Abstract
Sororin is a conserved protein required for accurate separation of sister chromatids in each cell cycle. Sororin is recruited to chromatin during DNA replication, protects sister chromatid cohesion in S and G2 phase, and regulates the resolution of sister chromatid cohesion in mitosis. Sororin binds to cohesin complex, but how Sororin and cohesin subunits interact remains unclear. Here we report that the C-terminus of Sororin, especially the last 12 amino acid (aa) residues, is important for Sororin to bind cohesin core subunit SA2. Deletion of the last 12aa residues not only inhibits the interactions between Sororin and SA2 but also causes precocious chromosome separation. Our data suggest that the C-terminus of Sororin functions as an anchor binding to SA2, which facilitates other conserved motifs on Sororin to interact with other proteins to regulate sister chromatid cohesion and separation.
Keywords: cell cycle, cohesin, SA2, sister chromatid cohesion, Sororin
Introduction
Accurate cohesion and separation of sister chromatids rely on the cohesin complex and its associated proteins. The cohesin complex is comprised of 4 core subunits: Smc1, Smc3, Rad21 and SA1/SA2. Several other proteins, such as Wapl, Pds5 and Sororin, are associated with the cohesin complex and regulate sister chromatid cohesion.1,2 The cohesin cycle, cohesin association and dissociation from chromosomes, consists of cohesin loading, sister chromatid cohesion generation and maintenance, as well as cohesin removal and its regeneration. Sororin is involved in the generation and maintenance of sister chromatid cohesion and regulates the removal of cohesin from chromatin.3 The cohesin complexes are loaded to chromatin by a heterodimeric protein complex, Scc2/NIPBL-Scc4/NAU2, in a replication licensing-dependent manner.4,5 Acetyltransferases, Eco1 vertebrate orthologs Esco1 and Esco2, are essential for acetylation of Smc3, a step required for the recruitment of Sororin and the generation of sister chromatid cohesion during DNA replication.6-9 The cohesin-associated Sororin maintains the sister chromatid cohesion established at S phase until Sororin is phosphorylated and dissociated from chromatids, when Wapl forms a complex with Pds5 to remove cohesin in prophase/prometaphase.7,10-15 In addition, Sororin plays a regulatory role in the resolution of sister chromatid cohesion during M-phase.3,15
Sororin can co-immunoprecipitate (co-IP) cohesin core subunits Rad21, Smc1, Smc3, and SA1/2, as well as cohesin-associated protein Pds5A/B and Wapl.7,16 The C-terminus of Sororin is known to be responsible for interaction with cohesin,17 but which cohesin core subunit Sororin binds remains unknown. Here we report that the last 12 amino acid (aa) residues of Sororin are required to interact with cohesin core subunit SA2, and that deletion of these 12aa residues not only inhibits the interaction of Sororin and cohesin, but also fails to rescue the precocious chromosome separation (PCS) caused by the silencing of endogenous Sororin.
Results
Sororin is required for the maintenance of sister chromatid cohesion
Silencing of Sororin by using siRNA in HeLa cells increased the mitotic index up to 50% of the cell population and slowed the cell cycle (Figure S1A–B). Despite no obvious differences in the formation of the mitotic spindle bodies, chromosomes in Sororin siRNA-treated cells had difficulty in aligning to the metaphase plate. Chromosomal morphology of these cells was reminiscent of the prometaphase cells, in which chromosomes are yet to be congregated on the cell plates (Figure S1B–C). These results are consistent with the previous observations.18 Unlike the control cells, in which the segregated chromosomes formed 2 compact round or oval-shaped nuclei that were pulled apart toward the opposite poles, the separated chromosomes in the Sororin-silenced cells were loose and dispersed, and they were not pulled apart to the opposite poles (Figure S1B). The overall chromosomal mass in the resulting daughter cells also appeared unequal in the Sororin knockdown cells but not in the control cells (Figure S1B), indicating a critical chromatid separation defect. After chromosomal decondensation occurred, most of the cells showed multi-nucleated cells (Figure S1C), demonstrating that Sororin plays critical roles in chromatid cohesion and separation. FISH analyses indicated that the vast majority (>95 %) of mitotic cells exhibited PCS (Fig. 1A–B).
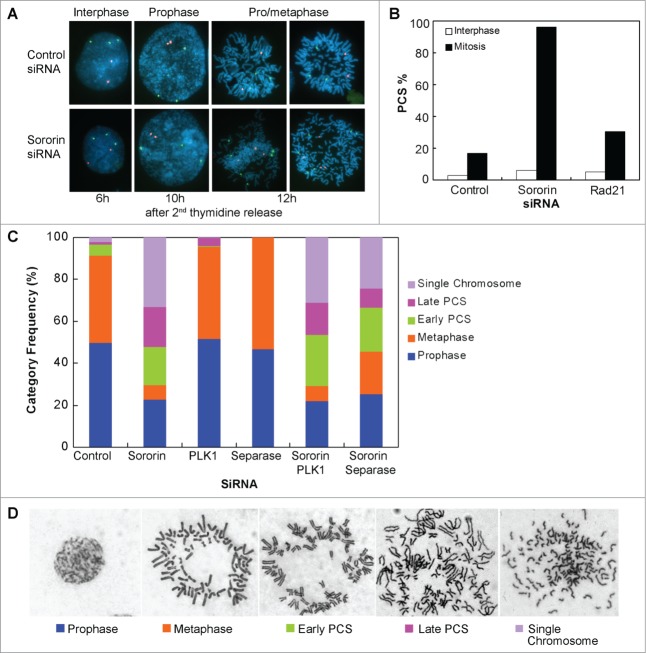
Figure 1.
Knockdown of Sororin results in precocious chromosome separation (PCS). (A and B) HeLa cells were synchronized with double thymidine. Sororin siRNA was transfected 4h after the cells were released from the first thymidine arrest. Cells were harvested and fixed at 2 h intervals after the 2nd thymidine release for 12 h. FISH was performed using probes CEP 4 (green) and CEP 9 (red). The images in (A) were shown 6, 10 and 12h after the 2nd thymidine release. The percentage of PCS was calculated in interphase cells (6 h after 2nd thymidine release) and in mitotic cells (12 h after 2nd thymidine release). Approximately 200 cells were counted in each treatment. Cells transfected with Rad21 siRNA were used as a positive control. (C). HeLa SA2 Tet-On cells were double thymidine arrested and transfected with one or 2 siRNAs as described above. 12 h after release from double thymidine arrest, the cells were fixed and metaphase spread was performed. Approximately 400 cells were counted for each treatment. (D). Representative pictures of the 5 categories of mitotic cells used for cytological analysis in (C).
To examine the distribution of mitotic stages of Sororin knockdown cells, we treated HeLa cells with siRNA. After the cells were released from double thymidine for 12 hours, metaphase spreads were performed (Fig. 1C–D). In control siRNA-treated cells, 90% of the mitotic cells were in the categories of prophase and prometaphase/metaphase. Contrariwise, in Sororin knockdown cells fewer than 30% of the mitotic cells were in the categories of prophase and prometaphase/metaphase, and cells with early PCS, late PCS, and single chromosome accounted for more than 70% of the spreads (Fig. 1C). Interestingly, compared to the control, the number of prophase cells were 27% fewer in Sororin siRNA-treated cells (Fig. 1C), suggesting that a fraction of the prophase cells had undergone PCS.
In higher eukaryotes, cohesins on the sister chromatids are removed in 2 steps. Protein kinases Plk1 and Aurora B are required for the non-proteolytic dissociation of cohesins from chromatid arms in prophase, whereas Separase is essential for the removal of centromeric cohesins at the onset of anaphase.19,20 Chromosome arms cannot be opened in Plk1-depleted cells until Separase cleaves cohesin at the onset of anaphase. To examine if knockdown of Plk1 could override the effect caused by Sororin siRNA, we transfected cells with Plk1 siRNA—alone or with Sororin siRNA. Knocking down Plk1 increased the population of cells at metaphase (Fig. 1C). However, combined treatment of Plk1 and Sororin siRNA rendered no significant differences in the level of PCS and single chromosomes compared to the treatment of Sororin siRNA alone (Fig. 1C), indicating that Plk1 knockdown cannot rescue the dissociation of cohesins from chromosomes caused by Sororin silencing.
We also examined cells treated with Separase siRNA in the presence and absence of Sororin siRNA. Separase is required for the separation of sister chromatid cohesion through cleavage of cohesin Rad21 at the onset of anaphase.19,21 When HeLa cells were treated with Separase siRNA alone, they were arrested at metaphase (Fig. 1C). However, most of the mitotic cells exhibited PCS when the cells were treated simultaneously with Separase siRNA and Sororin siRNA (Fig. 1C). These results strongly argue that Sororin is an essential structural component of the cohesin complex and plays an important role in the maintenance of sister chromatid cohesion. Furthermore, knockdown of effector molecules, such as Plk1 and Separase, which function on the cohesin subunits, cannot compensate for the loss of Sororin.
Sororin interacts with cohesin via SA2
Sororin physically interacts with the cohesin complex. Co-IP experiments indicated that Sororin could pull down cohesin core subunits, including Rad21, Smc3 and SA2 (Fig. 2A). To investigate which cohesin subunit directly binds to Sororin, we knocked down endogenous cohesin subunits and prepared cell lysates, which were incubated with Sororin-His-coated Ni-NTA beads or mock Ni-NTA beads. Immunoblotting demonstrated that Sororin pulled down SA2 irrespective of the Rad21 status (Fig. 2B), indicating that in our in vitro assay the interaction of Sororin with soluble SA2 is independent of Rad21.
Figure 2.
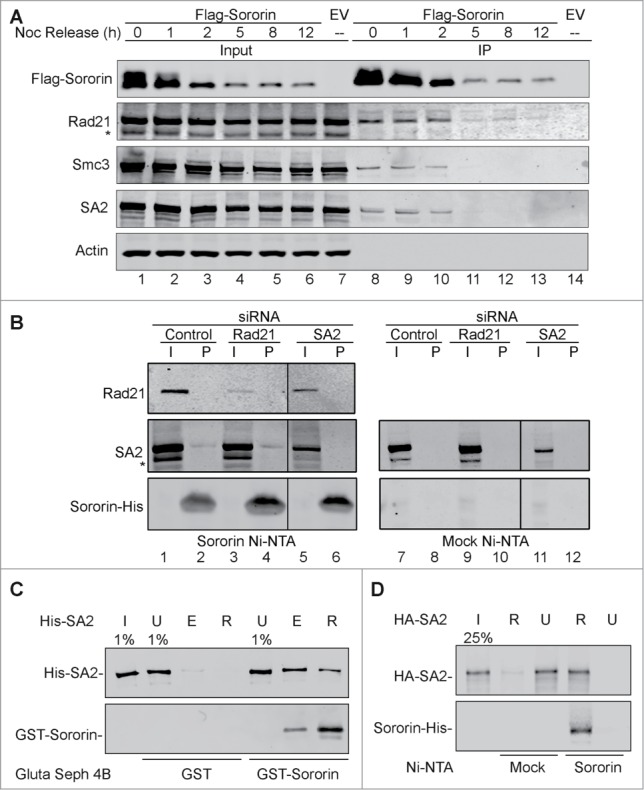
Interaction of Sororin and cohesin. (A) Thirty hours after Flag-Sororin was expressed, HeLa cells were arrested with nocodazole for 15 h. Cell samples were taken at various time points after nocodazole release. Flag-Sororin was immunoprecipitated, and the co-IP of cohesin subunits was visualized with immunoblotting. The Flag-empty vector (Flag-EV) was transfected; unsynchronized cells were used as control (lane 7, 14). (B) 293T cells were transfected with Rad21 or SA2 siRNA for 48 h; silencer negative control #1 siRNA was used as control. Whole cell lysate was made and incubated with Ni-beads coated with Sororin-His or mock Ni-beads. Cohesin subunits in input (I) and pull down (P) samples were immunoblotted with indicated antibodies. The vertical lines indicate the intervening lanes have been spliced out. (C–D) Glutathione Sepharose 4B beads coated with GST or GST-Sororin were incubated with His-SA2 for 4h (C), or Sororin-His-coated Ni-NTA agarose (Sororin) and Ni NTA-agarose (Mock) were incubated with HA-SA2 for 4 h (D). SA2 in input (I), unbound (U), eluted fraction with glutathione (E), and retained on beads (R) were immunoblotted with anti-SA2 and Sororin antibodies.
To further confirm the interaction of Sororin with SA2, we incubated the recombinant His-SA2 protein with either GST-Sororin or GST alone (mock) immobilized on Glutathione Sepharose 4B beads. GST-Sororin, but not GST alone, pulled down His-SA2 (Fig. 2C). We also performed another in vitro experiment to test the interaction of SA2 and Sororin. In this assay, Sororin-His immobilized on Ni-NTA beads could pull down recombinant HA-tagged SA2 (Fig. 2D). These results indicate that Sororin directly binds to SA2.
C-terminus of Sororin interacts with SA2
In order to identify which region of Sororin interacts with SA2, we made a series of Sororin truncated mutant constructs using a pFlag CMV2 vector. The constructs were engineered with progressive deletion of 150 nucleotides (50aa residues) from 5′ or 3′ends of Sororin cDNA (Fig. 3A). After these constructs were transfected into 293T cells, co-IP of endogenous SA2 by wild-type (WT) and mutant Flag-Sororin was examined. Immunoblotting results showed that Sororin truncated mutants lacking the last 52aa residues at C-terminus could not interact with SA2 (Fig. 3B, lanes 13–16), whereas the N-terminus from 1 to 200aa was not so critical for the interaction because the last 52aa residues of Sororin could efficiently pull down SA2 (Fig. 3B, lanes 17–20).
Figure 3.
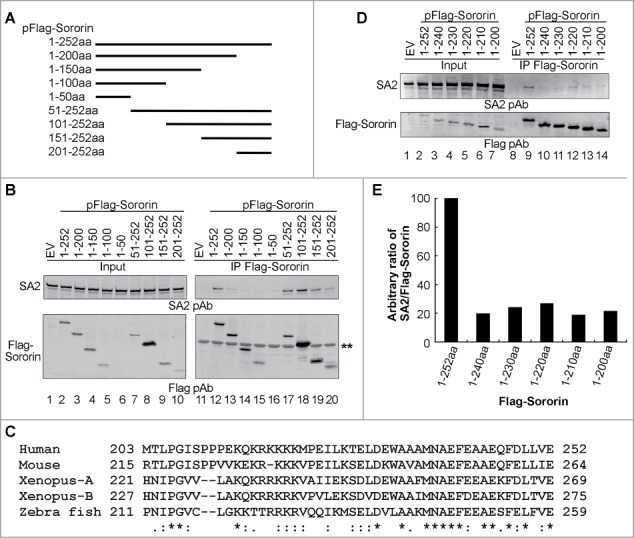
C-terminus of Sororin interacts with SA2. (A) Schematic illustration of wild-type and sequentially truncated Sororin. The cDNA of these Sororin proteins were cloned into a pFlag CMV2 vector. The expressed proteins are about 50aa residues different from either N-terminus or C-terminus of Sororin. (B) Flag-Sororin WT or mutants shown in (A) were expressed in 293T cells and immunoprecipitated with Flag mAb conjugated agarose beads. Co-IP of SA2 and Flag-Sororin were examined using immunoblotting. * nonspecific band. (C) Alignment of conserved C-terminus of vertebrate Sororin. Invariant, conserved, and semi-conserved residues are indicated by an asterisk (*), colon (:), and period (.), respectively. (D–E) Flag Sororin WT or mutants with about 10aa residues different were expressed in 293T cells. Co-IP of SA2 by Flag-Sororin was detected using immunoblotting (D). The amout of SA2 co-immunopreciptated by Flag-Sororin was determined, and the ratio of SA2/Sororin was calulated and ploted in bar graph (E).
The last 50aa residues of Sororin are highly conserved across vertebrates, which contain 2 domains consisting mainly of basic amino acids (213KQKRKKKK221) and acidic amino acids (239EFEAAEQFDLLVE252) (Fig. 3C). To further clarify which region within the last 50aa residues of Sororin is responsible for the interaction with SA2, we made constructs by progressively removing 10–12aa residues each time and performed co-IP experiments. Immunoblotting results demonstrated that deletion of the last 12aa residues reduced co-IP of endogenous SA2 by Sororin by 80% but that further deletion up to the complete removal of the 52aa residues of Sororin C-terminus did not enhance the effect (Fig. 3D–E). These studies indicate that the last 12aa residues of the Sororin molecule are important for its interaction with cohesin subunit SA2.
C-terminus of Sororin is important for sister chromatid cohesion
To determine the physiological effect of C-terminus deletion of Sororin on the sister chromatid cohesion, we expressed Flag-tagged Sororin WT or mutants (1–240aa, 1–200aa) with deletion of 12aa or 52aa residues on the Sororin C-terminus in HeLa cells. The endogenous Sororin was silenced with 3'UTR siRNA. Non-gene targeted siRNA was used as control. The expression level of Flag-tagged Sororin was similar in both control and Sororin siRNA-treated cells (Fig. 4A). According to the resolution status of sister chromatids in the metaphase chromosome spread, we categorized sister chromatids into arm-cohesed, arm-separated sister chromatids separated and single chromosomes (Fig. 4B). In control siRNA-treated cells, compared to the Flag-empty vector (EV), expression of Flag-Sororin WT increased the number of cells with arm-cohesed sister chromatids by approximately 15% and decreased the number of cells with arm-separated chromatids by approximately the same percentage, whereas Flag-Sororin 1–240aa and 1–200aa constructs did not affect the percentage of cells with arm-cohesed sister chromatids but slightly increased the percentage of cells with separated sister chromatids (sister separated) (Fig. 4C).
Figure 4.
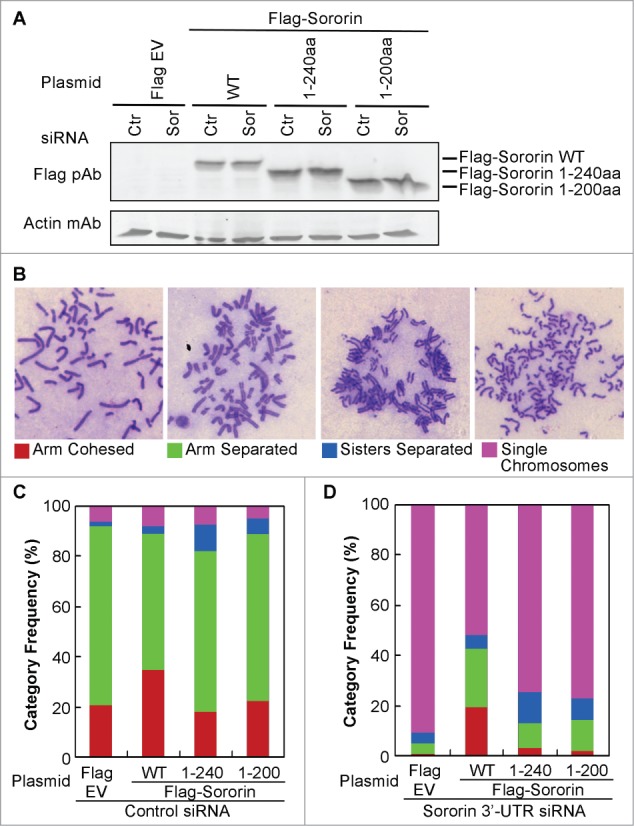
Deletion of Sororin C-terminus results in precocious chromosome separation. Sororin was knocked down using 3'-UTR siRNA at 24 h after Flag-Sororin WT or C-terminal deletion mutants (1–240aa, 1–200aa) were expressed in HeLa cells. pFlag CMV2 empty vector (EV) and Silencer siRNA control#1 were used as controls. Twenty-four hours after siRNA treatment, cells were harvested for immunoblotting and metaphase spread of chromosomes. (A) Immunoblotting showing the expression of Flag-Sororin WT and mutants. (B–D) Metaphase spread of chromosomes was performed after Flag-Sororin WT or mutants were expressed and endogenous Sororin was knocked down. More than 100 mitotic cells were randomly examined and categorized according to the resolution of sister chromatid cohesion shown in (B). The frequency of each category was plotted in (C) and (D).
Silencing of Sororin caused more than 90% of sister chromatids to become single chromosomes. The effect of Sororin silencing could be rescued up to 40% (arm-cohesed + arm-separated) by Flag-Sororin WT (Fig. 4D). However, the Sororin-silencing phenotype could be rescued up to only 10% by Flag-Sororin 1–240aa or 1–200aa constructs (Fig. 4D), suggesting that the last 12aa residues on the Sororin C-terminus are critical not only for its binding to SA2, but also for the maintenance of sister chromatid cohesion.
Discussion
Recruitment of Sororin to chromatin during DNA replication at S phase is dependent on the association of cohesin to chromatin and acetylation of Smc3.3,7, 22 However, it is unclear which cohesin subunit that Sororin binds to. We report herein that Sororin binds to cohesin-SA2 through its C-terminus and plays an important role in sister chromatid cohesion and separation.
Sororin orthologs from vertebrate species are highly conserved in their last 50aa residues, which contain an arginine/lysine-rich region from 214aa to 222aa and an acidic amino acid-rich stretch from 237aa to 252aa in the C-terminus of Sororin (Fig. 3C, numbering refers to the human Sororin sequence). The 16aa conserved residues toward the C-terminal end are critical for Sororin to interact with SA2 because a Sororin deletion mutant with only the last 52aa can co-IP SA2, whereas a mutant missing the 12aa of the 16aa conserved residues abolishes its interaction with SA2. The exact role of the arginine/lysine-rich region from 214 to 222aa residues remains to be defined because deletion of this region only slightly reduces Sororin association with chromatin and does not affect Sororin interaction with cohesin and sister chromatid cohesion.22
In addition to inhibiting interaction of Sororin and SA2, deletion of the last 12aa residues of Sororin also results in PCS, suggesting that this domain plays a crucial role in protecting or maintaining sister chromatid cohesion. However, the mechanism is not well elucidated. Published studies indicate that 2 other conserved domains on Sororin, [156(S/T)S(S/T)P160] and [161RxSxFGF(D/E)169], also play important roles in regulating sister chromatid cohesion and separation. [156(S/T)S(S/T)P160] is a Plk1-binding motif.3,15 The polo-box domain of Plk1 binds to the consensus motif S(S/T)P after the middle S/T is phosphorylated by protein kinases Cdk1/cyclin B in the early mitosis. Plk1 is required for the non-proteolytic arm cohesin removal via SA2 phosphorylation in prophase.19,23-26 Our published results suggest that after the Sororin ST159P motif is phosphorylated by Cdk1/cyclin B (becomes SpT159P) at prophase/prometaphase, Sororin serves as a docking protein for Plk1 to bind to the SpT159P motif so that the enzymatic catalytic domain of Plk1 is brought proximately to its substrate SA2 to phosphorylate.3,15
Sororin protects sister chromatid cohesion, and Wapl removes cohesins from sister chromatids. The [161RxSxFGF(D/E)169] domain contains a FGF motif that is conserved in Sororin orthologs across different taxa. Both Sororin and Wapl proteins contain FGF motifs and function against each other by competing with each other in binding to Pds5 via FGF motif.7 Wapl can remove cohesin from chromatin when it binds to Pds5 via FGF motif. When Sororin is recruited to cohesin, the FGF motif on Sororin binds to Pds5 and Wapl is displaced (but remains association with Pds5), resulting in the loss of Wapl's ability to remove cohesin; hence, sister chromatid cohesion is maintained. After the cell cycle progresses to prophase/prometaphase, protein kinases, such as Ckd1/cyclin B and Aurora B, phosphorylate Sororin, causing Sororin to dissociate from cohesin.13 Wapl1 regains the binding to Pds5 via its FGF motif, and cohesin is removed from chromatid arms.
In summary, the C-terminus of Sororin functions as an anchor that binds to cohesin core subunit SA2. Because of this anchorage, other Sororin motifs, such as the Pds5-binding FGF motif and the Plk1-binding ST159P motif, can regulate sister chromatid cohesion and separation at different stages of the cell cycle. The ability of Sororin to bind to SA2 depends on its phosphorylation status. Protein kinases phosphorylate Sororin, resulting in dissociation of Sororin from cohesin.
Methods and Materials
Cell culture: HeLa and 293T cells (ATCC, Manassas, VA) were grown in DMEM plus 10% FBS and maintained at 37oC, 95% humidity, and atmosphere of 5% CO2.
Antibodies: The sources of the antibodies used in this study were as follows. Rabbit polyclonal antibodies: Flag (Sigma, St. Louis, MO), human Rad21 (Pati et al., 2002), Smc3 (Bethyl Laboratories, Inc., Montgomery, TX). Goat polyclonal antibodies: Smc1, SA2 (Novus Biologicals, Littleton, CO). Mouse monoclonal antibodies: Flag (Sigma) and HA (Sigma).
Mutation and cloning of Sororin: Mutations of serine and threonine residues into alanine were performed using site-specific PCR. Sororin cDNA was cloned into EcoRI and EcoRV cloning sites of pFlag-CMV2 for expression in mammalian cells, or cloned into BamH1 and Xho1 sites of pET21a(+) for expression as a 6xHis epitope tagged protein in bacteria.
Transfection: The calcium phosphate method was used to transfect appropriate plasmids into 293T cells. EffecteneTM transfection reagent from Qiagen (Valencia, CA) was used to transfect HeLa cells. The medium was changed 16h after transfection.
siRNA and transfection: Sororin siRNA (Dharmacon), Plk1 siRNA (Qiagen), and Separase siRNA (Ambion) were transfected into cells using DharmaFECTTM 1 (Dharmacon). Silencer® Negative Control siRNA #1 (Ambion) was used as control.
Metaphase chromosome spread: Cells were trypsinized and spun down at 100 g for 6 min. The cells were mixed with 10 ml pre-warmed (37°C) hypotonic solution (0.075M KCl), which was added drop-wise with gentle agitation. The samples were incubated at 37oC for 15 min. One ml of fixative (freshly prepared 3:1 methanol:acetic acid) was added to each sample, followed by centrifugation at 100 g for 6 min. Ten ml fixative was added to the cells, which were incubated at room temperature for at least 30 min. The fixative was changed twice, and the cells were re-suspended in 0.5 ml fixative. Two to 3 drops of cell suspension were dropped from a height of 12 inches onto angled slides. After being air dried, the slides were stained with Giemsa solution for 5 min. and rinsed in running water for 10 min.
Fluorescence in situ hybridization (FISH). HeLa cells were transfected with Sororin, Rad21, or control siRNA as described above. Forty-eight hours after being transfected, the cells were detached with trypsin and fixed as described above. A spectrum green-labeled centromeric probe for chromosome 4 and a spectrum red-labeled, centromeric probe for chromosome 9 (Vysis) were used. Hybridization and detection were performed according to the manufacturer's protocols. For each experiment, 200 intact interphase nuclei were scored and captured for hybridization signals with Nikon E800 equipped with QUIPS imaging software (Applied Imaging).
Protein isolation, immunoprecipitation and Western blotting: Unless otherwise noted, cells were harvested 40–48h after being transfected. The protocol for protein isolation, immunoprecipitation and Western blotting has been described previously.27
Recombinant proteins: SA1 and SA2 were cloned into pFastBac1 vector (Invitrogen) with a 6-histidine tag on the N-terminal of both proteins. Both recombinant viruses were generated according to the Bac-to-Bac Baculovirus Expression System protocol (Version D, Invitrogen). Sf21 insect cells were grown in Grace's medium supplemented with 10% fetal bovine serum. SA1 and SA2 were expressed in Sf21 cells that were infected with recombinant viruses at a multiplicity of infection of 7. Sf21 cells were harvested 60 h post-infection and lysed by sonication. His-tagged recombinant proteins were pulled down by Ni-NTA beads (Qiagen) and eluted 250 mM imidazole in 20 mM Tris buffer (pH 7.5, containing 150 mM NaCl). The eluted proteins were further purified using a Hitrap Q column (GE healthcare, Piscataway, NJ) and gel filtration chromatography on a 24 ml Superose 6 column (GE healthcare).
Sororin cDNA was cloned into pGEX4T3 or pET21a(+) vector (Novagen) with GST or 6xHis tagged to Sororin N-terminus or C-terminus, respectively. Recombinant Sororin was expressed in E. coli BL21 codon plus. GST-Sororin was pulled down with glutathione Sepharose 4B and eluted with 10mM glutathione in 50mM Tris buffer (pH 8.0). Sororin-His was pulled down with Ni-NTA beads and eluted with 250mM imidazole.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Yunyun Jiang, Qilong Mao, Anil K. Panigrahi, and Pulivarthi H. Rao for their technical support. We also acknowledge the editorial assistance of Dr. Lee Ligon of the Center for Research, Innovation, and Scholarship, Department of Pediatrics, Baylor College of Medicine.
Funding
This study was supported by award 1RO1 CA109478 from the National Cancer Institute to D Pati.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Peters JM, Tedeschi A, Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev 2008; 22:3089-114; PMID:19056890; http://dx.doi.org/ 10.1101/gad.1724308 [DOI] [PubMed] [Google Scholar]
- 2. Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell DevBiol 2008; 24:105-29; PMID:18616427; http://dx.doi.org/ 10.1146/annurev.cellbio.24.110707.175350 [DOI] [PubMed] [Google Scholar]
- 3. Zhang N, Pati D. Sororin is a master regulator of sister chromatid cohesion and separation. Cell Cycle 2012; 11:2073-83; PMID:22580470; http://dx.doi.org/ 10.4161/cc.20241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol 2004; 6:991-6; PMID:15448702 [DOI] [PubMed] [Google Scholar]
- 5. Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol 2004; 14:1598-603; PMID:15341749; http://dx.doi.org/ 10.1016/j.cub.2004.07.053 [DOI] [PubMed] [Google Scholar]
- 6. Beckouet F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol Cell 2010; 39:689-99; PMID:20832721; http://dx.doi.org/ 10.1016/j.molcel.2010.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nishiyama T, Ladurner R, Schmitz J, Kreidl E, Schleiffer A, Bhaskara V, Bando M, Shirahige K, Hyman AA, Mechtler K, et al. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell 2010; 143:737-49; PMID:21111234; http://dx.doi.org/ 10.1016/j.cell.2010.10.031 [DOI] [PubMed] [Google Scholar]
- 8. Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell 2009; 33:763-74; PMID:19328069; http://dx.doi.org/ 10.1016/j.molcel.2009.02.028 [DOI] [PubMed] [Google Scholar]
- 9. Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 2007; 317:245-8; PMID:17626885; http://dx.doi.org/ 10.1126/science.1140637 [DOI] [PubMed] [Google Scholar]
- 10. Dreier MR, Bekier ME, Taylor WR. Regulation of sororin by Cdk1-mediated phosphorylation. J Cell Sci 2011; 124:2976-87; PMID:21878504; http://dx.doi.org/ 10.1242/jcs.085431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 2006; 16:2406-17; PMID:17112726; http://dx.doi.org/ 10.1016/j.cub.2006.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters JM. Wapl controls the dynamic association of cohesin with chromatin. Cell 2006; 127:955-67; PMID:17113138; http://dx.doi.org/ 10.1016/j.cell.2006.09.040 [DOI] [PubMed] [Google Scholar]
- 13. Nishiyama T, Sykora MM, Huis in 't Veld PJ, Mechtler K, Peters JM. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc Natl Acad Sci U S A 2013; 110:13404-9; PMID:23901111; http://dx.doi.org/ 10.1073/pnas.1305020110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shintomi K, Hirano T. Releasing cohesin from chromosome arms in early mitosis: opposing actions of Wapl-Pds5 and Sgo1. Genes Dev 2009; 23:2224-36; PMID:19696148; http://dx.doi.org/ 10.1101/gad.1844309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang N, Panigrahi AK, Mao Q, Pati D. Interaction of Sororin with polo-like kinase 1 mediates the resolution of chromosomal arm cohesion. J Biol Chem 2011; 286:41826-37; PMID:21987589; http://dx.doi.org/ 10.1074/jbc.M111.305888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol Cell 2005; 18:185-200; PMID:15837422 [DOI] [PubMed] [Google Scholar]
- 17. Wu FM, Nguyen JV, Rankin S. A conserved motif at the C-terminus of sororin is required for sister chromatid cohesion. J Biol Chem 2010; 286:3579-86; PMID:21115494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ. Regulation of centromeric cohesion by sororin independently of the APC/C. Cell Cycle 2007; 6:714-24; PMID:17361102; http://dx.doi.org/ 10.4161/cc.6.6.3935 [DOI] [PubMed] [Google Scholar]
- 19. Gimenez-Abian JF, az-Martinez LA, Waizenegger IC, Gimenez-Martin G, Clarke DJ. Separase is required at multiple pre-anaphase cell cycle stages in human cells. Cell Cycle 2005; 4:1576-84; PMID:16177575; http://dx.doi.org/ 10.4161/cc.4.11.2147 [DOI] [PubMed] [Google Scholar]
- 20. Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 2000; 103:399-410; PMID:11081627; http://dx.doi.org/ 10.1016/S0092-8674(00)00132-X [DOI] [PubMed] [Google Scholar]
- 21. Wirth KG, Wutz G, Kudo NR, Desdouets C, Zetterberg A, Taghybeeglu S, Seznec J, Ducos GM, Ricci R, Firnberg N, et al. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J Cell Biol 2006; 172:847-60; PMID:16533945; http://dx.doi.org/ 10.1083/jcb.200506119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lafont AL, Song J, Rankin S. Sororin cooperates with the acetyltransferase Eco2 to ensure DNA replication-dependent sister chromatid cohesion. Proc Natl Acad Sci USA 2010; 107:20364-9; PMID:21059905; http://dx.doi.org/ 10.1073/pnas.1011069107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hauf S, Roitinger E, Koch B, Dittrich CM, Mechtler K, Peters JM. Dissociation of Cohesin from Chromosome Arms and Loss of Arm Cohesion during Early Mitosis Depends on Phosphorylation of SA2. PLoS Biol 2005; 3:e69; PMID:15737063; http://dx.doi.org/ 10.1371/journal.pbio.0030069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 1998; 12:1986-97; PMID:9649503; http://dx.doi.org/ 10.1101/gad.12.13.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol 2000; 150:405-16; PMID:10931856; http://dx.doi.org/ 10.1083/jcb.150.3.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakajima M, Kumada K, Hatakeyama K, Noda T, Peters JM, Hirota T. The complete removal of cohesin from chromosome arms depends on separase. J Cell Sci 2007; 120:4188-96; PMID:18003702; http://dx.doi.org/ 10.1242/jcs.011528 [DOI] [PubMed] [Google Scholar]
- 27. Zhang N, Kuznetsov SG, Sharan SK, Li K, Rao PH, Pati D. A handcuff model for the cohesin complex. J Cell Biol 2008; 183:1019-31; PMID:19075111; http://dx.doi.org/ 10.1083/jcb.200801157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.