Abstract
AddAB and RecBCD-type helicase-nuclease complexes control the first stage of bacterial homologous recombination (HR) – the resection of double strand DNA breaks. A switch in the activities of the complexes to initiate repair by HR is regulated by a short, species-specific DNA sequence known as a Crossover Hotspot Instigator (Chi) site. It has been shown that, upon encountering Chi, AddAB and RecBCD pause translocation before resuming at a reduced rate. Recently, the structure of B.subtilis AddAB in complex with its regulatory Chi sequence revealed the nature of Chi binding and the paused translocation state. Here the structural features associated with Chi binding are described in greater detail and discussed in relation to the related E.coli RecBCD system.
Keywords: RecBCD, AddAB, Nuclease, Chi recognition, Homologous recomb-ination
Introduction
The accurate repair of double-strand DNA breaks (DSB) is crucial for viability and maintaining genomic integrity, so presents a continual challenge to cells. Homologous recombination (HR) is one of the major DSB repair pathways and offers high fidelity by using a homologous DNA copy as a template during repair. HR can be split into the interconnecting stages of resection, strand invasion, branch migration and resolution, which are conserved across all domains of life.1 During resection, the break is processed by helicase and nuclease activities to unwind the damaged DNA and generate a 3′ terminating single-stranded overhang that is coated with RecA or Rad51 recombinase proteins to form a filament. This filament initiates specific strand invasion of the homologous template as the first step in recombination repair.
In bacterial homologous recombination, a helicase-nuclease complex is responsible for controlling the resection process. There are 2 distinct, but related, systems (E.coli RecBCD and Bacillus subtilis AddAB) which have both been studied extensively (reviewed in refs. 2,3). RecBCD and AddAB unwind DNA with high processivity at speeds as fast as 1000-2000 bps−1 until encountering a short, species-specific DNA sequence known as a Crossover Hotspot Instigator (Chi) site.4-7 Chi sites are over represented in the bacterial genome and instigate a switch of activity from DNA degradation to repair, via recombination.4,8,9 Single molecule studies in vitro have shown that translocation of both RecBCD and AddAB pauses for up to 5 seconds upon encountering a Chi site in the 3′ terminating ssDNA strand.7,10-13 After this pause, both complexes resume translocation at a reduced rate and are unable to recognize additional Chi sites. Subsequent to the recognition event, there is a final cleavage a few bases downstream of Chi after which the 3′ tail, containing the Chi sequence, becomes protected from further digestion.14-18 Consequently, it is likely that the Chi sequence remains bound within the complex despite continued unwinding and translocation. Moreover, it has been proposed that the complex opens an alternate exit route, situated between the helicase and Chi-binding domains, to enable the newly unwound ssDNA to loop out for RecA loading.13,19-23
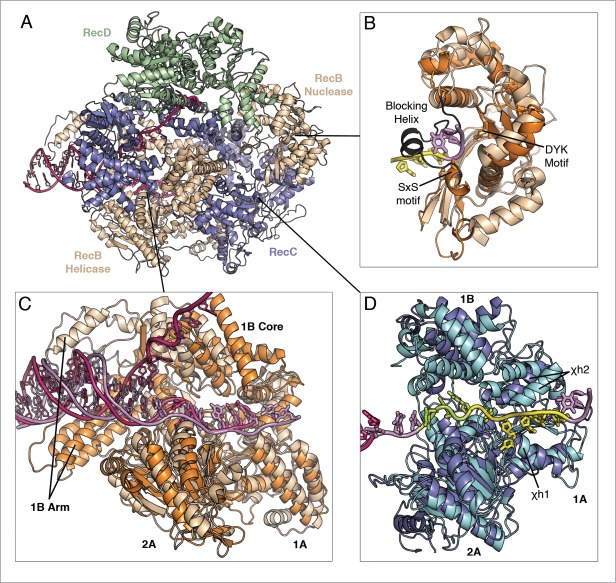
Crystal structures of RecBCD and AddAB have been determined in combination with a DNA substrate that mimics the complex bound to a broken DNA end.19,24,25 A simplified representation of their architectures is depicted in Figure 1. Briefly, RecBCD contains 2 independent SF1 helicase motors of opposite polarity,26,27 each of which is responsible for translocation along one of the ssDNA strands, together with one nuclease that intermittently cuts both strands. The core of the SF1 helicase folds contains the 2 motor domains (1A and 2A) that flank an ATP-binding site.28 Translocation of ssDNA is coupled to ATP binding and hydrolysis by conformational changes involving the 1A and 2A domains.29,30 By contrast, the AddAB complex utilises a single helicase motor to drive translocation along the 3′-terminating strand and contains 2 nuclease domains – one for each strand.31 The 3 nuclease domains (RecB, AddA, AddB) are all members of the diverse PD-(E/D)XK phosphodiesterase superfamily32 although the AddB nuclease domain contains an additional FeS cluster. The structure of one family member (lambda exonuclease) bound to DNA and Mg2+ has been determined recently, providing insight into the mechanism of these nucleases.33
Figure 1.
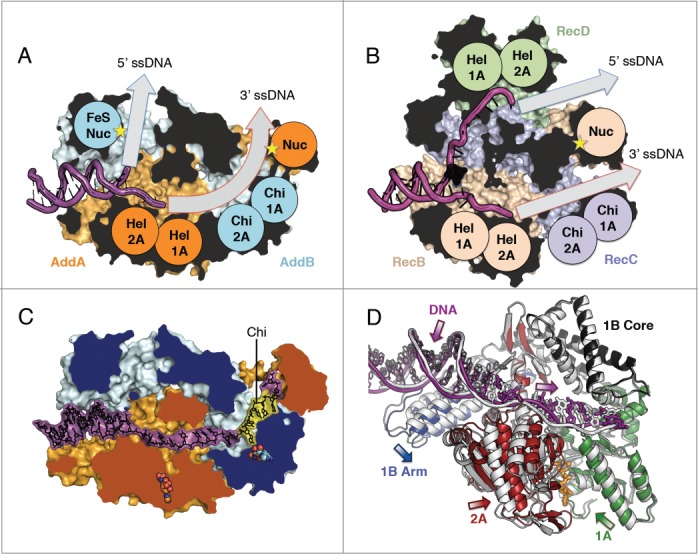
AddAB and RecBCD structural architectures. Clipped surface view of (A) AddAB initiation complex (pdb code 3u44) and (B) RecBCD initiation complex (pdb code 3k70) with helicase motor (Hel), Chi-scanning and nuclease (Nuc) domains labelled and respective ssDNA tunnels highlighted (grey arrows). The clipped interior surface is coloured black. (C) AddAB structure in complex with Chi (yellow) (pdb code 4cej) with 2 bound ADPNP molecules shown as spheres (orange AddA and cyan AddB respectively). The clipped surface is coloured by protein. (D) Translocation-like movements of AddA helicase domains upon binding of ADPNP. Overlay of the translocation-like binary (grey) (pdb code 4ceh) and ternary ADPNP-bound (coloured by domain) (pdb code 4cei) AddAB structures showing relative displacement of the helicase motor and 1B arm domains. The single-stranded portion of DNA (thick bonds) moves away from the junction, corresponding to the closing movement of the 2A domain, whilst movement of the duplex (thin bonds) is coupled to the arm domain. Bound ADPNP is shown in sticks with the Mg2+ ion as a sphere (orange). The structures were aligned based on the 1B core domain and the 2B domain, which is hidden for clarity.
In combination with mutational studies, the RecBCD and AddAB structures also allowed the putative assignment of a Chi-scanning domain (in RecC or AddB respectively) that has a SF1 helicase fold but lacks most of the associated conserved motifs. There are 7 key conserved SF1-family motifs that map around the ATP-binding site, many of which have critical roles in nucleotide binding or hydrolysis.34 The AddB Chi-scanning domain still retains many of the residues associated with ATP binding (such as the Walker A and B motifs) but residues that would normally promote ATP hydrolysis are not conserved.24 The corresponding region in RecC lacks all of the conserved helicase motifs and is not thought to bind ATP.
Recently, crystal structures of AddAB were published that trapped translocation-like states, using an extended DNA substrate that mimicked an unwound fork.35 These structures, obtained in the presence and absence of a non-hydrolysable ATP-analog (ADPNP), suggested a mechanism for duplex unwinding and translocation. Furthermore, the work also reported the first Chi-bound structure, with the ssDNA extending all the way through the complex to the 3′ nuclease domain as shown in Figure 1C. This structure reveals a key, intermediate stage that develops our understanding of the role of Chi in regulation of bacterial recombination. This article will expand upon and assess the implications of these structures, specifically in terms of the processes of Chi binding and Chi response in AddAB, with additional insights into the nature of Chi recognition in RecBCD.
Duplex unwinding and translocation
AddAB was crystallized in complex with a hairpin oligonucleotide with long, single-stranded tails that mimics an unwound fork.35 Two structures were obtained, with and without the non-hydrolysable ATP analog, ADPNP, thus trapping 2 steps in the translocation process. As seen with other helicases,29,30,36,37 the 1A and 2A motor domains of AddA close around a bound molecule of ADPNP (Figure 1D). In conjunction with the 2A domain, the ssDNA is pulled deeper into the protein resulting in a step of one base pair. Although there are likely to be additional intermediate stages to complete a full cycle of translocation, including ATP hydrolysis and product release steps, these snapshots provide insight into the unwinding mechanism. AddAB and RecBCD contain an extension of the 1B domain, termed the arm domain, which contacts the DNA duplex across the first helical turn. As the motor domains close, the arm moves in the opposite direction pulling the duplex away from the junction. This movement creates tension at the junction to assist in the unwinding of a base pair. Consistent with this proposal, the last paired base on the 5′ terminating strand becomes disordered in the ADPNP complex.
Recognition of Chi in AddAB
The structure of AddAB with a bound Chi sequence revealed the specific location and nature of the Chi-binding site, which straddles AddB and the interface with the AddA nuclease.35 The Chi sequence interacts with the protein with the bases facing toward the “helicase-like” AddB 1A and 2A domains as shown in Figure 2B. Each of the 5 Chi bases (AGCGG) is engaged by 2 or 3 specific hydrogen bonds with surrounding protein residues. This is contrary to the situation seen in the interaction between active helicase domains and ssDNA (such as in AddA - Fig. 2A), which involves only non-specific hydrophobic stacking interactions with the bases and polar contacts with the phosphodiester backbone.
Figure 2.
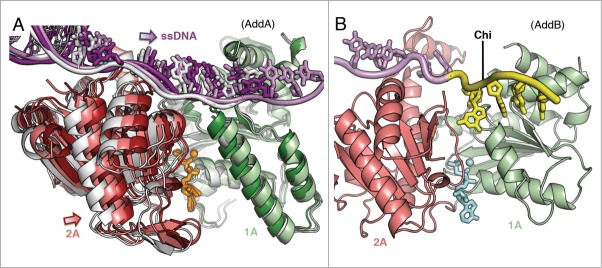
Alterations in the helicase domains upon binding of Chi. (A) Superposition of the AddA helicase motor domains with bound ADPNP (sticks), Mg2+ (spheres) and ssDNA (tube and sticks) from the binary (grey), ternary (coloured as in Fig. 1D) and Chi-bound (coloured by domain in lighter shades than the ternary structure) crystal structures. The Chi-bound 1A domain matches that in the binary structure whereas the 2A and ssDNA are in between the relative conformations of the binary and ternary states. (B) AddB helicase-like 1A and 2A domains from the Chi-bound complex. At the Chi sequence, the ssDNA flips by 180° to form specific interactions with the 1A domain.
Accompanying the publication of the first AddAB initiation complex structure was the identification of 7 mutations that affected the interaction with Chi.25 Of these mutations, 5 showed no detectable level of Chi binding and these all correspond to residues lining the Chi-binding pocket in the Chi-bound structure. As expected, residues T44, R70 and F210 form direct, specific interactions with the Chi bases. Two other residues (D41 and Q42) are located at the top turn of a stretched helix, labeled Chi-helix1 (χh1) in Figure 3B, which is pulled away to accommodate the second Chi residue. The guanine base inserts into the groove between turns, sandwiched below the aspartate and glutamine residues. With each residue appearing to participate in hydrogen bonds to neighbouring protein regions, D41 and Q42 may play an important role in forming/securing this distorted conformation in parallel with binding of the Chi sequence. Moreover, D41 interacts with S69, which is the residue immediately preceding a key Chi-interacting residue (R70). Consequently, mutation at this position could disrupt a wider section of the Chi-binding site. Interestingly, the 2 remaining mutations that showed a defect in Chi recognition (F68, W73) map to residues that may indirectly assist in coordinating binding of the Chi sequence. The side chain of F68 is located in the vicinity of the first base of the Chi sequence and appears to assist in correctly orientating both F210 and E217 for direct stacking and hydrogen bonding interactions, respectively, with the base. Additionally, W73, located on Chi-helix2 (χh2), caps a 3-tier planar stacking arrangement with R70 and the fourth base of the Chi sequence.
Figure 3.
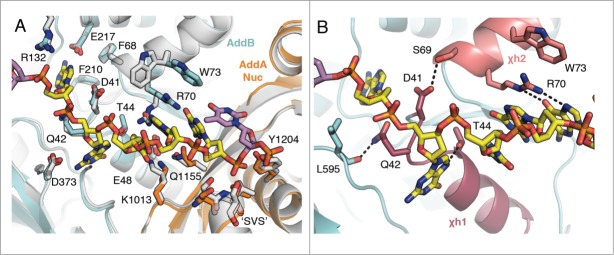
Summary of the specific Chi-binding interactions in AddB and the AddA nuclease. (A) The Chi-binding sites are superposed for the binary structure (grey) and the Chi-bound structure (coloured by subunit) to show relative changes in the presence of Chi. The site in the ternary complex is not shown as it is identical to the binary structure. The side chains of key residues that interact with Chi are shown as sticks. (B) Close-up view of the key interactions stabilising Chi located on the AddB 1A domain helices χh1 and χh2. Residues D41 and Q42 stretch the top turn of χh1 by interacting with S69 and L595 respectively. R70 and W73 of χh2 stack with 2 of the Chi bases.
The Chi-interacting residues are contributed in the main by the AddB 1A domain but with a few from the AddB 2A and AddA nuclease domains (Fig. 3A). The phosphate on the 3′-side of Chi is coordinated by a ‘SVS’ helical turn (AddA residues 1015-1017) before the DNA crosses into the nuclease active site. A similar feature is conserved in the structurally related lambda exonuclease where a ‘TAS’ helical turn (residues 33-35) is involved in orientating the DNA for cleavage.33 This motif is also conserved in the AddB (SVS, residues 791-793) as well as the RecB (SYS, residues 905-907) nuclease active sites. Coordination of the 3′ strand at the AddA SVS helix places the first and second base 3′ of Chi near the catalytic nuclease motif (DYK, residues 1172-1174). This is consistent with in vitro observations that the final cleavage predominantly occurs one nucleotide 3′ of Chi.18 When AddAB is co-crystallized with substrates lacking the Chi sequence, DNA is not ordered at the AddA SVS helix. This suggests that binding of the Chi sequence helps to position the 3′ tail to engage at the AddA nuclease active site. However, it should be kept in mind that all the published AddAB crystal structures are of a nuclease dead mutant form of the enzyme, which is unable to bind Mg2+. It is possible that the wild type enzyme may show stronger coordination of the 3′ terminating strand in the absence of Chi.
A molecule of ADPNP is bound to AddB, occupying the canonical ATP-binding site (Fig. 2B). The nucleotide analog sits in close proximity to the Chi sequence and is immediately parallel to χh1, which directly interacts with the 2nd base of Chi. The AddB 1A and 2A domains occupy a closed conformation that is unresponsive to binding of both nucleotide and Chi. It has been shown that the rate of dissociation of bound Chi is around 3-fold faster when the AddB ATP-binding site is disrupted.31 Despite this observation, the binding of nucleotide in tandem with Chi does not cause any changes that might explain this observation. It could be that the amino acid point mutation used to disrupt the binding site influences the stability of the complex more than simply the absence of nucleotide. Alternatively, there could be a conformational change linked to a slow ATP hydrolysis event in AddB that subsequently increases the half-life of the complex. Further work will be required to distinguish between these, or other, possibilities.
Chi scanning during translocation
A comparison of other AddAB structures with the Chi-bound structure reveals that the binding site is largely unaltered by binding of Chi except for four of the residues discussed above (D41, Q42, R70 and W73 (Fig. 3A)). The entrance to the AddA nuclease binding site - including the ‘SVS’ motif and the side chain of Y1204 – shows only a minimal adjustment moving from the translocation state to that with Chi bound. Consequently, the Chi-scanning region is set up to recognize its specific recognition sequence from the moment ssDNA begins to translocate through the site. With rapid translocation rates of up to 2000 bp/s this makes sense in that significant movements to recognize Chi would reduce the efficiency of the recognition process. In fact, Chi sites are only recognised with a frequency of around 30% in RecBCD14,38,39 and even less in AddAB.13
Chi binding pauses translocation
Looking outside of the Chi-scanning domain, there are additional structural changes upon Chi binding relating to the AddA helicase motor domains. These changes are highlighted by the superposition of the helicase domains from the Chi-bound structure with each of the 2 translocation-like states (Fig. 2A). Although there is an ADPNP molecule bound to the AddA subunit, the conformation of the 1A and 2A (motor) domains resembles that found in the binary structure that lacks bound nucleotide. The 2A domain and the ssDNA spanning the motor domains are both in a mixed, partially closed conformation between the open and closed states. It appears that when Chi binds, movement of the 1A domain becomes uncoupled from ADPNP binding in AddA. Most strikingly, a long helix, which contacts the ssDNA at one end and typically slides upwards to contact bound nucleotide via the side-chain of R132 at the other end, does not respond to ADPNP binding. This suggests that hydrolysis of ATP has become uncoupled from translocation of a base because this movement is a key part of the translocation mechanism. With the majority of the 1A domain unresponsive to ADPNP binding, the 2A domain presumably attempts to close but is left in a sort of no-man's land, unable to drive translocation by itself.
Single-molecule studies have shown that both AddAB and RecBCD pause at a Chi sequence before resuming translocation at a reduced speed.7,10-13 Moreover, AddAB also pauses at sequences that resemble Chi.13 However, after pausing briefly, translocation resumes and unlike a bona fide Chi recognition event, the rate of translocation is similar to that before the encounter. The studies also revealed that the nature of the pause at Chi may be a multi-step process.7,13 The Chi-bound crystal structure appears to represent the initial state in this series of events in which the complex has recognised Chi and paused translocation but has yet to respond. Consistent with this view, binding of Chi does not induce any significant conformational changes in the AddB protein apart from the local rearrangement of a few key residues that facilitate specific recognition of the Chi sequence. The lack of a distinct link between the Chi-binding site and the helicase motor domains of AddA may suggest that binding of the 3′ DNA end at Chi physically halts further translocation upstream, thereby inducing the pause in translocation. It is likely that the canonical movement of the helicase domains becomes recoupled to ATP binding after the pause because it was shown that the net efficiency of translocation of AddAB was the same before and after Chi recognition.7As such, it is evident that further conformational changes will be required (i.e. a response to Chi) for translocation to resume.
Structural comparison with RecBCD
The RecBCD complex has a different subunit composition to AddAB, with a second helicase (RecD) operating on the 5′ terminating strand (Fig. 4A).19 Despite this, the complexes share a range of similar biochemical properties when encountering and responding to Chi sites.2,3 Other than the additional RecD protein, the complexes share similar structural architectures with RecB being related to AddA whilst RecC is similar to AddB.
Figure 4.
Overview of the structural architecture of RecBCD in relation to AddAB. (A) Overview of the RecBCD structure (pdb code 3k70) coloured by subunit. (B) Close-up of the RecB nuclease domain aligned to that of AddA (orange) from the Chi-bound structure (pdb code 4cej) with DNA engaged at the SxS motif. The active site DYK and SxS motifs are shown in sticks to highlight their identical positions in both proteins. (C) Close-up of the RecB helicase domains aligned to those of AddA from the binary structure (pdb code 4ceh), aligned using the 1A and 2A domains. The 1B Arm and core domains occupy a different position in RecB relative to the DNA. (D) Close-up of the RecC helicase-like Chi-scanning domains aligned to hose of AddB from the Chi-bound structure, aligned using the 1A and 2A domains. The domains share a similar structural arrangement but with relative displacements of a section of 1A domain - containing χh2, the 1B and the 2B domains. In AddB about eight bases of the ssDNA span the length of the domains. The 3 bases 5′ of Chi (lime) may relate to the additional bases of the E.coli Chi sequence. In (C) and (D) the 2B domains are not shown for clarity.
The RecB helicase is aligned with AddA in Fig. 4C, both from structures of binary complexes without ADPNP. To date a crystal structure has not been obtained of RecBCD with bound nucleotide/nucleotide analog in the ATP-binding sites. The alignment shows that the core motor domains are largely similar suggesting a conserved mechanism of ssDNA translocation. RecB also contains the 1B domain extension known as the arm. However, relative to the rest of the helicase domains, it is rotated around the duplex by around 90° compared to its position in AddA. The adjustment of the arm domain could reflect an altered role in RecB or could result from the DNA hairpin in the substrate being closer to the junction in the RecBCD structures. In both complexes the 3′ ssDNA tail is fed toward the Chi-scanning domains.
A number of mutations in the RecC protein have been shown to influence Chi recognition.25,39-42 Handa et al.,42 generated a large set of point mutations to alanines, based on residues that line the ssDNA tunnel in the crystal structure of the RecBCD initiation complex.19 Although many of the mutations had little effect on Chi binding, eleven mutations did have an effect and were classified into 2 groups – Type I and Type II. Type I mutants were significantly impaired in their ability to recognize Chi, whereas Type II showed reduced specificity of Chi recognition, resulting in additional Chi-like responses at non-canonical Chi sequences. As in AddB, the predicted Chi-binding site in RecC also comprises an inactive SF1 helicase fold that is situated between the RecB helicase and nuclease domains (Fig. 4D). Overall, the sites display similar structural arrangements in terms of the domains, but with several rigid-body adjustments of secondary structure elements. One key structural variation between AddB and RecC is that the RecC subunit lacks all of the highly conserved SF1 helicase motifs, not just those responsible for ATP hydrolysis. Curiously, although the AddB SF1 fold responsible for Chi binding does indeed bind ATP, nucleotide binding does not induce any significant movement in the surrounding protein nor in the Chi-binding site. Consequently, the AddB 1A and 2A domains are in a conformation that is ready to bind Chi whether or not nucleotide is bound.
The current RecBCD structures are of the initiation complex,19,24 which is the state of the enzyme when bound initially to a DNA double-strand break. The RecB nuclease domain sits between 2 ssDNA tunnels exiting from RecD and RecC, respectively. However, in this initiation state the RecB nuclease active site is blocked by an α-helix (Fig. 4B). To progress to the translocating, Chi-scanning, state a conformational change must occur to allow activation of the nuclease active site. Additionally, there is a nuclease bypass tunnel between the 1A domain of RecC and the RecB nuclease domain that could direct the 3′ terminating strand out of the enzyme without being cleaved. Therefore, the minimum change that is required to reach the active, translocating RecBCD state would involve removing the blocking helix whilst closing the nuclease bypass route. The observation in vitro that the final 3′ strand cleavage in RecBCD occurs 4-6 bases downstream of Chi,14,15 rather than one base downstream as in AddAB,18 suggests that the nuclease domain and the bound Chi sequence occupy different positions relative to one another in RecBCD. In the current crystal structures the nuclease is actually slightly closer to the Chi-scanning domain than in AddAB. Consequently, it is likely that the nuclease adopts a different position in solution where it may be able to switch between the extruding ssDNA strands more readily.
Insights into chi binding in RecBCD
The Chi sequences recognised by the 2 complexes are different; E.coli RecBCD recognises the eight base sequence GCTGGTGG,43 whereas B.subtilis AddAB is regulated by AGCGG.9 Perhaps a poignant observation is the fact that, in all present structures, the SF1 helicase fold spans eight or nine DNA residues. As the AddAB Chi sequence binds at the 3′ end of the helicase-like fold spilling into the AddA nuclease, the additional 3 RecBCD Chi bases are likely to be present upstream of the bound AddAB Chi when overlaying the structures (Fig. 4D). Looking at Figure 5, the Type I and II RecC mutations are located in the 1A and 1B domains whereas in AddB the Chi interactions are almost exclusively provided by domain 1A. The RecC mutations in 1B, including the majority of the Type II mutants, would be positioned in the vicinity of where the additional 3 Chi bases would be expected to reside.
Figure 5.
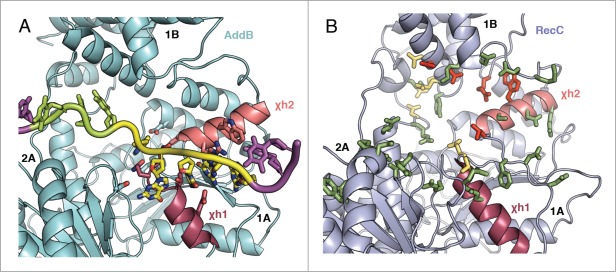
Comparison of the positions of Chi-interacting residues in AddB and RecC. The Chi-binding site of AddB from the Chi-bound crystal structure (pdb code 4cej) is summarised (A) with key residues shown with sticks. Alongside is the same region of RecC (B) with mutations from the work by Handa et al.42 shown as sticks. Mutations that didn't affect Chi recognition are shown in green, Type I mutations that abolished Chi-binding are in red and Type II mutations that increased the promiscuity of Chi-sequence recognition are in yellow-orange. The 2 Chi-helices χh1 and χh2 are coloured similarly in each structure.
The last 5 bases of the respective AddAB and RecBCD Chi sequences are highly similar, which may suggest a degree of conservation between the Chi-binding sites. The AddAB Chi sequence was mutated and superposed onto the equivalent position in RecC based on an alignment of the 1A and 2A domains (Fig. 6). The 2 helices χh1 and χh2, which play a significant role in the AddAB-Chi complex, are conserved in RecC but χh2 has swung away relative to its position in AddB. The side chains of D41 and Q42 in AddB, which stretch the top turn of χh1 to accommodate a base of the Chi sequence – even in the absence of Chi itself, are not conserved in RecC and the helix is not stretched. Residues T44 and E48, which directly contact 3 Chi bases in AddB, correspond to A43 and Q47 in RecC. Interestingly, these substitutions could fit with the RecBCD Chi sequence where a thymine substitutes for the cytosine base. Looking at χh2, the side chains of R70 and W73 in AddAB stack on top of Chi. Whilst both residues are conserved in RecC (W70 and R186) and are part of the set of Type I Rec mutations, the arginine is contributed by a different part of the sequence. The position corresponding to R70 is represented by S67 in RecC with the stacking R186 and W70 tucked away from χh1 in a groove at the 1A/1B domain interface.
Figure 6.

Modelling RecBCD-Chi binding at χh1 in relation to the interactions observed in AddAB. χh1 and χh2 are coloured as in Figure 5 with residues relating to key positions in the AddB-Chi complex as well as the nuclease SYS and DYK motifs shown in sticks. The DNA position and orientation is directly taken from the AddAB-Chi structure (pdb code 4cej) when superposed as in Figure 4D although the bases are mutated to match the RecBCD Chi sequence (lime green). The bases 3′ of Chi are coloured in hotpink and the potential nuclease-bypass exit tunnel is labelled. The nuclease blocking helix present in RecBCD structures is removed for clarity.
The altered location of χh2 in RecC suggests either a different orientation in the Chi binding site to that seen in AddB or that additional conformational changes associated with the transition from initiation to translocation states might make the sites more similar. When all of the RecC domains are modelled to fit directly onto AddB, the movements bring all of the Type I RecC mutants toward the location of Chi equivalent to the AddB binding site. However, it would still not bring R186 within reach of fulfilling the vacant Chi-stacking role lost by the substitution of S67. An additional, major contradiction to the idea of a conserved binding site is the indication that the RecB nuclease cuts further to the 3′ of Chi than in AddAB. In AddAB, part of the Chi-binding interactions at the 3′ end of the sequence come directly from the nuclease itself. Therefore, these particular interactions with Chi would be lost in RecBCD because the nuclease would have to be positioned further away from the terminal Chi residue. The groove containing the Type I RecC residues is on the opposite side of the Chi tunnel from the nuclease. If this does correspond to a different site of Chi binding in RecBCD then there would be more room to fit additional bases downstream of Chi before reaching the nuclease. This could explain the observation that a certain class of natural RecC variants (from the recC1004 strain) initiate recombination by recognising a longer, eleven-base Chi sequence GCTGGTGCTCG (Chi*).40,41,44
Summary and perspectives
The crystal structure of AddAB bound to its Chi sequence revealed the specific nature of the interactions between the Chi-scanning domain and each of the 5 bases. The Chi-binding site is mostly unaltered by Chi binding suggesting that the translocating complex constantly scans the 3′ terminating ssDNA strand. The similarity of the B.subtilis AddAB Chi sequence to the last 5 residues of the E.coli RecBCD Chi sequence raised the possibility of a conserved mechanism of recognition in the 2 systems. Furthermore, the predicted RecC Chi-scanning domain shares similar structural features to those observed in the AddB-Chi structure, but with some key differences. However, even when the structural components of RecC are aligned to match up with their counterparts in AddB it is evident that a number of the Chi interactions in AddAB are not conserved. The position of key Chi-binding residues, identified from mutational studies,23,42 may suggest an alternative path for the Chi-binding site within RecC. In conclusion, comparison of the structural and biochemical properties of RecBCD and AddAB suggests a divergence in the mechanism of Chi binding between the 2 complexes. This may involve a modified set of interactions with an altered nuclease location or it may extend to an alternative orientation of the Chi-binding interface toward the 1A/1B domain interface. The current RecBCD structures represent an initiation complex rather than one competent for translocation. Indeed, several conformational changes would be required for activation, not least of all at the nuclease active site. Further insights into the interaction between RecBCD and Chi will probably require structures of complexes trapped during translocation.
The AddAB-Chi structure inferred that ATP binding at AddA becomes uncoupled from translocation when Chi is bound, consistent with the pause in translocation observed in single-molecule studies.13 Now that this paused state has been visualised structurally, the question is how does the complex recover to re-couple nucleotide binding to translocation whilst keeping the Chi sequence sequestered? Currently, the best explanation is that the next stage of response to Chi binding involves a conformational change that would open up an alternative exit for the ssDNA between the AddA helicase and the AddB Chi-scanning domain to allow translocation and unwinding to resume.19-22 A site for a possible exit route guarded by a latch-like helix opening mechanism has been proposed in RecBCD, with a similar feature found in AddAB.23,25,35,42 In support of this proposal, the latch is structurally linked to the Chi sequence via the side-chain of R132 in AddAB.35 Furthermore, mutation of latch residues reduces the specificity of Chi recognition in RecBCD42 and increases the likelihood of canonical Chi recognition by 2-fold in AddAB.25 The paused Chi state does not explain these observations suggesting there is a further conformational change in response to Chi binding that may open an exit channel involving the latch.
Whatever the next stage entails, the trigger for leaving the paused state might involve cleavage of the 3′ terminating strand downstream of the Chi sequence or slow hydrolysis of ATP in the AddB inactive helicase fold which encases Chi (or a combination of both?). Cleavage of the 3′ end is necessary to generate a suitable substrate for strand extension by DNA polymerase after the strand invasion and exchange reactions. Therefore, it seems likely that this final cleavage event could play a key role in regulating the Chi response process. Regarding ATP hydrolysis in AddB, the catalytic motifs typically associated with ATP hydrolysis in helicases are absent and the rate of ATP turnover by AddAB is greatly reduced when the AddA ATP-binding site is disrupted.31 However, the complex where only the site in AddA was disrupted showed a slightly increased rate of ATP hydrolysis of 4.64 ± 0.08 min−1 compared to 2.6 ± 0.14 min−1 when both AddA and AddB sites were disrupted. However, such a role for ATP hydrolysis in AddB would not apply to the RecBCD system where ATP does not interact with the Chi-scanning domain (RecC). Now, our aim is to address some of these queries to start to understand the next stages of Chi recognition and response in AddAB to further unravel the complex process of resection in bacteria.
Funding Statement
This work is funded by Cancer Research UK.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Blackwood JK, Rzechorzek NJ, Bray SM, Maman JD, Pellegrini L, Robinson NP. End-resection at DNA double-strand breaks in the three domains of life. Biochem Soc Trans 2013; 41:314-20; PMID:23356304; http://dx.doi.org/ 10.1042/BST20120307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yeeles JTP, Dillingham MS. The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes. DNA Repair (Amst) 2010; 9:276-85; PMID:20116346; http://dx.doi.org/ 10.1016/j.dnarep.2009.12.016 [DOI] [PubMed] [Google Scholar]
- 3. Wigley DB. Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat Rev Microbiol 2013; 11, 9-13; PMID:23202527; http://dx.doi.org/ 10.1038/nrmicro2917 [DOI] [PubMed] [Google Scholar]
- 4. Lam ST, Stahl MM, McMilin KD, Stahl FW. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics 1974; 77:425-33; PMID:4415485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roman LJ, Kowalczykowski SC. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry 1989; 28:2863-73; PMID:2545238; http://dx.doi.org/ 10.1021/bi00433a018 [DOI] [PubMed] [Google Scholar]
- 6. Roman LJ, Eggleston AK, Kowalczykowski SC. Processivity of the DNA helicase activity of Escherichia coli recBCD enzyme. J Biol Chem 1992; 267:4207-14; PMID:1310990 [PubMed] [Google Scholar]
- 7. Gilhooly NS, Dillingham MS. Recombination hotspots attenuate the coupled ATPase and translocase activities of an AddAB-type helicase-nuclease. Nucleic Acids Res 2014; 42:5633-43; PMID:24682829; http://dx.doi.org/ 10.1093/nar/gku188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malone RE, Chattoraj DK, Faulds DH, Stahl MM, Stahl FW. Hotspots for generalized recombination in the Escherichia coli chromosome. J Mol Biol 1978; 121:473-491; PMID:353291; http://dx.doi.org/ 10.1016/0022-2836(78)90395-9 [DOI] [PubMed] [Google Scholar]
- 9. Chédin F, Noirot P, Biaudet V, Ehrlich SD. A five-nucleotide sequence protects DNA from exonucleolytic degradation by AddAB, the RecBCD analogue of Bacillus subtilis. Mol Microbiol 1998; 29, 1369-77; http://dx.doi.org/ 10.1046/j.1365-2958.1998.01018.x [DOI] [PubMed] [Google Scholar]
- 10. Spies M, Bianco PR, Dillingham MS, Handa N, Baskin RJ, Kowalczykowski SC. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell 2003; 114:647-54; PMID:13678587; http://dx.doi.org/ 10.1016/S0092-8674(03)00681-0 [DOI] [PubMed] [Google Scholar]
- 11. Handa N, Bianco PR, Baskin RJ, Kowalczykowski SC. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after chi recognition. Mol. Cell 2005; 17:745-50. [DOI] [PubMed] [Google Scholar]
- 12. Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD enzyme switches lead motor subunits in response to chi recognition. Cell 2007; 131:694-705; PMID:18022364; http://dx.doi.org/ 10.1016/j.cell.2007.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carrasco C, Gilhooly NS, Dillingham MS, Moreno-Herrero F. On the mechanism of recombination hotspot scanning during double-stranded DNA break resection. Proc Natl Acad Sci U S A 2013; 110:E2562-71; PMID:23798400; http://dx.doi.org/ 10.1073/pnas.1303035110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taylor AF, Schultz DW, Ponticelli AS, Smith GR. RecBC enzyme nicking at Chi sites during DNA unwinding: location and orientation-dependence of the cutting. Cell 1985; 41:153-63; PMID:3888405; http://dx.doi.org/ 10.1016/0092-8674(85)90070-4 [DOI] [PubMed] [Google Scholar]
- 15. Ponticelli AS, Schultz DW, Taylor AF, Smith GR. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell 1985; 41:145-151; PMID:3888404; http://dx.doi.org/ 10.1016/0092-8674(85)90069-8 [DOI] [PubMed] [Google Scholar]
- 16. Dabert P, Ehrlich SD, Gruss A. Chi sequence protects against RecBCD degradation of DNA in vivo. Proc Natl Acad Sci U S A 1992; 89:12073-7; PMID:1465442; http://dx.doi.org/ 10.1073/pnas.89.24.12073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taylor AF, Smith GR. Strand specificity of nicking of DNA at Chi sites by RecBCD enzyme. Modulation by ATP and magnesium levels. J Biol Chem 1995; 270:24459-67; PMID:7592661; http://dx.doi.org/ 10.1074/jbc.270.41.24459 [DOI] [PubMed] [Google Scholar]
- 18. Chédin F, Ehrlich SD, Kowalczykowski SC. The Bacillus subtilis AddAB helicasenuclease is regulated by its cognate Chi sequence in vitro. J Mol Biol 2000; 298, 7-20; http://dx.doi.org/ 10.1006/jmbi.2000.3556 [DOI] [PubMed] [Google Scholar]
- 19. Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 2004; 432:187-93; PMID:15538360; http://dx.doi.org/ 10.1038/nature02988 [DOI] [PubMed] [Google Scholar]
- 20. Spies M, Dillingham MS, Kowalczykowski SC. Translocation by the RecB motor is an absolute requirement for chi-recognition and RecA protein loading by RecBCD enzyme. J Biol Chem 2005; 280:37078-87; PMID:16041060; http://dx.doi.org/ 10.1074/jbc.M505521200 [DOI] [PubMed] [Google Scholar]
- 21. Wong CJ, Rice RL, Baker NA, Ju T, Lohman TM. Probing 3’-ssDNA loop formation in E. coli RecBCDRecBC-DNA complexes using non-natural DNA: a model for “Chi” recognition complexes. J Mol Biol 2006; 362:26-43; PMID:16901504; http://dx.doi.org/ 10.1016/j.jmb.2006.07.016 [DOI] [PubMed] [Google Scholar]
- 22. Yeeles JTP, van Aelst K, Dillingham MS, Moreno-Herrero F. Recombination hotspots and single-stranded DNA binding proteins couple DNA translocation to DNA unwinding by the AddAB helicase-nuclease. Mol Cell 2011; 42:806-16; PMID:21700225; http://dx.doi.org/ 10.1016/j.molcel.2011.04.012 [DOI] [PubMed] [Google Scholar]
- 23. Yang L. Handa N, Liu B, Dillingham MS, Wigley DB, Kowalczykowski SC. Alteration of χ recognition by RecBCD reveals a regulated molecular latch and suggests a channel-bypass mechanism for biological control. Proc Natl Acad Sci U S A. 2012; 109:8907-12; PMID:22603793; http://dx.doi.org/ 10.1073/pnas.1206081109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saikrishnan K, Griffiths SP, Cook N, Court R, Wigley DB. DNA binding to RecD: role of the 1B domain in SF1B helicase activity. EMBO J 2008; 27:2222-9; PMID:18668125; http://dx.doi.org/ 10.1038/emboj.2008.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saikrishnan K. Yeeles JT, Gilhooly NS, Krajewski WW, Dillingham MS, Wigley DB. Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J 2012; 31:1568-78; PMID:22307084; http://dx.doi.org/ 10.1038/emboj.2012.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 2003; 423:889-93; PMID:12815437; http://dx.doi.org/ 10.1038/nature01674 [DOI] [PubMed] [Google Scholar]
- 27. Dillingham MS, Spies M, Kowalczykowski SC. RecBCD is a bipolar DNA helicase. Nature 2003; 423:893-7; PMID:12815438; http://dx.doi.org/ 10.1038/nature01673 [DOI] [PubMed] [Google Scholar]
- 28. Subramanya HS, Bird LE, Brannigan JA, Wigley DB. Crystal structure of a DExx box DNA helicase. Nature 1996; 384:379-83; PMID:8934527; http://dx.doi.org/ 10.1038/384379a0 [DOI] [PubMed] [Google Scholar]
- 29. Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 1999; 184:75-84; http://dx.doi.org/ 10.1016/S0092-8674(00)80716-3 [DOI] [PubMed] [Google Scholar]
- 30. Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem 2007; 76:23-50; PMID:17506634; http://dx.doi.org/ 10.1146/annurev.biochem.76.052305.115300 [DOI] [PubMed] [Google Scholar]
- 31. Yeeles JTP, Gwynn EJ, Webb MR, Dillingham MS. The AddAB helicase-nuclease catalyses rapid and processive DNA unwinding using a single Superfamily 1A motor domain. Nucleic Acids Res 2011; 39, 2271-85; PMID:21071401; http://dx.doi.org/ 10.1093/nar/gkq1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Steczkiewicz K, Muszewska A, Knizewski L, Rychlewski L, Ginalski K. Sequence, structure and functional diversity of PD-(DE)XK phosphodiesterase superfamily. Nucleic Acids Res 2012; 40:7016-45; PMID:22638584; http://dx.doi.org/ 10.1093/nar/gks382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J, McCabe Ka, Bell CE. Crystal structures of lambda exonuclease in complex with DNA suggest an electrostatic ratchet mechanism for processivity. Proc Natl Acad Sci U S A 2011; 108:11872-7; PMID:21730170; http://dx.doi.org/ 10.1073/pnas.1103467108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hall MC, Matson SW. Helicase motifs : the engine that powers DNA unwinding. Mol Microbiol 1999; 34:867-877; PMID:1059481424670664 [DOI] [PubMed] [Google Scholar]
- 35. Krajewski WW. Fu X, Wilkinson M, Cronin NB, Dillingham MS, Wigley DB. Structural basis for translocation by AddAB helicase-nuclease and its arrest at χ sites. Nature 2014; 508:416-9; PMID:24670664; http://dx.doi.org/ 10.1038/nature13037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol 2002; 12:123-33; PMID:11839499; http://dx.doi.org/ 10.1016/S0959-440X(02)00298-1 [DOI] [PubMed] [Google Scholar]
- 37. Singleton MR, Wigley DB. Modularity and specialization in superfamily 1 and 2 helicases. J Bacteriol 2002; 184:1819-26; PMID:11889086; http://dx.doi.org/ 10.1128/JB.184.7.1819-1826.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dixon DA, Kowalczykowski SC. Role of the Escherichia coli recombination hotspot, chi, in RecABCD-dependent homologous pairing. J Biol Chem 1995; 270:16360-70; PMID:7608206; http://dx.doi.org/ 10.1074/jbc.270.27.16360 [DOI] [PubMed] [Google Scholar]
- 39. Arnold DA, Bianco PR, Kowalczykowski SC. The reduced levels of chi recognition exhibited by the RecBC1004D enzyme reflect its recombination defect in vivo. J Biol Chem 1998; 273:16476-86; PMID:9632715; http://dx.doi.org/ 10.1074/jbc.273.26.16476 [DOI] [PubMed] [Google Scholar]
- 40. Schultz DW, Taylor AF, Smith GR. Escherichia coli RecBC pseudorevertants lacking chi recombinational hotspot activity. J Bacteriol 1983; 155:664-80; PMID:6348024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arnold DA, Handa N, Kobayashi I, Kowalczykowski SC. A novel, 11 nucleotide variant of chi, chi*: one of a class of sequences defining the Escherichia coli recombination hotspot chi. J Mol Biol 2000; 300:469-79; PMID:10884344; http://dx.doi.org/ 10.1006/jmbi.2000.3861 [DOI] [PubMed] [Google Scholar]
- 42. Handa N, Yang L, Dillingham MS, Kobayashi I, Wigley DB, Kowalczykowski SC. Molecular determinants responsible for recognition of the single-stranded DNA regulatory sequence, χ, by RecBCD enzyme. Proc Natl Acad Sci U S A 2012; 109:8901-6; PMID:22603794; http://dx.doi.org/ 10.1073/pnas.1206076109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith GR, Kunes SM, Schultz DW, Taylor A, Triman KL. Structure of chi hotspots of generalized recombination. Cell 1981; 24:429-36; PMID:6453653; http://dx.doi.org/ 10.1016/0092-8674(81)90333-0 [DOI] [PubMed] [Google Scholar]
- 44. Handa N, Ohashi S, Kusano K, Kobayashi I. Chi-star, a chi-related 11-mer sequence partially active in an E. coli recC1004 strain. Genes Cells 1997; 2:525-36; PMID:9348042; http://dx.doi.org/ 10.1046/j.1365-2443.1997.1410339.x [DOI] [PubMed] [Google Scholar]