Abstract
Viruses often hijack cellular functions to facilitate their infection and replication. SIRT1, one of the most widely studied sirtuins, functions as both metabolic sensor and transcriptional regulator. SIRT1 has broad cellular functions including metabolic homeostasis, stress response, tumorigenesis and autophagy. The role of SIRT1 in the life cycle of viruses remains unclear. Like all herpesviruses, oncogenic gammaherpesvirus KSHV has both latent and lytic phases. In a recent study, we have shown that SIRT1 binds to the promoter and silence the expression of KSHV replication and transcription activator (RTA), a key activator of viral lytic replication. Chemical inhibition or knock down of SIRT1 is sufficient to initiate the lytic replication program by increasing active histone H3 trimethyl Lys4 (H3K4me3) mark and decreasing repressive histone H3 trimethyl Lys27 (H3K27me3) mark in the RTA promoter. SIRT1 also interacts with RTA and inhibits RTA transactivation of its own promoter and those of downstream target genes. Our findings reveal that SIRT1 regulates KSHV latency by inhibiting different stages of viral lytic replication, and link a metabolic sensor and transcriptional regulator SIRT1 to KSHV life cycle.
Keywords: Epigenetics, H3K4me3 and H3K27me3, Kaposi's Sarcoma-Associated Herpesvirus (KSHV), Kaposi's Sarcoma, KSHV Latency and Reactivation, Metabolism, Nicotinamide, Primary effusion lymphoma, SIRT1
Sirtuins have attracted significant attention since it was discovered that the homology of yeast sirtuin silent information regulator 2 (Sir2) could promote longevity through transcriptional silencing of mating-type loci, telomeres and ribosomal DNA.1 Sir2 was subsequently found to be an NAD+ dependent histone deacetylase (HDAC), indicating sirtuins could serve as both metabolic sensors and transcriptional regulators.2 The mammalian sirtuin family consists of 7 members, named SIRT1 to SIRT7, each with a discrete pattern of tissue specificity, subcellular localization, enzymatic activity, and substrate targets.3
SIRT1 is the most evolutionarily conserved and most widely studied sirtuin.3 It is mainly located in the nucleus but can shuttle between nucleus and cytosol. SIRT1 has broad cellular functions including stress response, metabolic homeostasis, cell proliferation, apoptosis, DNA repair, tumorigenesis and autophagy.3 The functions of SIRT1 depend on its diverse substrates. In the nucleus, SIRT1 deacetylases not only histones such as H3K9, H3K56 and H4K16 but also a number of transcriptional factors and their coactivators including FOXO1, STAT3, c-Myc, p53, NF-κB, PGC-1α, p300 etc.3 In the cytosol, SIRT1 deacetylases Notch I intracellular domain (NICD), cytosolic acetyl-CoA synthetase (AceCS-1), endothelial nitric oxide synthase (eNOS) and autophagy factors Atg5, Atg7 and Atg8.4
A number of studies have shown that SIRT1 might play an important role in the infection and replication of viruses. SIRT1 mediates human immunodeficiency virus (HIV) reactivation by directly deacetylating HIV protein Tat.5 SIRT1 is also a positive regulator of hepatitis B virus (HBV) replication.6 Chemical inhibition or knock down of SIRT1 suppresses HBV replication by targeting transcription factor AP-1. On the other hand, SIRT1 is a negative regulator for the reactivation of neurotropic viruses, which maintains dormancy in sensory neurons for years as a circular episome.7 SIRT1 overexpression or pharmacological up-regulation of SIRT1 reduced the reactivation of neurotropic viruses. However, it is unclear how SIRT1 suppresses the reactivation of neurotropic viruses. The role of SIRT1 in the infection and replication of herpesviruses remains unknown.
The life cycle of herpesviruses consists of latent and lytic phases.8 Following primary infection, herpesviruses establish life-long persistent latent infection in the hosts, often expressing a restricted number of viral latent proteins to avoid host immune surveillance. Upon stimulation by extracellular signals, the latent viruses can be reactivated, expressing viral lytic proteins and producing infectious virions.8 Lytic replication of herpesviruses is often associated with diseases. Kaposi's sarcoma-associated herpesvirus (KSHV) is an oncogenic γ2-herpesvirus. KSHV is etiologically associated with Kaposi's sarcoma (KS), a vascular tumor of proliferative endothelial cells presented in several epidemiologic forms.9 KSHV is also associated with lympho-proliferative diseases including primary effusion lymphoma (PEL) and a subset of multicentric Castleman's disease (MCD). Like all herpesviruses, KSHV has both latent and lytic phases. In KS tumors, most of the tumor cells are latently infected by KSHV expressing only a handful of latent genes that are involved in host cell survival, proliferation and manipulation of the tumor microenvironment.8,9 However, KSHV can be reactivated in a small number of the tumor cells albeit often without completing the full replication cycle, which promote the progression of KS, particularly in the early stage of the tumors. It has been demonstrated that epigenetic modifications of the KSHV genome play a critical role in the viral latent-lytic switch.10,11 A higher level of deposition of active histone 3 lysine 4 trimethylation (H3K4me3) mark is present in the KSHV latent locus while repressive histone 3 lysine 27 trimethylation (H3K27me3) mark is widespread in the KSHV lytic loci in latently infected PEL cells.10,11 KSHV immediate early (IE) gene replication and transcriptional activator (RTA) encoded by ORF50 is essential and sufficient for activating viral replication.12 The RTA promoter is associated with both active and repressive marks with the repressive marks dominating but the promoter is in a “poised” status for reactivation during viral latency.10,11 We have recently discovered that SIRT1 binds to the RTA promoter to epigenetically silence RTA expression during viral latency.13
To investigate the role of SIRT1 in KSHV life cycle, we treated latently KSHV-infected PEL cell line BCBL-1 with nicotinamide (NAM), a metabolic product of the NAD+ precursor and a general inhibitor of sirtuins.13 NAM treatment induced the expression of KSHV IE (RTA), early (ORF57 and ORF59) and late (ORF65) genes. The expression of KSHV lytic genes was accompanied with the production of infectious virions indicating the completion of full viral replication cycle. Similar effects were also observed with Sirtinol, another inhibitor of sirtuins, and in BCP-1 and BC-3 cells, two other latently KSHV-infected PEL cell lines.13 To confirm the results of chemical inhibitors, we found that knock down of SIRT1 was sufficient to induce full KSHV lytic replication program. Accordingly, inhibition of SIRT1 function with NAM or knock down of SIRT1 caused the reprograming of epigenetic marks in the RTA promoter by increasing the active H3K4me3 mark and decreasing the repressive H3K27me3 mark.13 Furthermore, we found that SIRT1 also interacted with RTA and inhibited RTA transactivation of its own promoter and those of downstream target genes.13 Together, our results indicate that SIRT1 is essential for maintaining KSHV latency and functions as a restriction factor for KSHV reactivation by inhibiting different stages of viral lytic replication (Fig. 1).
Figure 1.
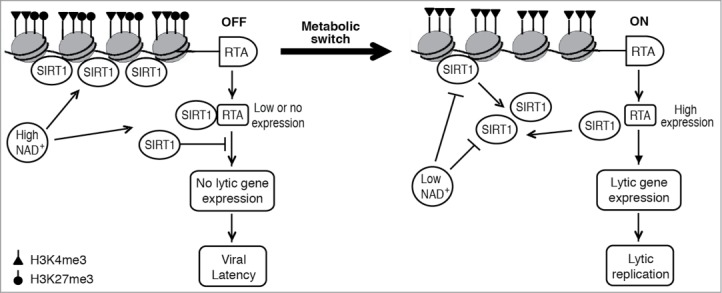
SIRT1 regulates KSHV life cycle. High NAD+ level promotes viral latency by activating SIRT1, resulting in the silencing of the RTA promoter and inhibition of RTA transactivation of downstream targets. Low NAD+ level disrupts viral latency by inhibiting SIRT1 function resulting in the induction of RTA expression and KSHV lytic replication program.
The function of SIRT1 depends on NAD+ as a cofactor for its enzymatic activity. Since NAD+ concentration reflects cellular metabolic and energy status, SIRT1 links cellular metabolic energy status to the adaptive transcriptional responses. A low-energy status that increases cellular NAD+ level, such as fasting, caloric restriction (CR), has been shown to increase SIRT1 activity; while metabolic switch to high-fat diet feeding decreases NAD+ level and reduces SIRT1 activity.14 SIRT1 modulates hepatic glucose and lipid metabolism, pancreatic insulin secretion, fat maturation and remodeling, and central nutrient sensing by regulating numerous transcription factors and co-factors such as TORC2, PPARα and SREBPs involved in systemic metabolic homeostasis. Therefore, SIRT1 is increasingly referred to as a master metabolic sensor and regulator.14 Viruses, as parasites, completely rely on host metabolism to supply the resources and energy for their replication. Our study, for the first time, links a metabolic sensor SIRT1 to the KSHV life cycle. Future studies are needed to investigate how SIRT1 mediates the infection and replication of herpesviruses under different metabolic conditions and whether other sirtuins are also involved in these processes, which could provide a scientific basis for targeting metabolic factors sirtuins for antiviral therapy.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from NIH (CA096512, CA124332, CA132637 and CA177377) to S-J Gao.
References
- 1. Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev 2000; 14:1021-6; PMID:10809662 [PubMed] [Google Scholar]
- 2. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000; 403:795-800; PMID:10693811; http://dx.doi.org/ 10.1038/35001622 [DOI] [PubMed] [Google Scholar]
- 3. Yuan H, Su L, Chen WY. The emerging and diverse roles of sirtuins in cancer: a clinical perspective. Onco Targets Ther 2013; 6:1399-416; PMID:24133372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Canto C, Auwerx J. Targeting Sirtuin 1 to Improve Metabolism: All You Need Is NAD(+)? Pharmacol Rev 2012; 64:166-87; PMID:22106091; http://dx.doi.org/ 10.1124/pr.110.003905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pagans S, Pedal A, North BJ, Kaehlcke K, Marshall BL, Dorr A, Hetzer-Egger C, Henklein P, Frye R, McBurney MW, et al. . SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol 2005; 3:e41; PMID:15719057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ren JH, Tao Y, Zhang ZZ, Chen WX, Cai XF, Chen K, Ko BC, Song CL, Ran LK, Li WY, et al. . Sirtuin 1 regulates hepatitis B virus transcription and replication by targeting transcription factor AP-1. J Virol 2014; 88:2442-51; PMID:24335313; http://dx.doi.org/ 10.1128/JVI.02861-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Picchione KE, Bhattacharjee A. Viral genome silencing by neuronal sirtuin 1. J Neurovirol 2011; 17:184-8; PMID:21165789; http://dx.doi.org/ 10.1007/s13365-010-0012-3 [DOI] [PubMed] [Google Scholar]
- 8. Ye F, Lei X, Gao SJ. Mechanisms of Kaposi's Sarcoma-Associated Herpesvirus Latency and Reactivation. Adv Virol 2011; 2011; PMID:21625290; http://dx.doi.org/ 10.1155/2011/193860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ganem D. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu Rev Pathol 2006; 1:273-96; PMID:18039116; http://dx.doi.org/ 10.1146/annurev.pathol.1.110304.100133 [DOI] [PubMed] [Google Scholar]
- 10. Gunther T, Grundhoff A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog 2010; 6:e1000935; PMID:20532208; http://dx.doi.org/ 10.1371/journal.ppat.1000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Toth Z, Maglinte DT, Lee SH, Lee HR, Wong LY, Brulois KF, Lee S, Buckley JD, Laird PW, Marquez VE, et al. . Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog 2010; 6:e1001013; PMID:20661424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A 1998; 95:10866-71; PMID:9724796; http://dx.doi.org/ 10.1073/pnas.95.18.10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Q, He M, Zhou F, Ye F, Gao SJ. Activation of Kaposi's sarcoma-associated herpesvirus (KSHV) by inhibitors of class III histone deacetylases: identification of sirtuin 1 as a regulator of the KSHV life cycle. J Virol 2014; 88:6355-67; PMID:24672028; http://dx.doi.org/ 10.1128/JVI.00219-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X. SIRT1 and energy metabolism. Acta Biochim Biophys Sin (Shanghai) 2013; 45:51-60; PMID:23257294; http://dx.doi.org/ 10.1093/abbs/gms108 [DOI] [PMC free article] [PubMed] [Google Scholar]


