocytes. Specific signals for Bub3 were detected in the overexpression group even after 10 h of culture (33/37), which were arrested at the Pro-MI/MI stage. In contrast, most of the control oocytes had entered anaphase and showed no signals of Bub3 (17/39) Fig. 3E. Detection of Bub3 signal indicates activation of the spindle assembly checkpoint. These results indicate that overexpressed Nuf2 inhibits the MI-AI transition and prevents homologous chromosomes from segregation by activating SAC.
Overexpressed Nuf2 inhibits metaphase-anaphase transition and chromatid segregation during meiosis II
Overexpression of Nuf2 can inhibit the MI-AI transition during meiosis I. To confirm whether it also functions in meiosis II, MII oocytes were injected with Myc6-Nuf2 mRNA or Myc6-control mRNA and cultured for 5 h to allow the expression. Then the oocytes were activated by using 10mM SrCl2 in Ca2+-free CZB. The results showed that the MII-AII transition was halted in the Myc6-Nuf2 mRNA injection group. However, most of the oocytes entered the AII stage in the control group (Fig. 4A). Figure 4B demonstrates the rates of oocytes arrested at the MII stage in the Nuf2 overexpression group (55.84 ± 6.01%, n = 57) and control group (26.01 ± 7.89%, n = 52, P < 0.05). These data show that Nuf2 participates in MII-AII transition during meiosis II.
Figure 4.
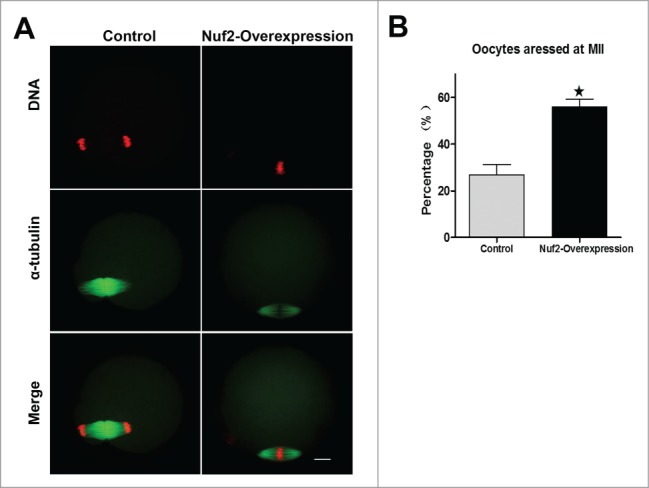
Overexpressed Nuf2 inhibits MII-AII transition and sister chromatid segregation. (A) MII oocytes were injected with Myc6-control or Myc6-Nuf2 mRNA and cultured for 2h after parthenogenetic activation. DNA (red); α-tubulin (green). Scale bars: 10 µm. (B) Percentages of oocytes arrested at MII in control and overexpression groups. Data are expressed as mean ± SEM of at least 3 independent experiments. *Significantly different (P < 0.05).
Overexpression of Nuf2 causes abnormal spindles and misaligned chromosomes
To further determine the reason for the metaphase-anaphase transition failure and SAC activation in Nuf2 overexpression oocytes, we examined spindle morphology and chromosome distribution using confocal microscopy. In the control group, most oocytes exhibited normal spindle morphology and chromosome alignment, but in the Nuf2 overexpression group, the oocytes showed different kinds of morphologically defective spindles and misaligned chromosomes. Chromosomes showed slight or severe misalignment, and some chromosomes were even observed at the poles of the spindle, or unattached to microtubules in overexpression oocytes (Figure 5A). The rate of abnormal spindles in the Myc6-Nuf2 mRNA injection group (49.86 ± 12.51%, n = 138) was significantly higher than that in the Myc6-control mRNA injection group (19.24 ± 1.01%, n = 129, Figure 5B) (P < 0.05). Moreover, as shown in Figure 5B, the rate of misaligned chromosomes significantly differed between the Nuf2 overexpression (54.61 ± 11.15%, n = 138) group and control group (24.93 ± 1.83%, n = 129) (P < 0.05).
Figure 5.
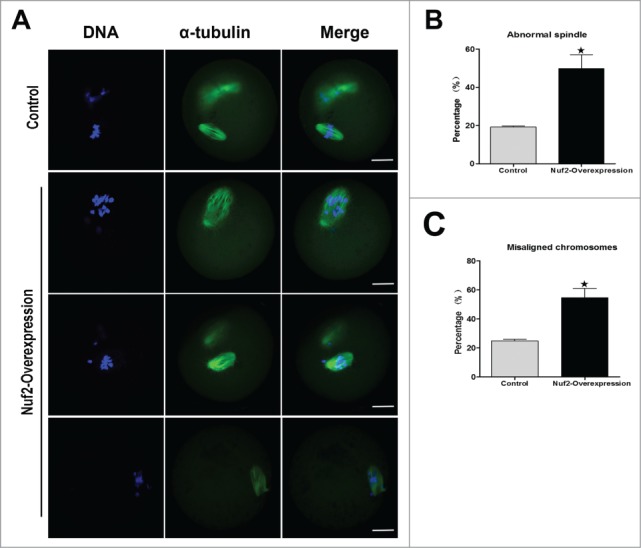
Overexpression of Nuf2 causes abnormal spindle and misaligned chromosomes in meiotic oocytes. (A) Oocytes microinjected with Myc6-Nuf2 or Myc6-control mRNA were incubated in M2 medium containing 2.5 μM milrinone for 5 h, and then transferred to milrinone-free M2 for 12 h, followed by immunostaining with α-tubulin antibody (green) and Hoechst 33342 (blue). In the Myc6-Nuf2 mRNA injection group, chromosomes showed slight or severe misalignment, and some chromosomes were even observed at the poles of the spindle; the spindle showed unattached microtubules and severe disruption. Scale bars: 10 µm. (B and C) The rates of oocytes with abnormal spindles or misaligned chromosomes in the Myc6-Nuf2 mRNA injection group and control group. Data are expressed as mean ± SEM of at least 3 independent experiments. *Significantly different (P < 0.05).
Live-cell imaging further revealed that in the control group, a clear bipolar spindle was visible and slowly migrated to the oocyte cortex followed by rapid polar body extrusion (Fig. 6A; Supplemental Video S1). However, in the Myc6-Nuf2 mRNA injection group, various morphologically defective spindles appeared, and chromosomes failed to align at the middle plate, and thus failed to separate. Oocytes remained at the Pro-MI/MII stage until about 14h. In addition, no first polar body extrusion was observed in the Nuf2 overexpression group (Fig. 6B; Supplemental Video S1).
Figure 6.
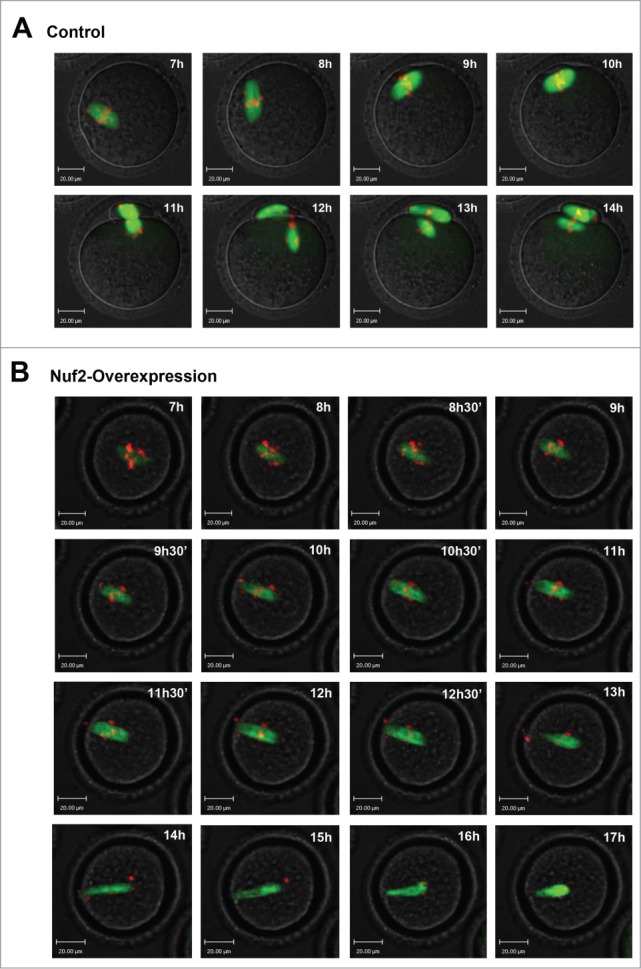
Overexpressed Nuf2 causes defects of meiotic spindle and chromosome alignment, and prevents the metaphase-anaphase transition of mouse oocytes as revealed by time-lapse live-cell imaging. (A) Oocytes co-injected with β5-tubulin-GFP mRNA and Myc6-control mRNA. Spindle (fluorescent tubulin) and DNA (red) images in a typical control oocyte during in vitro maturation. Time points indicate the time-lapse from about 3-4 h after GVBD. (B) Similar to (A), oocytes were co-injected with β5-tubulin-GFP mRNA and Myc6-Nuf2 mRNA. Shown are images of oocytes with abnormal spindles, aberrant chromosomes, unsuccessful chromosome segregation and failure of PB1 extrusion. Additionally, chromosomes become detached from microtubules during the impaired separation process. α-tubulin (green); DNA (red). Scale bars: 20 µm.
Depletion of Nuf2 causes chromosome misalignment, spindle disruption, precocious polar body extrusion and aneuploidy
To further explore the exact roles of Nuf2, the oocytes with Nuf2 RNAi were analyzed. Nuf2 siRNA or negative control siRNA was injected into GV stage oocytes, then the injected oocytes were maintained in 2.5 μM milrinone for 20 h. The RNAi efficiency was detected by real-time quantitative PCR. Compared with the control, the mRNA level of Nuf2 in siRNA injected oocytes was significantly decreased (33.73 ± 8.29 vs. 100%, n = 100, P < 0.05, Fig. 7A), implying successful Nuf2 down-regulation by RNAi. In addition, the rates of polar body extrusion were quantified at different times after Nuf2 RNAi. As shown in Figure 7B, in the Nuf2 RNAi group,11.79 ± 2.69%,24.79 ± 3.33% of the oocytes extruded polar bodies by 8, 9h, respectively, while in the control group, the polar body extrusion rates were significantly lower at the same time points (5.24 ± 1.99%, 14.22 ± 4.24%, respectively, P < 0.05). However, there was no significant difference between the control group and the siRNA microinjection group from 10 h to 12 h (35.57 ± 1.11%, 56.95 ± 11.02%, 66.67 ± 7.92% vs. 38.86 ± 5.98%, 55.03 ± 9.73%, 65.87 ± 14.59%). These results revealed that downregulation of Nuf2 may result in precocious anaphase, followed by precocious polar body extrusion.
Figure 7.
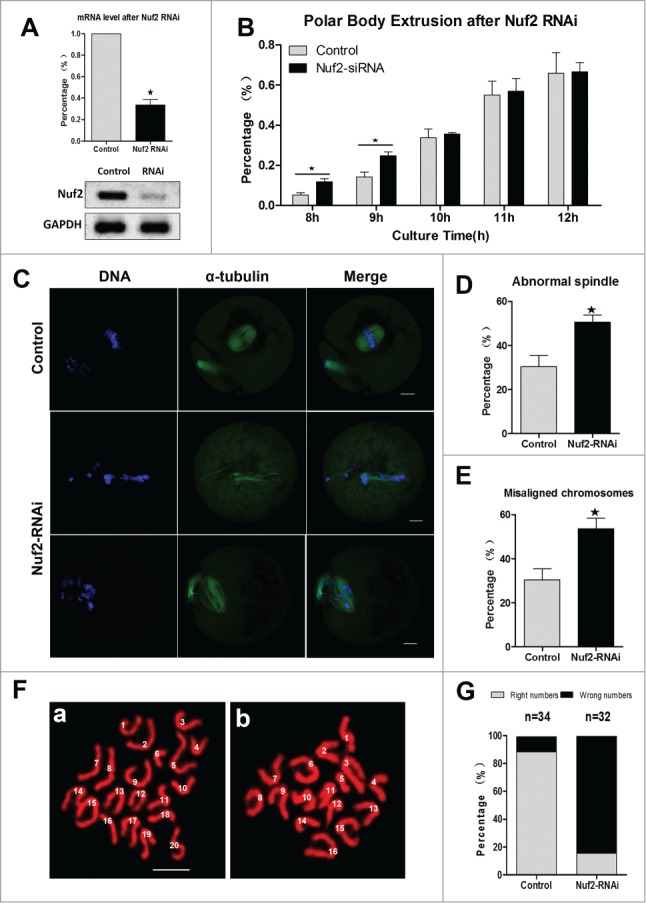
Nuf2-RNAi leads to misaligned chromosomes, precocious polar body extrusion, and aneuploidy. (A) Levels of Nuf2 mRNA in siRNA injected oocytes. GV stage oocytes were microinjected with negative control siRNA or Nuf2 siRNA and incubated for 24 h in M2 medium containing 2.5 μM milrinone before collecting the oocytes for real time quantitative PCR. (B) Rates of fist polar body extrusion in siRNA-injected oocytes cultured for various times after release from milrinone. Data are expressed as mean ± SEM of at least 3 independent experiments. *Significantly different (P < 0.05). (C) Spindle morphology and chromosome alignments in MII oocytes after microinjection of Nuf2 siRNA. In the control group, most oocytes showed normal chromosome alignment, while in the siRNA injection oocytes, most oocytes showed slight or severely misaligned chromosomes and different kinds of morphologically defective spindles. α-tubulin (green); DNA (blue). Scale bars: 10 µm. (D and E) The rates of oocytes with abnormal spindles or misaligned chromosomes in the siRNA injection and control group. Data are expressed as mean ± SEM of at least 3 independent experiments. *Significantly different (P < 0.05). (F and G) Chromosome spreading was performed in control and RNAi oocytes. The numbers of univalents in the oocytes in E a; b are 20; 16, respectively. Scale bars: 10 µm.
Oocytes were cultured for 12 h to examine the chromosome and spindle morphology using confocal microscopy. As shown in Figure 7C, compared with the control oocytes, the siRNA injection oocytes contained slight or severely misaligned chromosomes. Different kinds of morphologically defective spindles were observed in the Nuf2-RNAi group, including unattached microtubules and severely disrupted spindles. The rate of abnormal spindles in the Nuf2 siRNA injection group (50.47 ± 5.72%, n = 166) was significantly higher than that in the control group (30.40 ± 8.69%, n = 157) (P < 0.05) (Fig. 7D). Moreover, as shown in Figure 7E, the rate of misaligned chromosomes in the Nuf2 RNAi (53.57 ± 8.29%, n = 166) and control groups (30.40 ± 8.69%, n = 157) differed significantly (P < 0.05). Overall, these results showed that down-regulation of Nuf2 disrupted chromosome alignment and spindle formation during mouse oocyte meiosis.
To further prove the effects of Nuf2 RNAi, chromosome spreading was employed to determine the exact number of chromosomes in oocytes at the MII stage. The results showed that RNAi oocytes typically displayed incorrect numbers of univalents. As shown in Figure 7FA and B (“a” is control oocyte, “b” is RNAi oocyte), the numbers of chromosomes in the oocytes were 20 and 16, respectively, verifying the validity of RNAi-induced aneuploidy. Figure 7G showed the percentages of oocytes with aneuploidy in Nuf2-RNAi and control groups, respectively.
Discussion
The major components of the Ndc80 complex include Ndc80 (Hec1 in human), Nuf2, Spc24 and Spc25. It is generally accepted that the complex is important for kinetochore assembly and function.19,29-31 However, the specific role of Nuf2, particularly in mammalian meiosis, has only not been well explored. In this study, the expression and function of Nuf2 during mouse oocyte meiotic maturation were investigated. Both overexpression and RNAi demonstrate that Nuf2 is an important player in the regulation of meiotic cell cycle progression.
Subcellular localization by exogenous mRNA injection of mouse oocytes revealed that Nuf2 localized at kinetochores of chromosomes in meiotic oocytes, similar to the situation in mitotic cells.32-34 This result implies that Nuf2 may participate in the surveillance of kinetochore-microtubule attachment during oocyte meiotic maturation. However, the localization pattern of Nuf2 is different from that of 2 other core components of Ndc80 (Ndc80, Spc25), which were distributed on microtubules and chromosomes at all stages of meiosis.13,28 The localization of Nuf2 was similar to that of Bub3,7 implying that Nuf2 has a potential role during the metaphase-anaphase transition. Numerous studies showed that Nuf2 plays roles in the spindle checkpoint and kinetochore tension in mitotic cells.12,35,36
To characterize the functions of Nuf2 in oocyte meiotic maturation, overexpression and RNAi were carried out. Previous research confirmed that Nuf2 regulates the spindle assembly checkpoint in human mitosis and budding yeast meiosis.29,32,37,38 In this study, overexpressed Nuf2 clearly inhibited the homologous chromosome segregation, indicating that Nuf2 is involved in monitoring the MI-AI transition. Chromosome spreads showed that chromosomes were still in the bivalent state in Nuf2 overexpressed oocytes. In addition, overexpression of Nuf2 triggered a higher expression of cyclin B1, which indicates that the APC/C may be inhibited (Fig. 3D). A small proportion of oocytes in overexpression group extruded the first polar body, and this might be caused by less injected Myc6-Nuf2 mRNA or without injection of Myc6-Nuf2, an inevitable phenomenon in the process of microinjection. Indeed, compared to the oocytes reaching MII stage, the mRNA level of Nuf2 in MI-arresting oocytes was significantly increased (data not shown). During mammalian mitosis and meiosis, the spindle assembly checkpoint (SAC) serves as a correcting network to prevent anaphase onset until all kinetochores properly attach to the microtubules. Several studies revealed that SAC protein such as mitotic arrest-defiient-1 (Mad1), Mad2, Budding uninhibited by benzimidazole-1 (Bub1), Bub3, BubR1 are all involved in cell cycle progression.5,7,8 The results prompted us to explore whether overexpressed Nuf2 could regulate the spindle assembly checkpoint in meiosis after Nuf2 overexpression. Precise signals for Bub3 were detected in MI-arrested oocytes in the overexpressed group after 10 h of culture, while the control oocytes entered anaphase. Taken together, the data suggest that Nuf2 may be required for cell cycle progression by the regulation of proper SAC signaling in mouse oocyte meiosis. This is consistent with previous work showing that the KMN complex (including the Ndc80 complex) can bind microtubules and SAC proteins in corresponding checkpoints to “off” and “on”, respectively.13,34,39 On the other hand, down-regulating Nuf2 using RNAi showed that depletion of Nuf2 caused precocious polar body extrusion. The results further indicate that Nuf2 is involved in monitoring the MI-AI transition, which is consistent with findings obtained for mitosis.13,16,25 We also found that Nuf2 was involved in meiosis II by preventing sister chromatid segregation. Mammalian oocytes are arrested at the MII stage under control of the cytostatic factor (CSF),38,40 implying that Nuf2 function may be related to CSF in mouse oocytes.
Previous reports have indicated that Nuf2 is required for the organization of stable microtubule-kinetochore attachments in mitosis.10,17,26,29 Parallel to mitosis, a surveillance pathway is needed to safeguard fidelity of chromosome separation in oocyte meiosis. The present study clearly demonstrated that Nuf2-RNAi caused misalignment of chromosomes and unattached microtubules, which may indicate failure of microtubule attachments to chromosomes. Additionally, the study also revealed that Nuf2-RNAi oocytes typically displayed incorrect numbers of univalents. The missing or improper attachment caused congression failure of chromosomes at the metaphase plate during the MI stage, leading to aneuploidy.41,42 Remarkably, meiosis in humans is particularly error-prone in oocytes during the MI stage.43,44 Proper kinetochore-microtubule attachments are important to prevent chromosome segregation errors. Both overexpression and knockdown of Nuf2 caused misalignment of chromosomes and defects of spindles, which indicated that Nuf2 may be involved in kinetochore-microtubule interactions. Similar to previous reports regarding Ndc80 complex functions.13,14,28,45,46
In conclusion, our data indicate that Nuf2 is required for monitoring proper kinetochore-microtubule attachments and accurate chromosome segregation and thus chromosome fidelity in mammalian meiotic germ cells.
Materials and Methods
All chemicals and culture media were purchased from Sigma Chemical Company (St. Louis, MO) unless stated otherwise.
Antibodies
Mouse monoclonal anti-Myc6-FITC antibody was purchased from Invitrogen (R950–25, Carlsbad, CA); rabbit monoclonal anti-α-tubulin was obtained from Cell Signaling Technology (#2125, Beverly, MA); rabbit monoclonal anti-bub3 antibodies were purchased from Santa Cruz Biotechnology (SC-28258, Santa Cruz, CA). TRITC-conjugated goat anti-rabbit IgG was purchased from Zhongshan Golden Bridge Biotechnology Co, LTD (ZF-0316, Beijing). Rabbit polyclonal anti-cyclin B1 antibody was purchased from Santa Cruz (SC-752, Santa Cruz, CA) and mouse monoclonal anti-β-actin antibody was purchased from Santa Cruz Biotechnology (SC-8432, Santa Cruz, CA).
Oocyte collection and culture
Care and handling of 4–6 week-old ICR mice was conducted in accordance with policies promulgated by the Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences. Mouse oocyte collection and culture were performed as previously described.47 Oocytes were cultured in M2 medium supplemented with 2.5 μM milrinone to maintain them at the germinal vesicle (GV) stage. After specific treatments, oocytes were washed thoroughly and cultured in M2 medium to specific stages. The MII oocytes were released from the CSF arrest by using 10 mM SrCl2 in Ca2+/Mg2+−free CZB.
Nuf2 plasmid construction and mRNA synthesis
Total RNA was extracted from 200 mouse GV oocytes using RNeasy micro purification kit (Qiagen), and the first strand cDNA was generated with cDNA synthesis kit (Takara), using poly (dT) primers. The following 2 nested primers were used to clone the full length of Nuf2 cDNA by PCR. F1: GCTTT-CGGTTGGACGCTC, R1: AAGCAA-GTCAGTGTAGCAT, F2: GAAGGC-CGGCCAATGGAAACTTTGTC, R2: GTTGGCGCGCCTTGTTCAGACC-ATTC. The PCR products were purified, then digested using FseI and AscI (New England Biolabs, Inc.) and cloned into the pCS2+ vector, in which the Nuf2 sequence was linked to Myc6-tags at its N-terminus. For the synthesis of Myc6-tagged Nuf2 mRNA, the Nuf2-pCS2+, plasmids were linearized by NdeI and then purified. The SP6 mMESSAGE mMACHINE (Ambion) was used to produce capped mRNA, and mRNA was then purified using an RNeasy cleanup kit (Qiagen).
Microinjection of Myc6-Nuf2 mRNA or Nuf2 siRNAs
Microinjection of mRNAs and siRNAs were performed as previously described.48,49 Microinjections were performed using a Narishige microinjector and completed within 30 minutes. For Myc6-Nuf2 overexpression, 2.5 mg/ml (or 0.4 mg/ml for localization) Myc6–Nuf2 mRNA solution was injected into cytoplasm of the oocytes. For protein expression, oocytes were arrested at the GV stage in M2 medium supplemented with 2.5 μM milrinone for 4–6 hours. The same amount of RNase-free water or Myc6-control mRNA (virtually no discrepancy was observed between them) was injected as control. For MII oocyte overexpression experiments, 2.5 mg/ml Myc6-Nuf2 mRNA was injected into the cytoplasm and the oocytes were cultured for 5 hours before they were parthenogenetically activated with 10 mM SrCl2.
For knockdown experiments, small interfering RNAs (siRNAs) of Nuf2 (GenePharma) were microinjected into the cytoplasm to deplete Nuf2. The subsequent siRNAs were used at 25 μM, Nuf2 siRNA-1: GCUGAGCUUGUGGUUCAUATT; UAUGAACCACAAGCUCAGCTT. Nuf2 siRNA-2: GCUGGAGGUGCAGUUGUAUTT; AUACAACUGCACCUCCAGCTT. The same amount of negative control siRNA was used as control. After microinjection, the GV oocytes were cultured for 20 hours in M16 medium supplemented with 2.5 μM milrinone for the depletion of Nuf2.
Real-time quantitative PCR analysis
Total RNA was extracted from 100 oocytes using RNeasy micro purification kit (Qiagen), the first strand cDNA was generated with M-MLV first strand cDNA synthesis kit (Invitrogen), using oligo (dT) primers. A cDNA fragment of Nuf2 was amplified using the following primers: Forward: TCCCCAGATACAATGTAGCTGA; Reverse: CCGGACTCCATACACTAACTGT. GAPDH was selected as a reference gene. The SYBR Premix Ex Tag2 kit (Takara) was used in an ABI prism 7500 Sequence Detection System. Relative gene expression was calculated by the 2ΔΔCt method.
Immunofluorescence analysis and chromosome spread
Oocytes were fixed in 4% paraformaldehyde in PBS buffer for 30 minutes at room temperature. After being permeabilized with 0.5% Triton X-100 for 20 minutes, they were then blocked in 1% BSA-supplemented PBS for 1 hour at room temperature. For single staining of Myc6-Nuf2 and α-tubulin, oocytes were incubated overnight at 4°C with 1:100 anti-Myc6-FITC antibody (Invitrogen) or 1:200 anti-α-tubulin-FITC antibodies, respectively; DNA was stained with Hoechst 33342 for 15 minutes. For double staining of Myc6-Nuf2 and Bub3, oocytes were first stained with Bub3 (1:50) overnight at 4°C, and after 3 washes in washing buffer, oocytes were incubated with TRITC-conjugated goat anti-rabbit secondary antibody for 2 hours at room temperature, then washed 3 times and blocked again for 1 hour at room temperature and then incubated with anti-α-tubulin-FITC antibodies for 2 hours at room temperature.
Chromosome spreads were performed as described previously.50 Briefly, the oocytes were exposed to acid Tyrode's solution (Sigma) for 1–2 minutes at room temperature to remove the zona pellucida. After a brief recovery in M2 medium, the oocytes were transferred to glass slides and fixed in a solution of 1% paraformaldehyde in distilled H2O (pH 9.2) containing 0.15% Triton X-100 and 3 mM dithiothreitol. The slides were allowed to dry slowly in a humid chamber for several hours, and then blocked with 1% BSA in PBS for 1 hour at room temperature. The oocytes were then incubated with Bub3 (1:50), anti-Myc6 (1:50), primary antibodies overnight at 4°C. After brief washes with washing buffer, the slides were then incubated with the corresponding secondary antibodies for 2 hours at room temperature. The oocytes were finally stained with Hoechst 333342 after 3 washes in washing buffer and mounted on glass slides for immunofluorescence microscopy. The photos were taken with a confocal laser-scanning microscope (Zeiss LSM 710, Germany).
Immunoblotting analysis
Immunoblotting was performed as described previously.51 Briefly, a total of 200 mouse oocytes were collected in a 5 μL 2x SDS buffer and heated for 5 min at 100°C. The proteins were separated by SDS-PAGE and then transferred to PVDF membranes. Following transfer, the membranes were blocked in TBST containing 5% skimmed milk for 2 hour at room temperature, followed by incubation overnight at 4°C with mouse monoclonal anti-Myc6 antibody (1:1,000) and mouse monoclonal anti-β-actin antibody (1:1,000). After three washes in TBST buffer, 10 minutes each, the membranes were incubated with 1:1000 HRP-conjugated goat anti-mouse IgG, for 1 hour at 37°C. Finally, the membranes were processed using the enhanced chemiluminescence-detection system (Bio-Rad, CA).
Time-lapse live-imaging experiments
Microtubule and chromosome dynamics were filmed on a Perkin Elmer precisely Ultra VIEW VOX Confocal Imaging System. A narrow band passed EGFP and BFP filter sets and a 30% cut neutral density filter from Chroma. Exposure time was set ranging between 300–800 ms depending on the tubulin-GFP and Hoechst 33342 florescence levels. The acquisition of digital time-lapse images was controlled by IP Lab (Scanalytics) or AQM6 (Andor/Kinetic-imaging) software packages. Confocal images of spindles and chromosomes in live oocytes were acquired with a 20x oil objective on a spinning disk confocal microscope (Perkin Elmer).
Statistical analysis
Data (mean ± SE) were generated from replicates that were repeated at least 3 times per experiment and analyzed by ANOVA using SPSS software (SPSS Inc., Chicago, IL) followed by student-Newman-Keuls test. The number of oocytes observed was labeled in parentheses as (n). Difference at P < 0.05 was considered to be statistically significant and different superscripts indicate the statistical difference.
Funding
This study was supported by National Basic Research Program of China (No. 2012CB944404, 2011CB944501), Shenzhen R&D Basic Research Program (No. JCYJ20120618092558280) and the National Natural Science Foundation of China (Nos. 30930065, 31371451).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest are disclosed.
Acknowledgments
We are grateful to Yi Hou and Li-Juan Wang for their technical assistance. We also thank the other members in Dr. Sun's lab for their kind discussions about the experiments.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Welburn JP, Cheeseman IM. Toward a molecular structure of the eukaryotic kinetochore. Dev Cell 2008; 15:645-55; PMID:19000831; http://dx.doi.org/ 10.1016/j.devcel.2008.10.011 [DOI] [PubMed] [Google Scholar]
- 2.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J 2009; 28:2511-31; PMID:19629042; http://dx.doi.org/ 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musacchio A. Spindle assembly checkpoint: the third decade. Philos Trans R Soc Lond B Biol Sci 2011; 366:3595-604; PMID:22084386; http://dx.doi.org/ 10.1098/rstb.2011.0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 2007; 8:379-93; PMID:17426725; http://dx.doi.org/ 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- 5.Yin SWQ, Liu JH, Ai JS, Liang CG, Hou Y, Chen DY, Schatten H, Sun QY. Bub1 prevents chromosome misalignment and precocious anaphase during mouse oocyte meiosis. Cell Cycle 2006; 5:2130-7; PMID:16969117 [DOI] [PubMed] [Google Scholar]
- 6.Homer H, Gui L, Carroll J. A spindle assembly checkpoint protein functions in prophase I arrest and prometaphase progression. Science 2009; 326:991-4; PMID:19965510; http://dx.doi.org/ 10.1126/science.1175326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li MLS, Yuan J, Wang ZB, Sun SC, Schatten H, Sun QY. Bub3 is a spindle assembly checkpoint protein regulating chromosome segregation during mouse oocyte meiosis. Plos One 2009; 4:e7701; PMID:19888327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei LLX, Zhang QH, Li M, Yuan J, Li S, Sun SC, Ouyang YC, Schatten H, Sun QY. BubR1 is a spindle assembly checkpoint protein regulating meiotic cell cycle progression of mouse oocyte. Cell Cycle 2010; 9:1112-21; PMID:20237433 [DOI] [PubMed] [Google Scholar]
- 9.The Ndc80p Complex from Saccharomyces cerevisiae Contains Conserved Centromere Components and Has a Function in Chromosome Segregation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol 2002; 159:549-55; PMID:12438418; http://dx.doi.org/ 10.1083/jcb.200208159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 2002; 297:2267-70; PMID:12351790; http://dx.doi.org/ 10.1126/science.1075596 [DOI] [PubMed] [Google Scholar]
- 12.DeLuca JG, Howell BJ, Canman JC, Hickey JM, Fang G, Salmon ED. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr Biol 2003; 13:2103-9; PMID:14654001; http://dx.doi.org/ 10.1016/j.cub.2003.10.056 [DOI] [PubMed] [Google Scholar]
- 13.Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genome Res 2002; 12:47-56; PMID:11779830; http://dx.doi.org/ 10.1101/gr.203201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 2006; 127:969-82; PMID:17129782; http://dx.doi.org/ 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Riley DJ, Chen PL, Lee WH. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol Cell Biol 1997; 17:6049-56; PMID:9315664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng L, Chen Y, Lee WH. Hec1p, an evolutionarily conserved coiled-coil protein, modulates chromosome segregation through interaction with SMC proteins. Mol Cell Biol 1999. Aug; 19(8):5417-28; PMID:10409732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem 2005; 280:29088-95; PMID:15961401; http://dx.doi.org/ 10.1074/jbc.M504070200 [DOI] [PubMed] [Google Scholar]
- 18.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A 2005; 102:5363-7; PMID:15809444; http://dx.doi.org/ 10.1073/pnas.0501168102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang G, Kelstrup CD, Hu XW, Kaas Hansen MJ, Singleton MR, Olsen JV, Nilsson J. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci 2012; 125:3243-53; PMID:22454517; http://dx.doi.org/ 10.1242/jcs.104208 [DOI] [PubMed] [Google Scholar]
- 20.Nilsson J. Looping in on Ndc80 - how does a protein loop at the kinetochore control chromosome segregation? Bioessays 2012; 34:1070-7; PMID:23154893; http://dx.doi.org/ 10.1002/bies.201200096 [DOI] [PubMed] [Google Scholar]
- 21.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 2006; 127:983-97; PMID:17129783; http://dx.doi.org/ 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- 22.Wang HW, Long S, Ciferri C, Westermann S, Drubin D, Barnes G, Nogales E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J Mol Biol 2008; 383:894-903; PMID:18793650; http://dx.doi.org/ 10.1016/j.jmb.2008.08.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, Desai A, Milligan RA. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol 2008; 182:1055-61; PMID:18794333; http://dx.doi.org/ 10.1083/jcb.200804170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell 2004; 7:45-60; PMID:15239953; http://dx.doi.org/ 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- 25.Gillett ES, Espelin CW, Sorger PK. Spindle checkpoint proteins and chromosome-microtubule attachment in budding yeast. J Cell Biol 2004; 164:535-46; PMID:14769859; http://dx.doi.org/ 10.1083/jcb.200308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D, Ding X, Du J, Cai X, Huang Y, Ward T, Shaw A, Yang Y, Hu R, Jin C, et al.. Human NUF2 interacts with centromere-associated protein E and is essential for a stable spindle microtubule-kinetochore attachment. J Biol Chem 2007; 282:21415-24; PMID:17535814; http://dx.doi.org/ 10.1074/jbc.M609026200 [DOI] [PubMed] [Google Scholar]
- 27.Sun SC, Zhang DX, Lee SE, Xu YN, Kim NH. Ndc80 regulates meiotic spindle organization, chromosome alignment, and cell cycle progression in mouse oocytes. Microsc microanal 2011; 17:431-9; PMID:21600073; http://dx.doi.org/ 10.1017/S1431927611000274 [DOI] [PubMed] [Google Scholar]
- 28.Sun S-C, Lee S-E, Xu Y-N, Kim N-H. Perturbation of Spc25 expression affects meiotic spindle organization, chromosome alignment and spindle assembly checkpoint in mouse oocytes. Cell Cycle 2014; 9:4552-9; http://dx.doi.org/ 10.4161/cc.9.22.13815 [DOI] [PubMed] [Google Scholar]
- 29.Sundin LJ, Guimaraes GJ, Deluca JG. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol Biol Cell 2011; 22:759-68; PMID:21270439; http://dx.doi.org/ 10.1091/mbc.E10-08-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol 2012; 19:1161-7; PMID:23085714; http://dx.doi.org/ 10.1038/nsmb.2411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun Q. Ndc80 complex New evidence for the existence of spindle assembly checkpoint in mammalian oocyte meiosis. Cell Cycle 2011; 10:877-8; PMID:21346413 [PubMed] [Google Scholar]
- 32.Mikami Y, Hori T, Kimura H, Fukagawa T. The functional region of CENP-H interacts with the Nuf2 complex that localizes to centromere during mitosis. Mol Cell Biol 2005; 25:1958-70; PMID:15713649; http://dx.doi.org/ 10.1128/MCB.25.5.1958-1970.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bharadwaj R, Qi W, Yu H. Identification of two novel components of the human NDC80 kinetochore complex. J Biol Chem 2004; 279:13076-85; PMID:14699129; http://dx.doi.org/ 10.1074/jbc.M310224200 [DOI] [PubMed] [Google Scholar]
- 34.Janke COJ, Lechner J, Shevchenko A, Shevchenko A, Magiera MM, Schramm C, Schiebel E. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J 2001; 20:777-91; PMID:11179222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nabetani A, Koujin T, Tsutsumi C, Haraguchi T, Hiraoka Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: a link between the kinetochore function and the spindle checkpoint. Chromosoma 2001; 110:322-34; PMID:11685532; http://dx.doi.org/ 10.1007/s004120100153 [DOI] [PubMed] [Google Scholar]
- 36.Ciferri C, Musacchio A, Petrovic A. The Ndc80 complex: hub of kinetochore activity. FEBS Lett 2007; 581:2862-9; PMID:17521635; http://dx.doi.org/ 10.1016/j.febslet.2007.05.012 [DOI] [PubMed] [Google Scholar]
- 37.Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y. Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol Biol Cell 2005; 16:2325-38; PMID:15728720; http://dx.doi.org/ 10.1091/mbc.E04-11-0996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt A, Rauh NR, Nigg EA, Mayer TU. Cytostatic factor: an activity that puts the cell cycle on hold. J Cell Sci 2006; 119: 1213-18; PMID:16554437; http://dx.doi.org/ 10.1242/jcs.02919 [DOI] [PubMed] [Google Scholar]
- 39.Burke DJ, Stukenberg PT. Linking kinetochore-microtubule binding to the spindle checkpoint. Devel Cell 2008; 14:474-9; PMID:18410725; http://dx.doi.org/ 10.1016/j.devcel.2008.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masui Y. From oocyte maturation to the in vitro cell cycle: the history of discoveries of Maturation-Promoting Factor (MPF) and Cytostatic Factor (CSF). Differentiation 2001; 69:1-17; PMID:11776390; http://dx.doi.org/ 10.1046/j.1432-0436.2001.690101.x [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Wang L, Ma M, Song Z, Zhang J, Xu G, Fan J, Li N, Cram DS, Yao Y. A PGD pregnancy achieved by embryo copy number variation sequencing with confirmation by non-invasive prenatal diagnosis. J Genet Genomics 2014; 41:453-6; PMID:25160978; http://dx.doi.org/ 10.1016/j.jgg.2014.06.007 [DOI] [PubMed] [Google Scholar]
- 42.Jiang H, Wang L, Cui Y, Xu Z, Guo T, Cheng D, Xu P, Yu W, Shi Q. Meiotic chromosome behavior in a human male t(8;15) carrier. J Genet Genomics 2014; 41:177-85; PMID:24656237; http://dx.doi.org/ 10.1016/j.jgg.2014.01.005 [DOI] [PubMed] [Google Scholar]
- 43.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet 2001; 2:280-91; PMID:11283700; http://dx.doi.org/ 10.1038/35066065 [DOI] [PubMed] [Google Scholar]
- 44.Hunt PA HT. Sex matters in meiosis. Science 2002; 296:2181-3; PMID:12077403; http://dx.doi.org/ 10.1126/science.1071907 [DOI] [PubMed] [Google Scholar]
- 45.Wei RR, Schnell JR, Larsen NA, Sorger PK, Chou JJ, Harrison SC. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure 2006; 14:1003-9; PMID:16765893; http://dx.doi.org/ 10.1016/j.str.2006.04.007 [DOI] [PubMed] [Google Scholar]
- 46.Wu G, Wei R, Cheng E, Ngo B, Lee WH. Hec1 contributes to mitotic centrosomal microtubule growth for proper spindle assembly through interaction with Hice1. Mol Biol Cell 2009; 20:4686-95; PMID:19776357; http://dx.doi.org/ 10.1091/mbc.E08-11-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin F, Ma XS, Wang ZB, Wang ZW, Luo YB, Huang L, Jiang ZZ, Hu MW, Schatten H, Sun QY. Different fates of oocytes with DNA double-strand breaks in vitro and in vivo. Cell Cycle 2014; 13:2674-80; PMID:25486355; http://dx.doi.org/ 10.4161/15384101.2015.945375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen L, Ge ZJ, Wang ZB, Sun T, Ouyang YC, Sun QY, Sun YP. TGN38 is required for the metaphase I/anaphase I transition and asymmetric cell division during mouse oocyte meiotic maturation. Cell Cycle 2014; 13:2723-32; PMID:25486359; http://dx.doi.org/ 10.4161/15384101.2015.945828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo YB, Ma JY, Zhang QH, Lin F, Wang ZW, Huang L, Schatten H, Sun QY. MBTD1 is associated with Pr-Set7 to stabilize H4K20me1 in mouse oocyte meiotic maturation. Cell Cycle 2013; 12:1142-50; PMID:23475131; http://dx.doi.org/ 10.4161/cc.24216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hodges CA, Hunt PA. Simultaneous analysis of chromosomes and chromosome-associated proteins in mammalian oocytes and embryos. Chromosoma 2002; 111:165-9; PMID:12355205; http://dx.doi.org/ 10.1007/s00412-002-0195-3 [DOI] [PubMed] [Google Scholar]
- 51.Sun SC, Wei L, Li M, Lin SL, Xu BZ, Liang XW, Kim NH, Schatten H, Lu SS, Sun QY. Perturbation of survivin expression affects chromosome alignment and spindle checkpoint in mouse oocyte meiotic maturation. Cell Cycle 2009; 8:3365-72; PMID:19806029; http://dx.doi.org/ 10.4161/cc.8.20.9855 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.