Abstract
Overexpression of cyclin D1 and its catalytic partner, CDK4, is frequently seen in human cancers. We constructed cyclin D1 and CDK4 protein interaction network in a human breast cancer cell line MCF7, and identified novel CDK4 protein partners. Among CDK4 interactors we observed several proteins functioning in protein folding and in complex assembly. One of the novel partners of CDK4 is FKBP5, which we found to be required to maintain CDK4 levels in cancer cells. An integrative analysis of the extended cyclin D1 cancer interactome and somatic copy number alterations in human cancers identified BAIAPL21 as a potential novel human oncogene. We observed that in several human tumor types BAIAPL21 is expressed at higher levels as compared to normal tissue. Forced overexpression of BAIAPL21 augmented anchorage independent growth, increased colony formation by cancer cells and strongly enhanced the ability of cells to form tumors in vivo. Lastly, we derived an Aggregate Expression Score (AES), which quantifies the expression of all cyclin D1 interactors in a given tumor. We observed that AES has a prognostic value among patients with ER-positive breast cancers. These studies illustrate the utility of analyzing the interactomes of proteins involved in cancer to uncover potential oncogenes, or to allow better cancer prognosis.
Keywords: breast cancer, CDK4, cyclin D1, interactome, oncogenes, oncogenic signature
Abbreviations
- ACN
acetonitrile
- AES
aggregate expression score
- ATCC
American type culture collection
- DMEM
Dulbecco's Modified Eagle's medium
- FBS
fetal bovine serum
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- PPI
protein-protein interaction
- RPMI
Roswell Park Memorial Institute medium
- SCNA
somatic copy-number variation
- sicont
control small interfering RNA
- sicyclin D1
cyclin D1-specific small interfering RNA
- siFKBP4
FKBP4-specific small interfering RNA
- siFKBP5
FKBP5-specific small interfering RNA
- siRNA
small interfering RNA
- TCGA
the cancer genome atlas
- WB
immunoblotting
Introduction
Cyclin D1 belongs to the core cell cycle machinery. Once induced, it binds and activates the cyclin-dependent kinases CDK4 and CDK6. Cyclin D-CDK4/6 holoenzymes phosphorylate proteins governing cell cycle progression, such as the retinoblastoma protein, pRB.1 Overexpression of cyclin D1 is found in several human cancer types, including breast cancers2 colon cancers,4,5 squamous cell carcinomas,6 multiple myelomas,7 and mantle cell lymphomas.8 Many of these cancers contain amplification or rearrangements within the cyclin D1 (CCND1) locus. Indeed, CCND1 represents the second most frequently amplified gene across all human cancer types.9 Targeted overexpression of cyclin D1 using transgenic mouse models led to formation of malignant lesions, thereby providing a direct evidence for the causative role of cyclin D1 overexpression in oncogenesis.10-12 Moreover, the continued presence of cyclin D1 is required for maintenance of the malignant phenotype, as an acute ablation of cyclin D1 in breast cancer-bearing mice blocked cancer progression.13 Collectively, all these findings point to cyclin D1 as an attractive target for cancer therapy.14 Cyclin D1 also plays roles beyond cell cycle progression,15 and it is highly expressed in non-proliferating, senescent cells.16,17
Developments of high-throughput platforms and bioinformatic analyses have helped to reveal novel information about disease-causing proteins. Recently, we constructed a protein-protein interaction (PPI) network of cyclin D1 (cyclin D1 interactome) from 5 different human cancer cell lines representing mammary, squamous cell and colorectal carcinomas and mantle cell lymphoma.18 This oncogenic cyclin D1 network was composed of 132 proteins. Gene ontology analyses revealed that in cancer cells cyclin D1 interacts with proteins regulating cell cycle and proteins functioning in DNA repair pathways, both of which play roles in cancer formation.19
Because of a remarkable involvement of cyclin D1 overexpression in human cancers, we hypothesized that the cyclin D1 interactome may be enriched for cancer-causing proteins, and may allow identification of new oncogenes. We further hypothesized that the expression levels of cyclin D1 interactors (cyclin D1 interactome signature) may allow one to stratify cancer patients for prognostic reasons. To test these predictions, in this study we performed integrative analyses of cyclin D1 interactomes with the list of copy number alterations in human cancers, and with The Cancer Genome Atlas (TCGA)20 breast cancer dataset. In addition, we constructed a joint PPI network of cyclin D1 and its kinase partner, the cyclin-dependent kinase 4 (CDK4), in a human breast cancer cell line and identified novel cyclin D1- and CDK4-interacting proteins.
Results
An oncogenic cyclin D1-CDK4 interactome in breast cancer cells
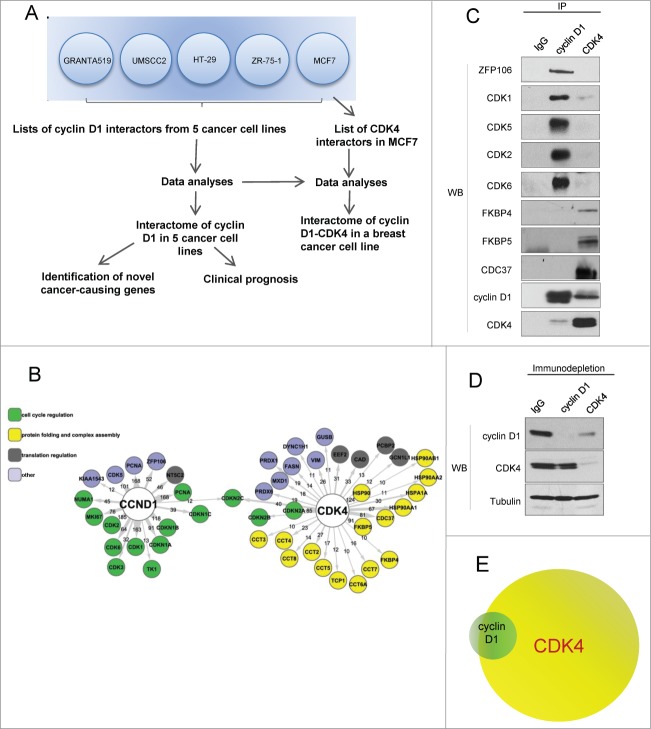
To determine the identity of cyclin D1 and CDK4 interactors in breast cancer cells, we expressed tandemly (Flag- and HA)-tagged versions of cyclin D1 and CDK4 in a human breast cancer cell line MCF7. We then used sequential immunoaffinity purifications with anti-Flag and anti-HA antibodies, followed by repeated rounds of liquid chromatography-tandem mass spectrometry (LC-MS/MS)18 to determine the identity of cyclin D118 and CDK4 interacting proteins. Integration of the results allowed us to construct breast cancer cyclin D1-CDK4 oncogenic network (Fig. 1A and B; Tables S1 and S2). Surprisingly, we found very little overlap between protein partners of cyclin D1 and those of CDK4. The great majority of cyclin D1 interactors were not found in CDK4 immunoprecipitates and vice versa (Tables S1 and S2). The only protein detected both in cyclin D1 and CDK4 immunoprecipitates was a cell cycle inhibitor p18INK4C (CDKN2C) (Fig. 1B).
Figure 1.
Oncogenic cyclin D1-CDK4 interactomes in human cancer cells (A) Flow chart of the interactome analyses. Cyclin D1 joint interactome was made from 5 human cancer cell lines and was already published.18 CDK4 interactome was obtained by analyzing a breast cancer cell line MCF7. (B) The oncogenic cyclin D1-CDK4 interactome in MCF7 cells. Colors indicate biological processes implicated for each of the interactors. Interactors are labeled by their gene names. Lines indicate whether a protein was detected as cyclin D1 (CCND1) or CDK4-associated protein; numbers on the lines represent number of spectra for a given protein detected in mass spectrometry analyses. The interactome of cyclin D1 was previously published.18 (C) Immunoprecipitation (IP) - immunoblot (WB) verification of cyclin D1 and CDK4 interacting proteins. Cell lysates were immunoprecipitated with either anti-cyclin D1 or anti CDK4 antibody, or isotype-matched mouse IgG (IgG). Immunoprecipitated products were resolved on SDS-PAGE gels and immunoblotted with the indicated antibodies. (D) Immunodepletion of cyclin D1 or CDK4. Lysates were depleted with anti-cyclin D1 or anti-CDK4 antibodies; isotype-matched IgG was used as a control. Supernatants were immunoblotted with the indicated antibodies. (E) Proposed stoichiometry of cyclin D1-CDK4 interaction in MCF7 cells. The majority of cyclin D1 molecules (green) interact with CDK4 (yellow), while only a small fraction of CDK4 forms complex with cyclin D1. Please note that the sizes of circles representing cyclin D1 and CDK4 are hypothetical, as we do not know the absolute number of cyclin D1 and CDK4 molecules per cell.
To verify the results of mass spectrometry analyses, cyclin D1 and CDK4 were immunoprecipitated from MCF7 cells, and immunoblotted with antibodies against various cyclin D1 and CDK4 interactors. We confirmed that cyclin D1-specific partners, including a novel interactor - zinc finger ZFP106 protein, were not found in anti-CDK4 immunoprecipitates (Fig. 1C). Conversely, CDC37, FKBP4, and FKBP5 proteins were confirmed as CDK4-specific interactors (Fig. 1C).
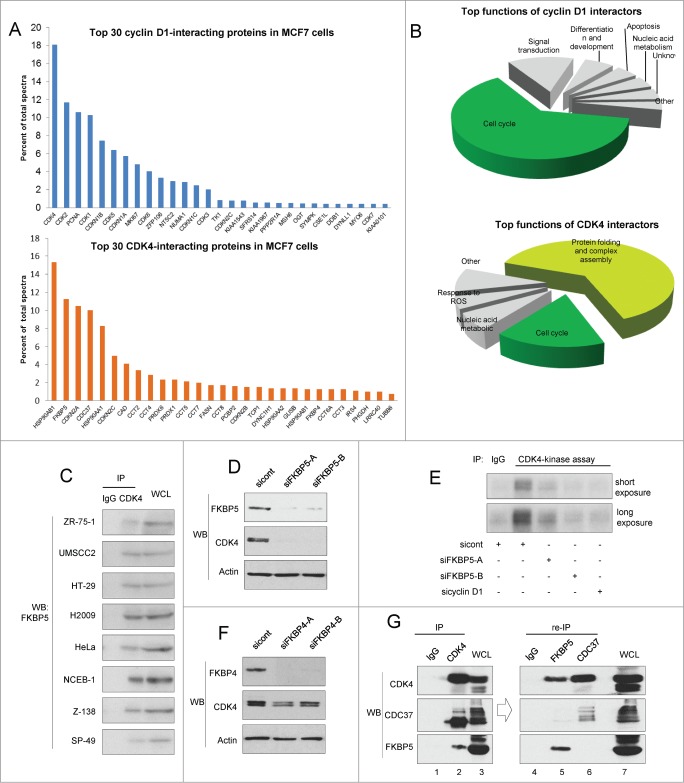
We next analyzed the biological function of cyclin D1 and CDK4 interactors with DAVID software.21,22 We found that the majority of cyclin D1 partners such as CDK4, CDK2, p21(CDKN1A), p27(CDKN1B) belonged to “cell cycle” category (Fig. 1B). In contrast, most of CDK4 interactors (67%) represented proteins involved in protein folding and complex assembly. Among CDK4 interactors, we observed several previously unknown partners, such as FKBP4, FKBP5, CAD, CCT2, CCT4, and PRDX6.
To better understand the stoichiometry of cyclin D1-CDK4 interaction, we performed immunodepletion experiments. First, we immunodepleted cyclin D1 from MCF7 lysates. We found that a nearly complete depletion of cyclin D1 eliminated only a small fraction of CDK4 (Fig. 1D, second lane). In contrast, when CDK4 was depleted from the lysates, the majority of cyclin D1 pool was also depleted (Fig. 1D, third lane). This indicates that in MCF7 cells, the great majority of cyclin D1 molecules interacts with CDK4 (the rest binds to other CDKs and to cyclin D1-specific partners, such as ZFP106, Fig. 1B). On the other hand, only a small portion of CDK4 molecules interacts with cyclin D1 and most of CDK4 molecules reside in protein complexes that are devoid of cyclin D1 (Fig. 1C and E). This cyclin D1-free pool of CDK4 interacts with proteins involved in protein folding and in complex assembly. We observed that proteins of the Hsp90 kinase chaperone complex, CDC37 and Hsp90, interacted with CDK4 outside of the cyclin D1-CDK4 complex (Figs. 1B, 2A and B).23-25 As shown in Fig. 1C, no interaction between CDC37 and cyclin D1 was observed in MCF7 cells (Fig. 1C).
FKBP5, a novel CDK4-binding protein, is required for full CDK4 expression and kinase activity
Our mass spectrometry analyses revealed that CDK4 interacts with several previously unknown partners representing chaperone proteins, such as members of the CCT complex, as well as with 2 members of FK506-binding family, FKBP4 and FKBP5 (Figs. 1B and 2A). The interaction of CDK4 with FKBP4 and FKBP5 was confirmed by immunoblotting (Fig. 1C). FKBPs represent a family of proteins that bind to immunosuppressive compounds such as FK506, rapamycin, and cyclosporin A. FKBPs are involved in several biochemical processes including protein folding, receptor signaling, and transcription.26,27 We verified the interaction between CDK4 and FKBP5 in a panel of human cancer cell lines. FKBP5 co-precipitated with CDK4 in every cell line tested, suggesting that the interaction is ubiquitous (Fig. 2C). Since FKBP5 appeared to be one of the most abundant CDK4-binding proteins, as judged by our mass spectrometry analyses (Fig 2A; Table S1), we tested whether FKBP5 is required for CDK4 function and stability. Depletion of FKBP5 using 2 independent siRNAs significantly reduced the levels of CDK4 protein (Fig. 2D), and diminished CDK4 kinase activity (Fig. 2E). Depletion of another CDK4 interactor, FKBP4 led to only modest reduction of CDK4 levels (Fig 2F). Collectively, these findings suggest that FKBP5 acts as CDK4 chaperonin and plays a role in controlling CDK4 levels.
Figure 2.
(See opposite page). Analyses of cyclin D1 and CDK4 interactors in breast cancer cells (A) Top 30 cyclin D1 and CDK4 interactors detected by mass spectrometry analyses. (B) Top biological processes of cyclin D1 and CDK4 interactors. (C) Physical interaction between CDK4 and FKBP5 in various human cancer cell lines. CDK4 was immunoprecipitated in the indicated cell lines, and immunoblots were probed with an anti-FKBP5 antibody. Isotype-matched IgG was used as a control. (D) The impact of FKBP5 depletion on CDK4 levels. MCF7 cells were transfected with 2 independent anti-FKBP5 siRNAs (siFKBP5-A and siFKBP5-B), or with control siRNA (sicont), and the levels of FKBP5 and CDK4 were gauged by immunoblotting (WB) using the indicated antibodies. Actin was used as a loading control. (E) In vitro CDK4 kinase assays. MCF7 cells were transfected with siRNAs against FKBP5 (siFKBP5-A and siFKBP5-B), cyclin D1 (sicyclin D1) or with a control non-targeting siRNA (sicont). CDK4 was immunoprecipitated (IP) and used for in vitro kinase reactions with the retinoblastoma protein as a substrate. Immunoprecipitation with isotype-matched IgG (IgG) was used as a negative control. Short exposure: 30 min, long exposure: 3 hrs. (F) The impact of FKBP4 depletion on CDK4 levels. The experiment was performed as in panel D, except that 2 independent anti-FKBP4 siRNAs were used (siFKBP4-A and siFKBP4-B). (G) CDK4 immunoprecipitation followed by re-immunoprecipitation of either FKBP5 or CDC37. Flag-tagged CDK4 was immunoprecipitated (IP) from MCF7 cells using an anti-Flag antibody; complexes were then eluted with 3XFlag peptide. Ten percent of the eluent was resolved on an SDS-PAGE gel and immunoblotted with the indicated antibodies (left panel). The remaining eluent was split into 3 equal parts. One part was subjected to re-immunoprecipitation with anti-FKBP5 antibody (lane 5), the second with anti-CDC37 antibody (lane 6), the third with IgG (for control, lane 4). Co-immuprecipitated proteins were then detected with the indicated antibodies (right panel). IgG was also used for control immunoprecipitation in lane 1. WCL; whole cell lysate (lanes 3 and 7).
Previous reports indicated that CDC37 physically interacts with CDK4 and stabilizes CDK4 protein levels.23 Given our observation that FKBP5 appeared to play a similar role, we investigated whether FKBP5 is a part of the CDK4-CDC37 complex. Using MCF7 cells expressing Flag-tagged CDK4, we immunoprecipitated CDK4 with anti-Flag antibody, eluted the complex from the beads using the Flag peptide, re-immunoprecipitated the complex with anti-FKBP5 (Fig. 2G, lane 5) or CDC37 (Fig. 2G, lane 6) antibodies, and immunoblotted with antibodies against FKBP5 and CDC37. We found that CDK4 interacts with FKBP5 or with CDC37, but it does not form a ternary CDK4-FKBP5-CDC37 complex. These observations suggest that FKBP5 stabilizes CDK4 level, via a CDC37-independent mechanism.
Integrative analysis of cyclin D1 interactome and somatic copy number alterations in human cancers uncovers a potential oncogene, BAIAP2L1
Previously, we have identified cyclin D1 interactomes from 5 human cancer cell lines, MCF7, ZR-75–1, UMSCC2, HT-29, and Granta519.18 Such interaction networks have been shown to represent a good source for uncovering biological functions of the participating proteins. One of the approaches, called “guilt-by-association” principle,28 predicts a function of a novel protein based on known functions of its interactors. We applied a reverse logic to the cyclin D1 interactome. Given the fact that cyclin D1 represents a well-established oncogene, we asked whether cyclin D1 partners are enriched for proteins implicated in oncogenesis. To this end, we interrogated the cyclin D1 interactome for the presence of genes associated with tumorigenesis, as defined by Gene Census data (n = 513 genes) obtained from the COSMIC database.29 Among cyclin D1 interactors we observed 12 proteins that have been causally implicated in cancer development (CDK6, FANCD2, CDKN2C, NUMA1, NONO, RB1, CDK4, XPO1, PPP2R1A, MSH6, BRCA2, and IKZF1). This number is significantly larger than that expected by a random chance (p = 1.15X10−5, Fisher Exact test). These observations suggested to us that the cyclin D1 interactome may contain additional, currently unknown cancer-relevant proteins.
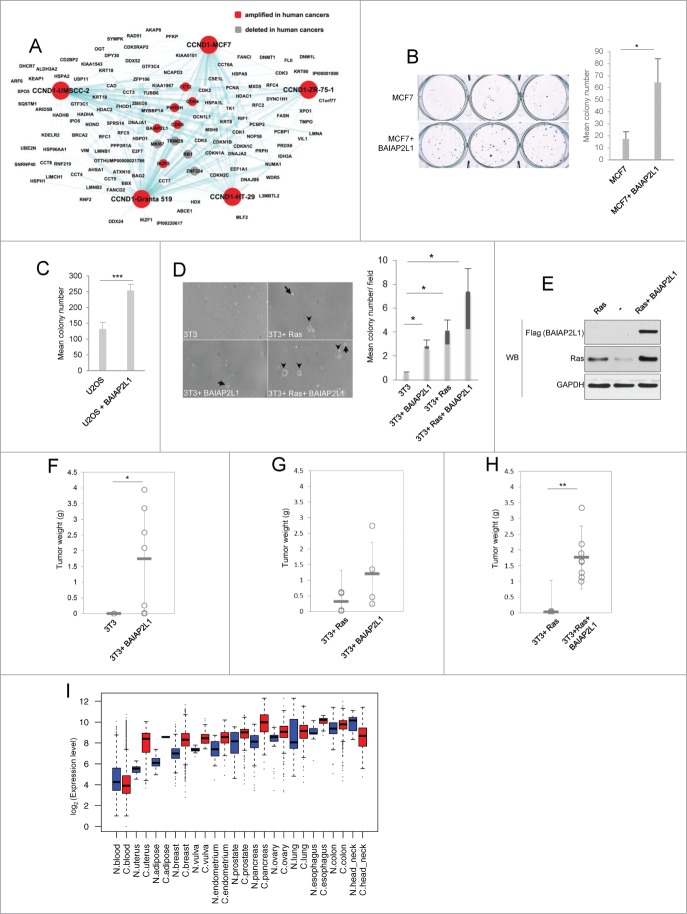
To identify these proteins, we intersected our interactome with a comprehensive list of genomic regions found to be frequently amplified or deleted in a large-scale analysis of over 3,000 of human cancers.9 We searched for interactors whose genes map to commonly amplified or deleted regions in the human cancer genome. Genes encoding 6 cyclin D1 interactors: CDK4, CDK6, IKZF3, PHGDH, CCT2, and BAIAP2L1 map to peaks of commonly amplified chromosomal cancer regions (Fig. 3A, highlighted in red), whereas 4 cyclin D1 interactor genes: RB, TRIM28, ZNF324, and MKI67 are located within the peak deleted areas (Fig. 3A, highlighted in gray). These numbers indicated a significant enrichment for amplified and deleted genes within the cyclin D1 interactome (p = 0.0015, Fisher Exact Test) and suggested that some of these genes may encode previously unknown cancer-relevant proteins.
Figure 3.
(See opposite page). Analyses of cyclin D1 interactome to identify novel cancer-relevant genes (A) Cyclin D1 interactome from 5 cancer cell lines (this interactome was already presented in Jirawatnotai et al.18) Nodes represent gene symbols of cyclin D1 interactors. Lines depict interactions found by our mass spectrometry analyses. Genes found to be amplified in human cancers9 are highlighted in red, genes deleted in cancers9 in gray. (B) MCF7 colony formation assay. MCF7 cells stably expressing BAIAP2L1 (MCF7 + BAIAP2L1) or cells transfected with an empty vector (MCF7) were seeded at a low density, and colonies were stained with crystal violet and enumerated after 3 weeks. Right panel shows mean colony numbers, error bars represent standard deviation (n = 3), *; P < 0.05. (C) U2OS colony formation assay. U2OS cells were engineered to stably express BAIAP2L1 (U2OS + BAIAP2L1), or were transfected with an empty vector (U2OS) and used for assays as in panel B. Bar graphs represent mean colony numbers, error bars standard deviation (n = 3); ***, P < 0.005. (D) Soft agar assay of murine 3T3 cells stably expressing BAIAP2L1 (3T3+ BAIAP2L1), Ras (3T3 + Ras), Ras + BAIAP2L1 (3T3 + Ras + BAIAP2L1), or transfected with an empty vector (3T3). Left panel: microscopic images of representative fields. Right panel: Bar graphs showing mean total colony numbers; dark gray bars show mean numbers of large colonies ( 200 cells or more). Error bars, standard deviation; *, P < 0.05 (E) Protein levels of Ras and BAIAP2L1 in Ras- and in Ras + BAIAP2L1 transduced 3T3 cells, determined by immunoblotting (WB). Anti-Flag antibody was used to detect ectopically expressed Flag-BAIAP2L1. (F) Weights of BAIAP2L1-induced tumors at 8 weeks post inoculation. Mice were injected subcutaneously with 3T3 cells (3T3) or with 3T3 cells stably expressing BAIAP2L1 (3T3 + BAIAP2L1). Each circle corresponds to a separate tumor, horizontal lines depict mean values; *, P < 0.05 (G) Weights of Ras- or BAIAP2L1-induced tumors at 6 weeks post inoculation. Mice were injected subcutaneously with 3T3 cells stably expressing activated Ras (3T3 + Ras) or BAIAP2L1 (3T3 + BAIAP2L1). Each circle corresponds to a separate tumor, horizontal lines depict mean values. (H) Weights of Ras- or Ras + BAIAP2L1-induced tumors at 4 weeks post inoculation. Mice were injected subcutaneously with 3T3 cells stably expressing activated Ras (3T3 + Ras) or Ras plus BAIAP2L1 (3T3 + Ras + BAIAP2L1). Each circle corresponds to a separate tumor, horizontal lines depict mean values; **, P < 0.001 (I) Boxplots of log-2 transformed expression levels of BAIAP2L1 transcripts in 13 tissue types with differential expression in cancer versus in normal tissue. Source of material is described on the x-axis: N- denotes normal and C- denotes cancer.
To test this notion, we focused on a novel cyclin D1 interacting protein with relatively unknown function, BAIAP2L1 (BAI1-associated protein 2-like 1), a phosphorylation substrate for insulin receptor.30 Since the gene encoding BAIAP2L1 is amplified in human cancers (please see above), we hypothesized that this protein may have oncogenic properties. Prior evidence suggested that BAIAP2L1 may play a role in cancer formation. Thus, a fusion protein between a growth factor receptor FGFR3 and BAIAP2L1 was reported to be expressed in bladder cancer cells.31 Moreover, BAIAP2L1 was also shown to play a role in cell migration.32 We tested the impact of BAIAP2L1 on cell proliferation by ectopically expressing it in MCF7 cells. We observed that overexpression of BAIAP2L1 significantly increased colony forming ability of MCF7 cells (Fig. 3B). BAIAP2L1 overexpression also enhanced colony formation in another human cancer cell line, an osteosarcoma U2OS cells (Fig. 3C).
To test the impact of BAIAP2L1 expression on anchorage-independent growth, we overexpressed BAIAP2L1 in immortalized murine 3T3 cells, or in 3T3 cells engineered by us to express activated Ha-Ras. Ectopic expression of BAIAP2L1 endowed 3T3 cells with the ability to grow in soft agar, and enhanced anchorage-independent growth of Ras-expressing 3T3 cells (Fig 3D). BAIAP2L1 expression not only increased the total number of foci, but it also increased the number of large foci in Ha-Ras-transformed 3T3 cells (Fig. 3D). Surprisingly, immunoblot analyses revealed that ectopic expression of BAIAP2L1 significantly augmented the levels of Ha-Ras in Ras-transformed cells (Fig 3E).
We also tested the impact of BAIAP2L1-overexpression on the ability of cells to form tumors in immunocompromised mice. To this end, we injected BAIAP2L1-overexpressing 3T3 cells under the skin of nude mice. While none of the mice subcutaneously implanted with 3T3 cells developed any tumors after 8 weeks, 6 out of 7 (85.57%) mice injected with BAIAP2L1-expressing 3T3 cells formed tumors (Fig. 3F; Table 1). BAIAP2L1 appeared to be a stronger transformation inducer for the murine 3T3 cells than Ras. Thus, at 6 weeks post-inoculation, tumors triggered by BAIAP2L1 expression were significantly larger than those induced by Ras (Fig. 3G; Table 1). Moreover, cells expressing BAIAP2L1 formed tumors in a higher fraction of mice than cells expressing Ras (85.57% vs. 66%, Table 1). Co-expression of Ras and BAIAP2L1 in 3T3 cells synergistically enhanced the ability of cells to form tumors, and increased both tumor weight as well as the fraction of mice with tumors (Fig. 3H; Table 1). Tumors co-expressing Ras and BAIAP2L1 grew so rapidly, that the endpoint of the experiment had to be shortened to only 4 weeks (Fig. 3H; Table 1). These analyses indicate that BAIAP2L1 has several properties of an oncogene.
Table 1.
Tumor formation in nude mice
| Cells | Number of mice with a tumor at endpoint | Percent of mice with a tumor at endpoint | Time to endpoint |
|---|---|---|---|
| 3T3 |
0/7 |
0 |
N/A |
| 3T3 + Ras |
8/12 |
66.7 |
>6 weeks |
| 3T3 + BAIAP2L1 |
6/7 |
85.7 |
4-8 weeks |
| 3T3 + Ras + BAIAP2L1 | 8/8 | 100 | 4 weeks |
Numbers and percentages of mice with tumors at the endpoints are indicated. Mice injected with 3T3 cells were observed for 8 weeks, and no tumors were detected. N/A, not applicable.
We next compared the levels of BAIAP2L1 transcripts in various cancer types, versus in corresponding normal tissues using GENT (Gene Expression Across Normal and Tumor Tissue) database.33 We were able to query BAIAP2L1 expression across 32 tumor types and in normal tissues from 21,434 samples (17,931 tumor samples and 3,503 normal tissue samples). Mean expression level of BAIAP2L1 in tumor samples was higher than that seen in normal tissues (303.80 ± 443.85 vs 235.93 ± 386.38 [mean± SD], p = 1.6019 × 10−17, log-rank test). Levels of BAIAP2L1 expression in tumor samples were then compared to the corresponding types of normal tissues using linear mixed model, which allows each type of tissue to have different baseline expression of BAIAP2L1. Among the 32 types of tumors and normal tissues, the cancer-normal matched samples were available for analyses of 25 tumor types (see Materials in Methods). In these 25 tumor types, BAIAP2L1 expression levels were 1.fold17- higher in tumor samples than in normal tissues (95%CI from 1.12 to 1.24, p = 4.25x10−10). Thirteen tumor-normal matched types showed statistically significant differences in BAIAP2L1 expression levels between tumors vs. normal samples (p < 0.002) (Fig. 3I; Table S3). This list includes malignancies of blood, uterus, adipose tissue, breast, vulva, endometrium, prostate, pancreas, ovary, lung, esophagus, colon, and head and neck. In all pairs, except blood and head-neck cancers, BAIAP2L1 was expressed at higher levels in cancer samples than in normal counterparts (Fig. 3I, boxplots). These observations are consistent with a growth-promoting function for BAIAP2L1 in cancer cells, and support the notion that BAIAP2L1 likely represents a bona fide oncogene.
A gene signature generated from cyclin D1 interactome holds prognostic value for survival of breast cancer patients
Since the cyclin D1 interactome is enriched for potential cancer-relevant genes, we hypothesized that expression levels of cyclin D1 interactors (cyclin D1 interactome signature) might contain prognostic value for survival of cancer patients. To test this, we focused on estrogen receptor-positive breast cancers due to the availability of clinical data that utilizes large cohorts of patients, and given the well-established overexpression of cyclin D1 in this breast cancer subtype.34 We interrogated the TCGA breast cancer data set and imputed the data to retain only individuals with confirmed ER-positive mammary carcinomas and with known survival status (n = 45).
We first asked whether cyclin D1 expression correlates with patient survival. We observed no such correlation in our cohort (data not shown). Next, we used Aggregate Expression Score (AES) (as described in Materials and Methods), to assess the prognostic value of cyclin D1 interactome levels. Briefly, AES summarizes the overall expression level of the interactome in a given tumor, by summing up the number of genes expressed one standard deviation above (+1) and one standard deviation below (−1) of the mean expression levels seen across all cancer samples (please see Materials and Methods). We then stratified the patients into AES high and AES low groups to assess if interactome expression level correlates with survival. We found that high AES of cyclin D1 interactome correlated with poor survival rate in patients bearing ER-positive breast cancers (p = 0.042, log-rank test) (Fig. 4A). In contrast, the interactome of CDK4 had no predictive value (data not shown). To verify our observation in an independent cohort, we repeated the same analysis using the TRANSBIG data set,35 which contains a large number of patients with ER-positive breast cancers (n = 134). Again, we observed that the cyclin D1 interactome AES had a predictive value for patients survival; with high AES corresponding to poor survival (KM-plot, p =0.032, log-rank test) (Fig. 4B).
Figure 4.
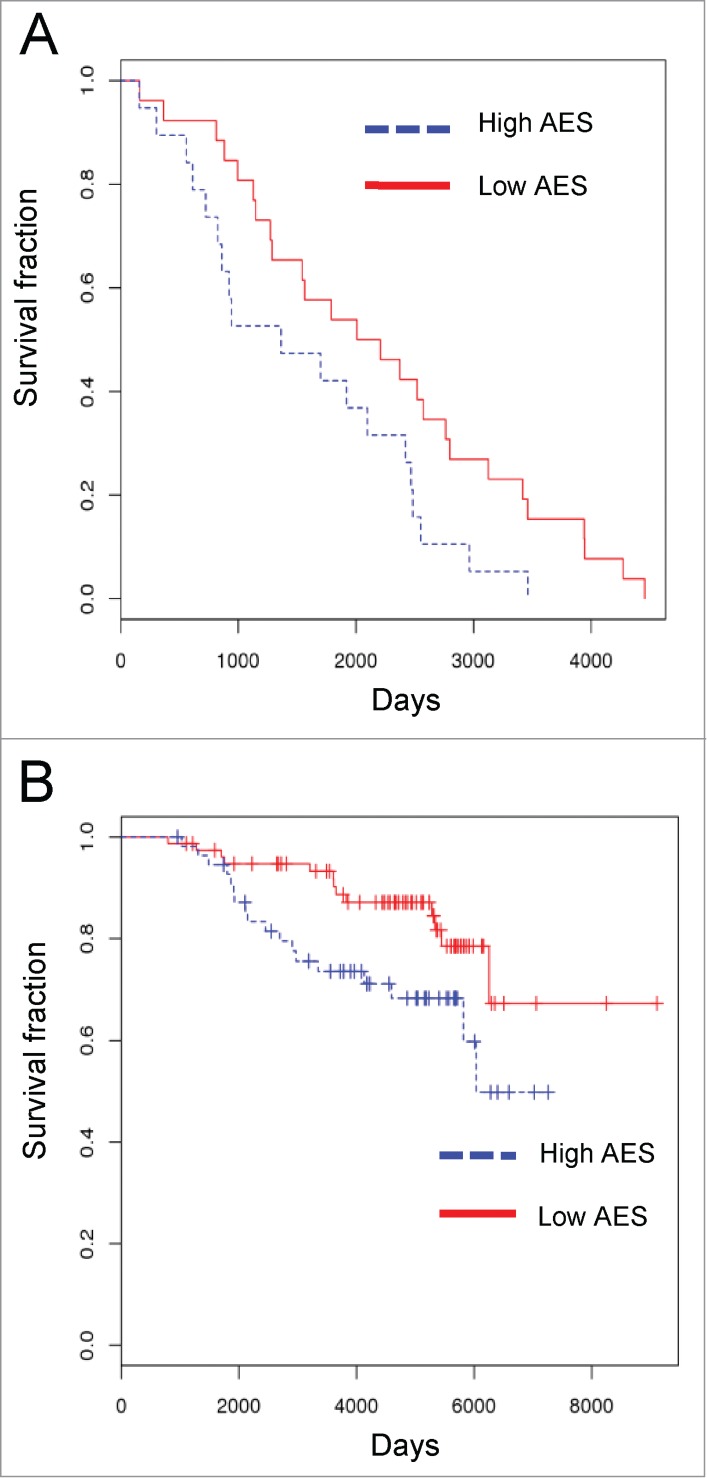
Prognostic value of cyclin D1 interactome-based signature(A) ER-positive breast cancers from TCGA data set were stratified by cyclin D1 interactome Aggregated Expression Score (AES). Red and blue lines indicate survival of patients with low and high AES scores, respectively. p = 0.042 (B) Patients with ER-positive breast cancers from TRANSBIG cohort were stratified and analyzed as in Fig. A. Red and blue linesindicate survival of patients with low and high AES scores, respectively. p = 0.032.
Discussion
Recently, we have generated an integrated oncogenic cyclin D1 interactome from several human cancer cell lines.18 In the current study, we analyzed the cyclin D1 interactome in conjunction with other data sets, namely in the context of the CDK4 interactome, somatic copy-number alterations (SCNA), gene expression, and clinical data sets from human cancer patients to extract additional contextual biological information.
Our analyses of the CDK4 interactome revealed that a member of the FKBP family (FKBP5) represents a novel chaperone regulating CDK4 protein levels and kinase activity. Several studies postulated that FKBP5 may play a role in tumorigenesis. Thus, FKBP5 was shown to stimulate androgen-dependent transcriptional activation and to promote prostate cancer growth.36 Ectopic overexpression of FKBP5 was demonstrated to increase radio-resistance of melanoma cells.37 However, other studies implicated FKBP5 as a negative regulator of cell growth, by inhibiting the AKT signaling pathway. Specifically, overexpression of FKBP5 was shown to decrease the activating AKT-Ser473 phosphorylation, whereas depletion of FKBP5 had an opposite effect.38 We observed that FKBP5 functions to stabilize CDK4. We found that depletion of FKBP5 led to decreased CDK4 protein levels and decreased CDK4 kinase activity. Given the well-established role of CDK4 hyperactivation in tumorigenesis, it seems likely that FKBP5 promotes oncogenesis in part by stabilizing CDK4.
Analyses across several thousands of human tumors led to delineation of regions commonly amplified (or commonly deleted) in cancers.9 However, these regions are usually large (median length of 1.8 Mb, range 0.5 kb-85 Mb), and hence they contain a substantial number of genes.9,39 One of the main challenges is to identify “driver” cancer causing genes that reside in the commonly amplified or deleted regions. We hypothesized that by overlaying copy number alteration data with cancer cell interactome of a well-established oncogene (cyclin D1), we might uncover novel cancer-causing genes. Indeed, we demonstrated that cyclin D1 interactome is enriched for known oncogenes and tumor suppressor genes. Moreover, we used the intersection of the interactome and copy number alteration analyses to identify BAIAP2L1 as a potential novel oncogene.
We found that BAIAP2L1 has several characteristics of a bona fide oncogene. First, the BAIAP2L1 gene is amplified in several cancer types.9 Second, the gene is expressed at higher levels in cancer samples, as compared to normal, non-transformed counterparts. Third, ectopic overexpression of BAIAP2L1 increased colony formation of MCF7 and U2OS cancer cells. Fourth, BAIAP2L1 overexpression was sufficient to trigger anchorage-independent growth of 3T3 cells, and endowed 3T3 cells with an ability to form tumors in nude mice. Further studies are needed to decipher the exact molecular mechanism of BAIAP2L1 oncogenic role. As stated above, we observed that BAIAP2L1 expression increased protein levels of the oncogenic Ras. Of note, it was recently reported that BAIAP2L1 can activate EGFR/ERK signaling pathway and promote cell proliferation of hepatocellular carcinoma.40
Our study examined the utility of using the interactome of an oncogene as a prognostic tool in cancer patients. We devised an AES scoring method to represent the interactome expression level in a given tumor, and then evaluated the prognostic value of AES to predict survival using the TCGA breast cancer cohort. The recentness of this cohort substantially limited the power of survival analysis, yet the score of cyclin D1 interactome correlated with survival of ER-positive breast cancer patients. In contrast, the levels of cyclin D1, on its own, had no any predictive value. This suggests that the interactome adds substantial prognostic information. We further confirmed our findings by analyzing the TRANSBIG cohort35 as a testing set. To the best of our knowledge, this is the first instance of using aggregate gene expression scoring of an oncogenic interactome to construct a prognostic signature. While there are several published multigene signatures, mostly derived from expression profiling,41-45 our method yielded comparable statistically significant prognostic value despite its simplicity.35
Since the cyclin D1 interactome AES was built from 4 diverse types of human tumors, it may have a predictive value also in other malignancies. With the rapidly growing amount of high-throughput data from various cancer types, in the future one will be able to study the prognostic value of the signature in other types of neoplastic diseases.
Materials and Methods
Cell lines and nuclear extracts
MCF7, HEK239, HeLa, ZR-75–1, HT-29, U2OS, and Z138 cells were purchased from the American Type Culture Collection (ATCC). UMSCC2 cells were purchased from University of Michigan. NCEB-1 and SP-49 cells were a gift from Dr. Jiri Bartek, the Danish Cancer Society Research Center. H2009 cells were a gift from Novartis. Mouse 3T3 cells were generated as described.46 All cell lines were maintained in high glucose DMEM with 10% fetal bovine serum (FBS) plus penicillin/streptomycin, except Z138 and U2OS cells that were grown in RPMI1640 and McCoy's medium, respectively, with 10% FBS plus and penicillin/streptomycin,
Tandemly tagged CDK4 was generated by cloning human CDK4 cDNA into Xho I and Not I sites of pOZ-FH-N expression vector.47 Nuclear extractions were performed as described,48 using 2 ×107 cells expressing tagged CDK4 as a source of material. Nuclear extracts were then used for tandem immunoaffinity purification18 of CDK4-containing complexes. In parallel, we obtained nuclear extracts from cells expressing empty pOZ-FH-N vectors, which were used for control (“mock”) purifications.
Mass spectrometry analysis and generation of cyclin D1 and CDK4 interactomes
Cyclin D1 interactome from 5 cancer cell lines, including MCF7 cells, was from our previous report.18 Mass spectrometry analysis of CDK4 interacting protein partners from MCF7 cells was performed as follows. Purified CDK4-containing complexes from MCF7 cells were subjected to 3 independent mass spectrometry runs (each using a sample containing approximately 300 ng of CDK4). “Mock” purified samples were analyzed in parallel.
Sample preparation for mass spectrometry analysis was as described.18 A sample that contained at least 300 ng of tagged CDK4 (or the corresponding amount of material prepared from “mock” purified samples) was TCA precipitated, and digested for 5-6 hours at 37°C in a reaction mixture consisting of 50 mM ammonium bicarbonate, pH 8.0, 10 % Acetonitrile (ACN) and 400 ng modified trypsin (Promega). The digestion mixture was quenched with 50 % ACN, 5 % formic acid (FA), and lyophilized to dryness. Dried peptides were then desalted using Empore C18 solid phase extraction disks (3M) as previously described18. Samples were resuspended in 5 % ACN, 5 % FA prior to analysis by liquid chromatography and tandem mass spectrometry.
Construction of high-confidence CDK4 and cyclin D1 interactomes
CDK4 interacting proteins were selected as high confidence interacting proteins, if they fulfilled all of the following criteria. (1) The protein was detected by more than 10 independent peptides in CDK4 immunopurifications. (2) The number of spectra seen for this protein in CDK4 immunopurifications was over 20 times higher than that observed in mock purified samples. (3) The protein had a substantially higher probability of being detected in CDK4 immunopurifications than in the “mock” purifications (p ≤ 0.01 using Chi2 test).
Using these criteria, we identified 30 high-confidence interactors of CDK4.
The same criteria were previously applied to identify high-confidence cyclin D1 interactors from 5 cancer cell lines, including MCF7 cells.18 In our previous report18 we identified 17 high-confidence interactors for cyclin D1 in MCF7 cells; these interactors are shown in Fig. 1B.
Statistical analysis of cyclin D1 interactome
The list of genes for which somatic mutations have been implicated in tumorigenesis was obtained from the COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) and used to determine the enrichment of cancer-causing genes within the list of high confidence cyclin D1 interacting proteins. In addition, cyclin D1 interacting proteins were annotated for somatic copy-number alterations as determined by Beroukhim et al.9 Genes within the top 30% of the GISTIC q-values were used. Enrichment was determined by Fisher Exact test.
Co-immunoprecipitation and immunodepletion analyses
Cell lysates were prepared in ELB buffer (160 mM NaCl, 50 mM HEPES pH 7.5, 5 mM EDTA pH 8.0 and 0.1% NP-40) supplemented with Roche cocktail proteinase inhibitor. Four μg of mouse anti-cyclin D1 antibody (Ab1 or Ab3, Lab Vision) or mouse anti-CDK4 antibody (Ab1, Lab Vision) were incubated with 5 mg of lysates. Protein G beads were then added, and immunoprecipitated proteins were detected by immunoblotting with the antibodies against: CDK1 (A17.1), from Lab Vision, ZFP106 (A301–527A), FKBP4 (A301–426A), FKBP5 (A301–430A) from Bethyl Laboratories, CDK2 (M2), CDK4 (C-22), CDK5 (DC-17), CDK6 (C-21), CDC37 (H-271) from Santa Cruz Biotechnologies, Flag-M5 (F4042) from Sigma Aldrich, Ras (3965) from Cell Signaling Technology. Anti-β-actin antibody (AC-15) from Sigma or anti-GAPDH antibody from Cell Signaling were used to control for loading.
Immunodepletion was performed similarly to immunoprecipitation except that 10 μg of the indicated antibodies were used. The supernatants were examined by immunoblotting using the indicated antibodies.
Immunoprecipitation-re-immunoprecipitation was performed as follows. MCF7 cells were transfected with p3X Flag-CMV-10-CDK4 mammalian expression vector (E7658, from Sigma-Aldrich). Cell lysates were prepared in ELB buffer. CDK4 was immunoprecipitated using anti-Flag-M2 affinity beads (F2426, Sigma- Aldrich). Immunoprecipitated proteins were eluted by addition of 200 mM of 3X FLAG® Peptide (F7499, Sigma-Aldrich) in ELB and divided into 3 equal parts. The first part was subjected to an anti-FKBP5 immunoprecipitation, the second to an anti-CDC37 immunoprecipitation, and the third to immunoprecipitation with control IgG. Immunoprecipitated complexes were analyzed by immunoblotting using anti-CDK4, FKBP5 and CDC37 antibodies.
Immune complex kinase assays
The immune complex kinase assays were performed as described.49 Briefly, 106 cells were lysed at in 300 μl of ELB buffer supplemented with Roche proteinase inhibitor cocktail, 10 mM β-glycerophosphate, 1 mM NaF, and 0.1 mM sodium orthovanadate (Sigma Aldrich) and sonicated at 4°C. Lysates were incubated with protein G-sepharose beads pre-coated with saturating amounts of anti-CDK4 antibody (Ab1, Lab Vision). The beads were suspended in kinase buffer (50 mM HEPES [pH 7.5], 10 mM MgCl2, 1 mM DTT) containing 0.5 μg of GST-RB fragment as a substrate (sc-4112, Santa Cruz Biotechnologies) plus 2.5 mM EGTA, 10 mMβ-glycerophosphate, 0.1 mM sodium orthovanadate, 1 mM NaF, 20 μM ATP, and 10 μCi of γ-32P ATP. After incubation for 30 min at 30°C, the samples were resolved on SDS-PAGE gels, and analyzed autoradiography.
Depletion of FKBP4 and FKBP5
Knock-down of FKBP5 and FKBP4 was performed using 2 independent anti-FKBP5 siRNAs: siFKBP5-A (FKBP5–5, SI02780372) and siFKBP5-B (FKBP5–6, SI02780414), and 2 anti-FKBP4 siRNAs: siFKBP4-A (FKBP4–5, SI02780365), and siFKBP4-B (FKBP4–6, SI02780407). As a control, non-targeting siRNA (Allstars Negative Control siRNA, SI03650318) was used. All siRNAs were from Qiagen.
Ectopic expression of BAIAP2L1
BAIAP2L1 cDNA was obtained by reverse transcription (SuperScript III reverse transcriptase, Invitrogen) from total RNA isolated from U2OS cells, followed by PCR amplification using BAIAP2L1 forward primer: 5′ATATGCGGCCGCATCCCGGGGCCCGAG3′, and the reverse primer: 5′ATGGTACCTTCATCGAATGATGGGTGCCGA3′. BAIAP2L1 cDNA was then cloned into p3X FLAG-CMV-10 expression vector. The resulting p3X FLAG-CMV-10 BAIAP2L1 plasmid was transfected using Lipofectamine 2000 (Invitrogen). Cells stably expressing BAIAP2L1 were selected with neomycin (500 μg/ml).
Colony formation and soft agar assays
MCF7 or U2OS cells stably overexpressing BAIAP2L1 or transfected with an empty vector were seeded into 6 well plates in triplicate at 100, 200 or 500 cells per well and cultured for 10–12 d. Cells were stained with 0.1% crystal violet. Colonies that contained more than 25 cells were counted.
To generate murine 3T3 cells stably expressing activated v-Ha-Ras, cells were transduced with the retrovirus expressing activated Ras (pBabe-puro-RasV12, Addgene), or for control, with an empty vector. Cells were selected with puromycin (2 μg/ml) for 5 d. To overexpress BAIAP2L1, cells were transfected with p3X FLAG-CMV-10 BAIAP2L1 plasmid as above, and selected in G418 sulfate (500 μg/ml). To perform the soft agar assay, 2 ml of 1.2% noble agar (Becton Dickson) was prepared in DMEM to form the bottom layer of the plate. The top layer of the agar (0.8% noble agar in DMEM) contained cells at the concentration of 5 × 103- 5 × 104 of cells per 2 ml. All visible colonies were counted under 4X-20X magnification after 4 weeks.
In vivo tumor growth assays
All experiments were performed in accordance with the guidelines established by Dana-Farber Cancer Institute's Institutional Care and Use Committee. 4-6 weeks old female nude mice [Nu/Nu (CD-1), from Charles River Laboratories] were injected with 106 cells in 0.2 ml PBS and sacrificed 4-8 weeks after injection, when the tumors reached 2 cm in diameter.
Expression of BAIAP2L1 in normal vs. cancer samples
We searched the GENT database,33 which curates over 40,000 expression profiles measured by Affymetrix U133A or U133plus2 platform for BAIAP2L1 expression. The raw expression levels in cancers vs. in normal samples in 32 tissue types were compared using simple linear regression. Among the 32 tumor types available, only 25 had both cancer and normal samples (the remaining 7 had only tumor samples). These 25 cancer/normal sets were included in the subsequent analysis. The raw expression data were log-transformed and normalized to control for differences in baseline levels between different samples, e.g. cancers vs. normal samples. Then, a random intercept model was used to test for the differences in the expression levels between cancer vs. normal samples in each tissue type.
The difference between cancer vs. normal samples within each tissue type was later evaluated using simple linear regression. Differential expression within the given tissue type was considered statistically significant when p < 0.002 (0.05/25), using Bonferroni correction for multiple testing to control for type I error rate.
Construction of cyclin D1 interactome Aggregate Expression Score and survival analysis
Level 3 breast cancer expression data with clinical annotation was obtained from the NIHTCGA portal (https://tcga-data.nci.nih.gov/tcga/). Patients with ER-positive breast cancers for which with full molecular and survival data was available were used for analyses. We designed the Aggregate Expression Score (AES) to represent overall gene expression level of the interactome as follows:
and
where i represents gene i in the interactome, and sd and U indicate the standard deviation and mean gene expression of i, respectively. Cyclin D1 “up” and “down” expression was defined similarly as 1 standard deviation (SD) away from the mean. For every tumor, each of 132 cyclin D1 interactors was assigned a value of +1 (if the expression level of this interactor in this tumor was one SD or more above the mean expression level for this interactor seen in all breast cancer samples), −1 (if the expression level of this interactor in this tumor was at least one SD below the mean expression value for this interactor in all breast cancer samples), or “0” if none of the above 2 criteria were fulfilled. Subsequently, values for all interactors in a given tumor were added up, and AES was obtained. As “high AES,” we defined tumors with AES values in the upper 50% across all tumor samples; "low AES" had values are those in the bottom 50%.
Survival curves were estimated using the Kaplan–Meier method for patients partitioned into AES high and AES low groups. The association of AES with survival status was evaluated through a Cox proportional hazard model. All analysis was performed with the R software (http://www.R-project.org).
Funding Statement
This study was supported by Siriraj Research Development Fund and by R01 CA083688 and P01 CA080111 (to P.S.). S.J. is supported by “Chalermphrakiat” Grant and Siriraj Foundation D003421, Faculty of Medicine Siriraj Hospital, Mahidol University and the Thailand Research Fund RSA5580018. Y.E.W is supported through the CCCB and the Dana-Farber Strategic Plan Initiative.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev 2004; 18:2699-711; PMID:15545627; http://dx.doi.org/ 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 2. Gillett C, Fantl V, Smith R, Fisher C, Bartek J, Dickson C, Barnes D, Peters G. Amplification and overexpression of cyclin D1 in breast cancer detected by immunohistochemical staining. Cancer Res 1994; 54:1812-7; PMID:8137296. [PubMed] [Google Scholar]
- 3. Bartkova J, Lukas J, Muller H, Lutzhoft D, Strauss M, Bartek J. Cyclin D1 protein expression and function in human breast cancer. Int J Cancer 1994; 57:353-61; PMID:8168995; http://dx.doi.org/ 10.1002/ijc.2910570311. [DOI] [PubMed] [Google Scholar]
- 4. Bartkova J, Lukas J, Strauss M, Bartek J. The PRAD-1/cyclin D1 oncogene product accumulates aberrantly in a subset of colorectal carcinomas. Int J Cancer 1994; 58:568-73; PMID:8056453; http://dx.doi.org/ 10.1002/ijc.2910580420. [DOI] [PubMed] [Google Scholar]
- 5. Bartkova J, Lukas J, Strauss M, Bartek J. Cyclin D1 oncoprotein aberrantly accumulates in malignancies of diverse histogenesis. Oncogene 1995; 10:775-8; PMID:7862456. [PubMed] [Google Scholar]
- 6. Jiang W, Kahn SM, Tomita N, Zhang YJ, Lu SH, Weinstein IB. Amplification and expression of the human cyclin D gene in esophageal cancer. Cancer Res 1992; 52:2980-3; PMID:1533816. [PubMed] [Google Scholar]
- 7. Kobayashi H, Saito H, Kitano K, Kiyosawa K, Gaun S, Aoki K, Narita A, Watanabe M, Uchimaru K, Motokura T. Overexpression of the PRAD1 oncogene in a patient with multiple myeloma and t(11;14)(q13;q32). Acta Haematol 1995; 94:199-203; PMID:8610478; http://dx.doi.org/ 10.1159/000204010. [DOI] [PubMed] [Google Scholar]
- 8. Tsujimoto Y, Jaffe E, Cossman J, Gorham J, Nowell PC, Croce CM. Clustering of breakpoints on chromosome 11 in human B-cell neoplasms with the t(11;14) chromosome translocation. Nature 1985; 315:340-3; PMID:3923362; http://dx.doi.org/ 10.1038/315340a0. [DOI] [PubMed] [Google Scholar]
- 9. Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010; 463:899-905; PMID:20164920; http://dx.doi.org/ 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature 1994; 369:669-71; PMID:8208295; http://dx.doi.org/ 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- 11. Bodrug SE, Warner BJ, Bath ML, Lindeman GJ, Harris AW, Adams JM. Cyclin D1 transgene impedes lymphocyte maturation and collaborates in lymphomagenesis with the myc gene. EMBO J 1994; 13:2124-30; PMID:8187765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lovec H, Grzeschiczek A, Kowalski MB, Moroy T. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. Embo J 1994; 13:3487-95; PMID:8062825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi YJ, Li X, Hydbring P, Sanda T, Stefano J, Christie AL, Signoretti S, Look AT, Kung AL, von Boehmer H, et al. The requirement for cyclin D function in tumor maintenance. Cancer Cell 2012; 22:438-51; PMID:23079655; http://dx.doi.org/ 10.1016/j.ccr.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer; 11:558-72; PMID:21734724; http://dx.doi.org/ 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 15. Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology 2004; 145:5439-47; PMID:15331580; http://dx.doi.org/ 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 16. Leontieva OV, Lenzo F, Demidenko ZN, Blagosklonny MV. Hyper-mitogenic drive coexists with mitotic incompetence in senescent cells. Cell Cycle 2012; 11:4642-9; PMID:23187803; http://dx.doi.org/ 10.4161/cc.22937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leontieva OV, Demidenko ZN, Blagosklonny MV. MEK drives cyclin D1 hyperelevation during geroconversion. Cell Death Differ 2013; 20:1241-9; PMID:23852369; http://dx.doi.org/ 10.1038/cdd.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jirawatnotai S, Hu Y, Michowski W, Elias JE, Becks L, Bienvenu F, Zagozdzon A, Goswami T, Wang YE, Clark AB, et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature 2011; 474:230-4; PMID:21654808; http://dx.doi.org/ 10.1038/nature10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jirawatnotai S, Hu Y, Livingston DM, Sicinski P. Proteomic identification of a direct role for cyclin d1 in DNA damage repair. Cancer Res 2012; 72:4289-93; PMID:22915759; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455:1061-8; PMID:18772890; http://dx.doi.org/ 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44-57; PMID:19131956; http://dx.doi.org/ 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 22. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37:1-13; PMID:19033363; http://dx.doi.org/ 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stepanova L, Leng X, Parker SB, Harper JW. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev 1996; 10:1491-502; PMID:8666233; http://dx.doi.org/ 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- 24. Dai K, Kobayashi R, Beach D. Physical interaction of mammalian CDC37 with CDK4. J Biol Chem 1996; 271:22030-4; PMID:8703009; http://dx.doi.org/ 10.1074/jbc.271.26.15451. [DOI] [PubMed] [Google Scholar]
- 25. Lamphere L, Fiore F, Xu X, Brizuela L, Keezer S, Sardet C, Draetta GF, Gyuris J. Interaction between Cdc37 and Cdk4 in human cells. Oncogene 1997; 14:1999-2004; PMID:9150368; http://dx.doi.org/ 10.1038/sj.onc.1201036. [DOI] [PubMed] [Google Scholar]
- 26. Tropschug M, Wachter E, Mayer S, Schonbrunner ER, Schmid FX. Isolation and sequence of an FK506-binding protein from N. crassa which catalyses protein folding. Nature 1990; 346:674-7; PMID:1696687; http://dx.doi.org/ 10.1038/346674a0. [DOI] [PubMed] [Google Scholar]
- 27. Jiang W, Cazacu S, Xiang C, Zenklusen JC, Fine HA, Berens M, Armstrong B, Brodie C, Mikkelsen T. FK506 binding protein mediates glioma cell growth and sensitivity to rapamycin treatment by regulating NF-kappaB signaling pathway. Neoplasia 2008; 10:235-43; PMID:18320068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oliver S. Guilt-by-association goes global. Nature 2000; 403:601-3; PMID:10688178; http://dx.doi.org/ 10.1038/35001165. [DOI] [PubMed] [Google Scholar]
- 29. Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res 2010; 38:D652-7; PMID:19906727; http://dx.doi.org/ 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J Cell Sci 2007; 120:1663-72; PMID:17430976; http://dx.doi.org/ 10.1242/jcs.001776. [DOI] [PubMed] [Google Scholar]
- 31. Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 2013; 22:795-803; PMID:23175443; http://dx.doi.org/ 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen G, Li T, Zhang L, Yi M, Chen F, Wang Z, Zhang X. Src-stimulated IRTKS phosphorylation enhances cell migration. FEBS Lett 2011; 585:2972-8; PMID:21840312; http://dx.doi.org/ 10.1016/j.febslet.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 33. Shin G, Kang TW, Yang S, Baek SJ, Jeong YS, Kim SY. GENT: gene expression database of normal and tumor tissues. Cancer Inform 2011; 10:149-57; PMID:21695066; http://dx.doi.org/ 10.4137/CIN.S7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barnes DM, Gillett CE. Cyclin D1 in breast cancer. Breast Cancer Res Treat 1998; 52:1-15; PMID:10066068; http://dx.doi.org/ 10.1023/A:1006103831990. [DOI] [PubMed] [Google Scholar]
- 35. Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d'Assignies MS, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res 2007; 13:3207-14; PMID:17545524; http://dx.doi.org/ 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 36. Ni L, Yang CS, Gioeli D, Frierson H, Toft DO, Paschal BM. FKBP51 promotes assembly of the Hsp90 chaperone complex and regulates androgen receptor signaling in prostate cancer cells. Mol Cell Biol 2010; 30:1243-53; PMID:20048054; http://dx.doi.org/ 10.1128/MCB.01891-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Romano S, D'Angelillo A, Pacelli R, Staibano S, De Luna E, Bisogni R, Eskelinen EL, Mascolo M, Cali G, Arra C, et al. Role of FK506-binding protein 51 in the control of apoptosis of irradiated melanoma cells. Cell Death Differ 2010; 17:145-57; PMID:19696786; http://dx.doi.org/ 10.1038/cdd.2009.115. [DOI] [PubMed] [Google Scholar]
- 38. Wang L. FKBP51 regulation of AKT/protein kinase B phosphorylation. Curr Opin Pharmacol 2011; 11:360-4; PMID:21498116; http://dx.doi.org/ 10.1016/j.coph.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Solomon E, Borrow J, Goddard AD. Chromosome aberrations and cancer. Science 1991; 254:1153-60; PMID:1957167; http://dx.doi.org/ 10.1126/science.1957167. [DOI] [PubMed] [Google Scholar]
- 40. Wang YP, Huang LY, Sun WM, Zhang ZZ, Fang JZ, Wei BF, Wu BH, Han ZG. Insulin receptor tyrosine kinase substrate activates EGFR/ERK signalling pathway and promotes cell proliferation of hepatocellular carcinoma. Cancer Lett 2013; 337:96-106; PMID:23693078; http://dx.doi.org/ 10.1016/j.canlet.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 41. Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, Lehr HA, Hengstler JG, Kolbl H, Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res 2008; 68:5405-13; PMID:18593943; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 42. van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002; 415:530-6; PMID:11823860; http://dx.doi.org/ 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 43. Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004; 351:2817-26; PMID:15591335; http://dx.doi.org/ 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 44. Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 2006; 98:262-72; PMID:16478745; http://dx.doi.org/ 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 45. Weigel MT, Dowsett M. Current and emerging biomarkers in breast cancer: prognosis and prediction. Endocr Relat Cancer 2010; 17:R245-62; PMID:20647302; http://dx.doi.org/ 10.1677/ERC-10-0136. [DOI] [PubMed] [Google Scholar]
- 46. Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature 2001; 411:1017-21; PMID:11429595; http://dx.doi.org/ 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 47. Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol 2003; 370:430-44; PMID:14712665; http://dx.doi.org/ 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- 48. Bienvenu F, Jirawatnotai S, Elias JE, Meyer CA, Mizeracka K, Marson A, Frampton GM, Cole MF, Odom DT, Odajima J, et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature 2010; 463:374-8; PMID:20090754; http://dx.doi.org/ 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matsushime H, Quelle DE, Shurtleff SA, Shibuya M, Sherr CJ, Kato JY. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol 1994; 14:2066-76; PMID:8114738; http://dx.doi.org/ 10.1128/MCB.14.3.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.