Abstract
Apoptosis is a primary characteristic in the pathogenesis of liver disease. Hepatic apoptosis is regulated by autophagic activity. However, mechanisms mediating their interaction remain to be determined. Basal level of autophagy ensures the physiological turnover of old and damaged organelles. Autophagy also is an adaptive response under stressful conditions. Autophagy can control cell fate through different cross-talk signals. A complex interplay between hepatic autophagy and apoptosis determines the degree of hepatic apoptosis and the progression of liver disease as demonstrated by pre-clinical models and clinical trials. This review summarizes recent advances on roles of autophagy that plays in pathophysiology of liver. The autophagic pathway can be a novel therapeutic target for liver disease.
Keywords: autophagy, apoptosis, liver injury, cross-talk, mechanism
Abbreviations
- Atg
autophagy-related gene
- Bcl-2
B-cell lymphoma-2
- Beclin-1
Bcl-2-interacting protein-1
- BH3
Bcl-2 homology domain-3
- Bcl-xL
B-cell lymphoma extra long
- ER stress
endoplasmic reticulum stress
- CSE
cigarette smoke extract
- DISC
death-inducing signaling complex
- TNFα
tumor necrosis factor-α
- FADD
Fas-associated protein with death domain
- DRAM
damage regulated autophagic modulator
- ALT
alanine aminotransferase
- mTOR
mammalian target of rapamycin
- PCD
programmed cell death
- TUNEL
terminal deoxynucleotidyl transferase dUTP nick-end labeling
- ATP
adenosine triphosphate
- DNA
DNA
- RNA
ribonucleic acid
- siRNA
small interfering RNA
- LAMP-2
lysosome-associated membrane protein 2
- LD
lipid droplets
- HCV
hepatitis C virus
- Drp1
dynamin-related protein 1
- HBV
hepatitis B virus
- HBx
hepatitis B X protein
- FFA
free fatty acids
- HCC
hepatocellular carcinoma
- HSC
hepatic stellate cells
- MDBs
Mallory-Denk bodies
- MOMP
mitochondrial outer membrane permiabilization
- APAP
N-acetyl-p-aminophenol
- ROS
reactive oxygen species
- Microtubule LC3
microtubule light chain 3
- PI3KC3
phosphatidylinositol-3-kinase class-3
- Vps34
vacuolar protein sorting-34
- UVRAG
UV-resistance-associated gene
- AMBRA-1
activating molecule in Beclin-1-regulated autophagy
- Barkor
Beclin-1-associated autophagy-related key regulator
- BNIP
Bcl-2/adenovirus E1B 19 kd-interacting protein
- c-FLIP
cellular FLICE-like inhibitor protein
Cross-Talk Between Autophagy and Different Modes of Cell Death
Autophagy is a self-digestive process with different types in mammalian cells. The macroautophagy is a predominant form that is often referred to as autophagy. Autophagy takes part in diverse activities such as nutrient starvation, destruction of intracellular pathogens and degradation of damaged organelles. The autophagy is the unique mechanism to decompose large organelles and protein aggregates. Therefore, autophagy can maintain cell homeostasis and ensures cell survival under stressful conditions. Multifunctional roles of autophagy exert its potential for both adaptive and harmful outcomes. The malfunction of autophagy plays a pathogenic part in human diseases such as microbial infection, neurodegeneration and cancerogenesis.1-3 There are considerable cross-talks between the autophagy and different modes of cell death (Fig. 1). The latter includes apoptosis, pyroptosis, necroptosis, and necrosis. Apoptosis as one of terminal paths of cell death is a typical form of programmed cell death (PCD), which is involved in morphogenesis during embryonic development and elimination of aged or harmful cells to maintain adult tissue homeostasis. The deficiency of apoptosis can lead to developmental defects, neurodegeneration and carcinogenesis. In liver, chronic disease accompanies a lot of hepatocyte apoptosis. Hepatic apoptosis is considered to be a prominent pathological feature in most forms of liver injury. Interventions in hepatic apoptosis can delay disease progression and reduce the morbidity of liver disease. However, no therapeutic approaches are currently satisfied in clinical practice. The responsible mechanisms of hepatic apoptosis are still under investigation.4 Autophagy contributes to bulk degradation of cytoplasm and mitochondrion.5,6 Autophagy influences mitochondrial recycle and can thus modulate hepatic apoptosis via mitochondrial pathway. Inflammasome-dependent caspase-1 activation promotes pyroptotic cell death and the secretion of proinflammatory cytokines.7 Autophagy is also associated with caspase-independent cell death, which leads to necrosis and necroptosis.8 Furthermore, proapoptotic roles of autophagy have been reported.9,10 Future studies may focus on the dynamic equilibrium among autophagy, apoptosis, and necrosis in the context of disease-related pathogenesis.11,12 A good understanding of these relationships would be essential in the development of therapeutics targeting the autophagy pathway for the treatment of relevant diseases.
Figure 1.
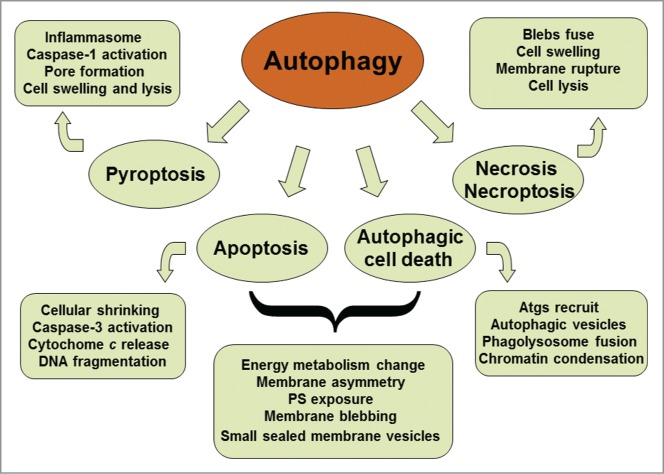
Schematic representation of the cross-talk between autophagy and different modes of cell death. Both autophagy and apoptosis can lead to cell death. They can act independently in parallel pathways. However, autophagy and apoptosis are partners, one may influence the other. Autophagic process is an early adaptive response prior to apoptotic cell death, placing it upstream of apoptosis. Cytoprotective autophagy may suppress apoptosis, and vice versa.
Interconnection Between Autophagy and Apoptosis
Autophagy plays important roles in cell survival as well as in the regulation of cell death, especially apoptosis-signaling pathways. The evidence that autophagy regulates apoptosis includes (i) mitophagy. Autophagy can selectively degrade damaged mitochondria and maintains mitochondrial homeostasis. Mitochondrion-dependent (intrinsic) pathway is well known to be important machinery for apoptotic cell death; (ii) morphological similarity of final products. Autophagy itself can cause cell death or autophagic cell death.13-15 The autophagic cell death as one form of PCD is accompanied with the formation of autophagosomes and mediated by autophagy proteins.16,17 The activation of autophagic cell death through the JNK/Beclin 1 pathway has been investigated in the treatment of liver cancer.18 There is a similarity between autophagosomes (autophagic cell death) and apoptotic bodies (general apoptosis) in morphology. Study on the metamorphosis of moths has revealed a novel form of PCD that is different from T cell death during negative selection in the mouse thymus. The cell death of moth at the end of metamorphosis is a classic example of autophagic cell death.19,20 The autophagic activity probably is a survival mechanism to protect dying cells from death, but the causative relationship between autophagic response and cell death has not been confirmed by histological and biochemical studies; (iii) interaction between Beclin-1 and antiapoptotic Bcl-2 family members. Bcl-2 proteins not only counteract the activity of proapoptotic proteins to downregulate apoptosis, but interact with Beclin-1 to impede autophagy as well. Bcl-2 members such as Bcl-2, Bcl−XL and Bcl−B can bind to Beclin-1, prevent the association of Beclin-1 with PI3KC3 complex and inhibit autophagy.21-23 BNIP3 can cause apoptosis through sequestering Bcl-2 family proteins, promoting Bax/Bad-dependent release of proapoptotic mediators, and disrupting the interaction between Bcl-2 and Beclin-1.24 Autophagy and apoptosis can be coordinately regulated by Bcl-2 family proteins. Moreover, Beclin-1 is cleaved and inactivated by caspases during activation of apoptosis. Apoptosis-effector molecules may suppress autophagy;25 (iv) role of p53. p53 can co-regulate autophagy and apoptosis. p53 modulates the expression of apoptosis-related genes (i.e. Bcl-2 and Apaf1) and autophagy-related pathways (i.e., AMPK/mTOR and Bmf/Beclin-1).26,27 Furthermore, p53 targets the expression of DRAM and can stimulate both autophagy and apoptosis.28 The cross-talk between apoptosis and autophagy is critical to the cell fate, but complicated by their contradictory roles under some conditions. Both autophagy and apoptosis as partners affect each other.
Autophagy May be an Adaptive Stress Response Prior to Apoptotic Cell Death
Basal level of autophagy ensures the physiological turnover of aged and/or damaged organelles. Autophagy maintains cell survival subsequent to stressful factors. The massive accumulation of autophagic vacuoles may stand for either an ultimate attempt for cells to survive by adapting to stress or an alternative pathway of cell death. An increased formation of autophagosomes is often coincident in cells that are dying, which represents a possibility that the adaptive mechanism fails and cells are dying under crucial conditions.29 Autophagic response is earlier than apoptotic cell death, placing autophagy upstream of apoptosis. An excess activation of autophagy may contribute to apoptotic cell death through unchecked degradative processes. Autophagy involves the apoptotic process as reflected by morphological and biochemical features.30 Autophagy participates in ATP-dependent events and shows certain morphological changes such as membrane asymmetry and blebbing. Autophagic cell death also is similar to apoptosis in their final products as small sealed membrane vesicles. However, autophagic cell death follows an increase in autophagic vesicles such as autophagosomes and autophagolysosomes.31,32 Only partial chromatin condensation is found in autophagy-mediated cell death, but DNA fragmentation is identified in apoptotic cell death. Because the distinction between autophagy and apoptosis remains to be defined, 2 processes may occur simultaneously in the same cell type. Both autophagy and apoptosis may act independently in parallel pathways, or one may influence the other.
Autophagy May Enable Apoptosis
The autophagic activation beyond a certain threshold may incur the collapse of cellular function, resulting in autophagic cell death directly or the execution of apoptotic cell death via common regulators such as Bcl-2 family proteins.30,33 The genetic manipulation of autophagic pathway has suggested that the autophagy may be a protagonist of apoptosis. In a toxicological model of cigarette smoke extract (CSE) exposure to epithelial cells, the cells die of the apoptosis-extrinsic pathway that involves the activation of Fas-dependent death-inducing signaling complex (DISC) and downstream activation of caspases −8, −9, and −3.9,10 The CSE exposure increases autophagosome formation in epithelial cells and cell processing of LC3B-I to LC3B-II.9,10 Knockdown of autophagy proteins Beclin-1 or LC3B could inhibit CSE-induced apoptosis, suggesting that an enhanced autophagy is associated with apoptotic death in epithelial cells. LC3B is a regulator of apoptosis-extrinsic pathway engaged with the Fas complex. CSE exposure induces a rapid dissociation of LC3B from Fas and the activation of apoptosis signaling.9,10 Perhaps, the nature of foreign substrate (e.g. tar) causes a toxic autophagy and alters the functionality of the autophagic response, which may differ from starvation-induced physiological autophagy. In addition, p53-dependent autophagy through upregulation of DRAM is coincidental with upregulation of apoptosis.34-36 Interestingly, DRAM/mitophagy signaling mediates apoptosis only in mild hepatosteatosis, whereas p53/BAX pathway induces apoptosis mainly in severe hepatosteatosis.37 TNFα can induce autophagy in trophoblasts leading to the activation of apoptosis-intrinsic pathway. Knockdown of Atg5 prevents TNFα-dependent activation of proapoptotic caspases.38 Autophagy regulates human neutrophil apoptosis and mediates the early pro-apoptotic effect of TNFα in neutrophils.39 Another study demonstrated that the deletion of Atg5 was also shown to protect cells from pro-death environmental stimuli. However, this resistance may result from compensatory activation of chaperone-mediated autophagy, rather than inhibition of macroautophagy as such.40 The deprivation of growth factors activates autophagy followed by apoptotic cell death. Autophagy precedes apoptosis through caspase-mediated cleavage that abrogates the autophagic function of Beclin-1 as well as generates a Beclin-1-C fragment. The purified Beclin-1-C fragments can promote a release of cytochrome c and HtrA2/Omi from mitochondria. Caspase-dependent generation of Beclin-1-C creates an amplifying loop and further stimulates apoptosis subsequent to growth factor withdrawal.41 Of note, the genetic knockdown of one autophagy-related factor cannot establish whether autophagy is protective or not in any context, since a down-regulation of the target may arouse other signaling pathways that are independent of autophagy, or compensatory activation of different types of autophagy.
Autophagy Can Antagonize Apoptosis
Autophagy and apoptosis are 2 distinct processes. Autophagy acts to create a cellular milieu in which survival is favored. Actually, autophagy triggers the pro-survival mechanism. Thus, autophagy counteracts apoptotic cell death via cell survival pathway. Mechanisms mediating the counter-regulation of apoptosis through autophagic pathway are still under investigation. Autophagy can attenuate apoptosis by (i) removing damaged debris or denatured subcellular constituents. Autophagy selectively eliminates damaged organelles and dangerous pathogens that are proapoptotic factors. Degradation of damaged mitochondria impedes apoptotic pathways by preventing MOMP and the subsequent release of pro-apoptotic molecules such as cytochrome c and Smac/Diablo;42,43 (ii) maintaining genomic integrity in the face of variously disturbing conditions such as metabolic stress, drug toxicity or radiation damage.44-46 Autophagic process or mitophagy can scavenge depolarized mitochondria that are a source for genotoxic ROS.43 The absence of autophagy causes DNA damage, gene amplification and chromosomal abnormalities following metabolic stress in tumor cells.45,47 The autophagy defect synergized with altered apoptotic activity may facilitate tumor malignant differentiation, which results in a more aggressive cancer cell phenotype and poor prognosis of HCC;48,49 (iii) catabolizing cellular organelles and macromolecules to provide a source of nutrients and energy for the starved cell. Autophagy is a mechanism to maintain cellular energy balance and preserve cellular function. The limited self-eating can provide cells with metabolic substrates to meet their energetic demands as shown during periods of starvation in adult mice and in the feeding adaptation period of neonatal mice;50-52 (iv) limiting ER stress through the degradation of unfolded protein aggregates. ER stress caused by disturbances in the structure and function of the ER incurs the accumulation of misfolded proteins. Autophagy can recycle protein aggregates and misfolded proteins to maintain ER function. Therefore, autophagy is able to suppress the ER stress response and subsequent apoptosis.53-57 Particularly, autophagy promotes survival only in apoptosis-competent cells. When the ER stress is prolonged in Bax/Bak−/− cells that are defective in apoptosis, autophagy is associated with increased necrotic cell death;58,59 (v) facilitating cell growth and proliferation. An inhibition of autophagy in human pancreatic tumor cells, leukemia cells, and malignant glioma cells can enhance the death response of these cells to anticancer therapy.60-62 Likewise, the inhibition of autophagy can increase the antineoplastic potency of the histone deacetylase inhibitor SAHA in imatinib-resistant primary CML cells and the antiangiogenic effects of kringle 5 in endothelial cells, resulting in apoptotic cell death.63,64 Furthermore, autophagy can protect non-transformed epithelial cells from anoikis. The loss of matrix attachment induces apoptotic cell death.65,66 Collectively, autophagy antagonizes apoptosis and promotes cell survival via a series of response to damaged organelles, ER stress, DNA stability, or loss of nutrient and growth factor signaling pathways. Remarkably, autophagy can enable and/or antagonize apoptosis. Contrary effects of autophagy are regulated by a complex network of signal transducers, most of which participate in non-autophagic signaling cascades as well.30,67,68
Cross-Talk Mechanism Between Autophagy and Apoptosis
Autophagy and apoptosis are 2 interconnected mechanisms subsequent to cellular stress. The molecular interplay between 2 mechanisms is not fully understood. It is well known that autophagy serves a cytoprotective role under physiological condition. The cytoprotective function of autophagy involves negative modulation of apoptosis and vice versa.25,69 Cross-talks between autophagy and apoptosis have been demonstrated by recent advances, which are manifested by regulatory genes that are shared with common pathways. These regulatory genes include p53, Atg5, Bcl-2, and so on. Stimuli that lead to the activation or suppression of these genes will affect cell fate via particular signaling pathway. Mechanisms of the cross-talk between autophagy and apoptosis include (i) autophagic degradation of active caspases (Fig. 2). By employing advantage of Bax(−/−) Hct116 cells that are TRAIL-resistant, a recent study reveals the expression pattern and subcellular localization of active caspase-8 in TRAIL-mediated autophagy and its role in the autophagy-to-apoptosis shift upon autophagy inhibition.70 The TRAIL-mediated autophagic activity counter-balances the TRAIL-mediated apoptotic response by the continuous sequestration of the large caspase-8 subunit in autophagosomes and its subsequent elimination in lysosomes.71 Caspase-8 can also cleaves autophagic protein Atg3, leading to Atg3 degradation.72 This event is inhibited by a pan-caspase inhibitor (Z-VAD-fmk) or a caspase-8-specific inhibitor (zIETD-fmk).73 Atg3 cleavage plays a regulatory role during TNFα-induced apoptosis. Atg3 becomes a novel link between apoptosis and autophagy. Other caspase-dependent events have also implicated in antiautophagy or proautophagy reactions. Caspase-3 cleaves Atg4D into DeltaN63 Atg4D that displays an increased activity against the Atg8. Caspase-cleaved Atg4D triggers mitochondria-targeted apoptosis.74 Proapoptotic caspase-3 can down-regulate autophagy by interacting with Beclin-1. Caspase processing of Atg4D is pro-autophagy, whereas caspase processing of Beclin-1 is anti-autophagy. Caspase-mediated cleavage inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria.41 Inhibition of the cytoprotective autophagy shifts the cells toward apoptosis.70 Apoptotic signaling, in turn, serves to inhibit autophagy.69 Autophagic degradation of active caspases is a crosstalk mechanism between autophagy and apoptosis; (ii) interactions between Beclin-1 and Bcl-2/Bcl-xL. Mammalian Beclin-1 as a key protein in autophagic pathway is a novel BH3-only member of the Bcl-2 family. A deletion of mouse Beclin-1 alleles leads to early death during embryogenesis.75 Beclin-1 functions as a platform by binding to PI3KC3/Vps34, UVRAG, AMBRA-1 and/or Barkor to assemble the PI3KC3 complex during initiation of autophagosome formation.76-80 Beclin-1 utilizes the BH3 domain to mediate its interaction with the anti-apoptotic proteins Bcl-2 and Bcl-xL.81,82 Binding of Bcl-2 family proteins to Beclin-1 inhibits autophagy by preventing the association of Beclin-1 with the class III PI3K complex.82,83 Both autophagy and apoptosis are regulated by Bcl-2 proteins.25 Beclin-1 may have a role in the convergence between autophagy and apoptosis in response to specific signals; (iii) dual function of certain Atg proteins. Some Atg proteins play dual roles in the regulation of autophagy. The autophagic protein Atg5, well known in autophagy, contributes to autophagic cell death. Atg5 can also interact with FADD to stimulate extrinsic apoptosis pathways and induces apoptotic death that can be blocked by pan caspase inhibitor Z-VAD-fmk.84 Atg12, the binding partner for Atg5, is required for autophagosomal elongation. Atg12 also is a positive mediator associated with anti-apoptotic Bcl-2 family members to promote mitochondrial apoptosis.85 Atg12 may bind and inactivate Bcl-2 proteins (i.e. Bcl-2 and Mcl-1) through its unique BH3-like motif, and thereby act as a proapoptotic regulator as well; (iv) calpain-mediated cleavage of Atg protein switches autophagy to apoptosis. Atg5 is required for the formation of autophagosomes and regulates autophagy. Atg5 can enhance susceptibility toward apoptotic stimuli as well. Apoptosis was associated with calpain-mediated Atg5 cleavage, which is independent of the cell type and the apoptotic stimulus. The calpain-dependent Atg5 cleavage generates an Atg5 truncation product. The truncated Atg5 is translocated from the cytosol to mitochondria, which inhibits the anti-apoptotic Bcl-xL and provokes cytochrome c release and caspase activation.86 Moreover, following calpain-mediated Atg5 cleavage, doxorubicin treatment shifts protective autophagy induced by deficiency of sphingosine-1-phosphate phosphohydrolase 1 into apoptosis in human breast cancer cells.87 Calpain-mediated pathway also dominates cisplatin-induced apoptosis in human lung adenocarcinoma cells. Inactivating calpain or silencing Bid delays cytochrome c release, caspase-3 activation and subsequent cell death;88 (v) role of c-FLIP. The protein c-FLIP (cellular FLICE-like inhibitory protein) can suppress death receptor-induced caspase 8 activation. Thus, the c-FLIP is an apoptosis-inhibitor of the extrinsic apoptotic pathway. Recently, c-FLIP is also found to involve an inhibition of autophagy.89 c-FLIP(L)-deficient T cells exhibits an enhanced autophagy. c-FLIP(L) is an important player to regulate apoptosis and autophagy in T lymphocytes. The c-FLIP can act to prevent the binding of Atg3 to LC3. Cellular and viral FLIPs limit the Atg3-mediated step of LC3 conjugation to regulate autophagosome biogenesis.36 Anti-apoptotic c-FLIP is a negative regulator of autophagy. Furthermore, the FLIP-derived short peptides induce growth suppression and cell death. FLIP-mediated autophagy controls the regulation of cell death;36 (vi) p53 co-regulates autophagy and apoptosis. The p53 regulates the expression of Bcl-2 proteins (e.g., Bax, Bid) and apoptosis-related gene targets (e.g. Apaf1, HSP90 and GRP75).26 p53 thus is a regulator of cell cycle progression and apoptosis. An activation of p53 inhibits mTOR activity and regulates its downstream target such as autophagy. The mechanism by which p53 regulates mTOR involves the upregulation of AMP kinase.27 p53 and mTOR signaling machineries can coordinately regulate cell growth, proliferation, and death. Knockdown or pharmacological inhibition of p53 can induce autophagy. Different autophagy-inducers promote the proteasome-mediated degradation of p53 through the HDM2/E3 ubiquitin ligase pathway.90 The deacetylation of p53 induces autophagy by reducing the interaction of Beclin-1 and Bcl-2. The nuclear p53 targeted DRAM pathway mediates both autophagy and apoptosis.28 Definitive mechanisms that control the cross-talk between autophagy and apoptosis remain unknown. Analysis of any single regulatory component (via siRNA silencing) for its potential to cross-regulate autophagy and/or apoptosis is hard to answer how these pathways in response to specific stimuli. Alternative or compensatory mechanisms must be considered. Thus, an integrative approach is needed to understand how the entire molecular machinery of apoptosis and autophagy are coordinated to influence cell fate.
Figure 2.
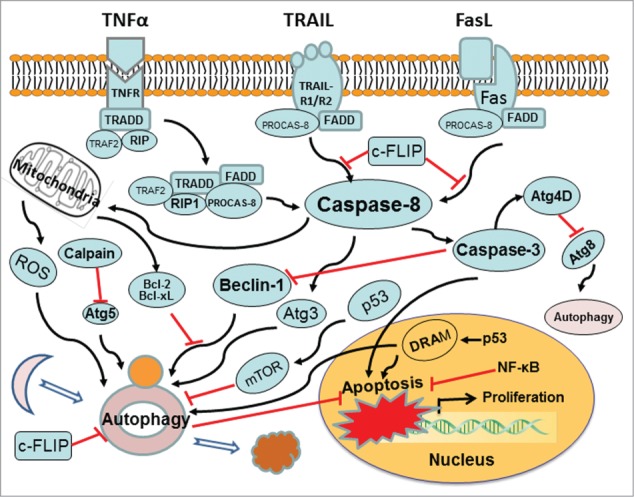
Cross-talk mechanism between autophagy and apoptosis. Molecular signaling pathways affect both autophagy and apoptosis. Atg levels are mediated by caspase, calpain, c-FLIP, Bcl-2, p53, and so forth. Red arrows ending in perpendicular lines denote inhibition of the reaction.
Autophagy and Aged Liver
Autophagic activity is declined in the aging process, which is reflected by a progressive accumulation of non-functional cellular components and a weak role of housekeeping mechanisms. The aging-related disadvantage can be improved by the condition such as caloric restriction that activates autophagy. Indeed, the stimulation of autophagy significantly enhances the life span of multiple organisms, e.g., yeast, C elegans, mice and primates.91–93 The relationship between autophagy decline and hallmarks of aging has been known for a long time and has been carefully studied in livers. The autofluorescent pigment lipofuscin is the oldest and simplest biomarker of declining autophagy and represents undigested material inside hepatocytes. In livers of aging humans and mice, relative levels of many autophagy-related genes and their proteins (i.e., Atg7, Atg5, Atg4B, Beclin-1 and Lamp-2) are dramatically reduced.94,95 Aged liver tissues have a diminished activity of autophagy. Autophagy has been associated with a growing number of pathological conditions. There is an increased liver injury in aged mice compared to young mice, which is likely due to the elevated apoptosis as demonstrated by an enhancement of capase-3 activity and TUNEL positive cells. For example, Lamp-2A expression is downregulated in aged livers, which results in decreased chaperone-mediated autophagy as well as impaired liver function. When Lamp-2A level of the mouse liver is restored using an inducible-transgenic technique, the apoptosis is significantly decreased in mouse livers and the detoxification rate of xenobiotics is increased in the aged animal liver.95 Enforcement of autophagy in aging liver improves hepatic function. Moreover, ischemia/reperfusion-induced liver injury is much severe in aged mice. After autophagy activity is increased through an overexpression of either Atg4B or Beclin-1, the onset of mitochondrial permeability transition is blocked and ischemia/reperfusion-induced liver injury is significantly alleviated.94 Taken together, the failure of autophagy with aging may be caused by multiple factors. Energy metabolism is an important initiator (Fig. 3). Autophagic dysregulation further reinforces the situation. Degradation of autophagosomes relies on fusion with lysosomes that are key organelles in the aging process due to their involvement in autophagy and cell homeostasis. Accumulation of defective mitochondria is critical in the progression of aging. Inefficient removal of damaged mitochondria by lysosomes represents a major issue in the aging process. Deficient or impaired autophagy contributes to aging. Potential mechanisms that result in a failed autophagy with aging include (a) energy compromise. With aging, there is a decline in energy demand and production, which thus result in energetic compromise of the aging cells and the age-dependent decline in autophagy;96 (b) insufficient activation of autophagy. The autophagic response needs a full activation or enhancement by hormone (i.e. glucagon) stimulation.97 The stimulatory effect of glucagon on autophagy is decreased in aged animals.98 The glucagon can maintain negative signaling through the insulin receptor. The insulin signaling is activator of mTOR, a known repressor of autophagy. The formation of autophagosomes in old cells may be due to malfunction of glucagon pathway; (c) negative regulation by insulin receptor. Aging can potentiate the inhibitory signaling of insulin receptor. Insulin/IGF-1 signaling is up-regulated via the insulin receptor tyrosine kinase in aging cells. The insulin/IGF-1 pathway activates mTOR and inhibits autophagy;99-101 (d) mitophagy decline. The turnover of mitochondria via mitophagy is decreased in aging cells. If mitophagy cannot catch up with the demand for the clearance of old and damaged mitochondria, high levels of ROS would damage cell molecules including proteins, membrane lipids, and nucleus DNA.102-104 A decline of autophagy thus is responsible for cell aging via insufficient turnover of damaged mitochondria; (e) fusion deficiency between autophagosomes and lysosomes. Autophagic vacuoles are accumulated in the liver with age. This may result from fusion problem between the autophagosomes and the lysosomes. Autophagosomes are unable to fuse with lysosomes and further to degrade the sequestered content;105,106 (f) weak function of lysosome enzymes. Autophagic vacuoles such as lipofuscin-loaded lysosomes are accumulated in the liver with age.107,108 This may result from inactive or weak function of lysosome enzymes with aging.
Figure 3.
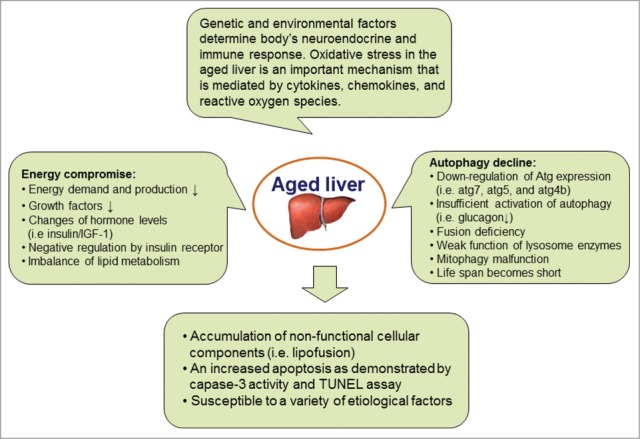
Pathophysiological role of autophagy in aged livers. The aged liver can be estimated by multiple biomarkers, such as autofluorescent pigment lipofuscin in hepatocytes, an enhancement of hepatocyte apoptosis and the increased susceptibility to different etiological factors. Oxidative stress modulated by genetic and environmental factors is an important mechanism via signaling pathway mediated by cytokines, chemokines, and reactive oxygen species. Change in energy metabolism that is regulated by autophagy is an essential initiator.
Modulation of Autophagy Affects the Progression of Liver Injury
Cells sequester and degrade their own intracellular contents via autophagy. Owing to high biosynthetic activity and role in protein and carbohydrate storage, hepatocytes may be particularly dependent on basal autophagy. In the liver, autophagy suppresses protein aggregate, lipid accumulation, oxidative stress, chronic cell death, and inflammation.5 Autophagy is targeted for drug-induced liver injury as demonstrated in acetaminophen (APAP) overdose (Fig. 4). APAP induces necrotic liver injury. Rapamycin treatment can inhibit APAP-induced necrosis in cultured primary hepatocytes and in mouse livers respectively. The mechanism for rapamycin to suppress necrosis is likely through the induction of autophagic activity, not via APAP metabolism.8 Autophagy removes damaged mitochondria, termed as mitophagy. The mitophagy can reduce mitochondria-derived ROS formation and the release of pro-cell death factors from mitochondria. Really, APAP-induced ROS are decreased by rapamycin but exacerbated by chloroquine treatment.8 The post-treatment of rapamycin may be a therapeutic intervention for APAP-overdose patients. Efavirenz also induces hepatotoxicity via mitochondrial damage. The moderate level of efavirenz stimulates autophagy as a cell survival mechanism, but high concentrations block the autophagic flux and result in autophagic stress and cell damage. Pharmacological inhibition of autophagy neutralizes the effect of efavirenz on cell survival/proliferation and triggers apoptosis.109 Autophagy can selectively degrade lipid droplets via a process entitled lipophagy.110 Cellular lipids are surrounded by a phospholipid monolayer in a lipid droplet and generally are stored as triglycerides (TG). Autophagy can regulate hepatic TG contents as demonstrated by (i) the genetic manipulation of autophagic pathway. siRNA knockdown of Atg5 enhances TG contents in fatty acid-treated hepatocytes. Liver-specific Atg7 knockout results in an increased hepatic TG and cholesterol contents in high-fat fed mice or starved mice;110 (ii) pharmacological inhibition of autophagic pathway. Autophagy inhibitor 3-methyladenine targets the mammalian PI3-kinase vps34 and increases TG contents in hepatocytes cultured with an unsaturated oleic acid.110 Lysosomal inhibitor chloroquine raises the lysosomal pH that represses both fusion of autophagosome with lysosome and lysosomal degradation, which also elevates TG contents in hepatocytes treated with fatty acids.111,112 Several potential chemicals have been used as a pharmacological approach to modulate hepatocyte autophagy in mouse liver. Moreover, many small molecules that mediate autophagy have been conducted to benefit patients with liver diseases. Pharmacological modulation of autophagy in the liver can be an effective strategy for improving alcoholic and nonalcoholic fatty liver diseases.113 Fatty acid-induced lipotoxicity is also regulated by autophagy and apoptosis. Lipotoxicity refers to cellular toxicity in the presence of excessive free fatty acids. Unsaturated oleic acid (OA) can promote the formation of TG-enriched lipid droplets and induces autophagy with minimal effects on apoptosis. Saturated palmitic acid (PA), on the other hand, promotes lipid droplet formation poorly and suppresses autophagy but significantly stimulates apoptosis. OA-induced autophagy is independent of mammalian target of rapamycin but dependent on ROS and PI3-kinase. PA-induced apoptosis inhibits autophagy by inducing caspase-dependent Beclin-1 cleavage, indicating cross-talk between apoptosis and autophagy. There are differential effects of unsaturated and saturated fatty acid-induced lipotoxicity through the regulation of autophagy and apoptosis.111 Autophagy exerts its role in lipid breakdown by removing lipid droplets. A pharmacological modulation of autophagic pathway may be an effective approach to alleviate fatty liver injury. Alcohol consumption causes a series of hepatic changes including mitochondrial damage, oxidative stress, accumulation of fatty acids and so on. Most of these events can be regulated by autophagy. Recent studies investigate the role and mechanisms of autophagy in alcoholic liver disease (ALD). Resultant data indicate the possibility that ALD can be treated through modulating autophagy.113,114 Autophagic flux can be suppressed by bile acids during cholestasis. In the development of cholestatic liver injury, high concentrations of cytotoxic bile acids (BAs) induce liver cell death, inflammatory response, and perhaps tumorigenesis. Autophagic flux is impaired in livers of farnesoid X receptor (FXR) knockout mice. BAs suppress autophagic flux in hepatocytes by impairing autophagosomal-lysosomal fusion, which may be implicated in liver tumorigenesis of FXR knockout mice.115 Liver tissues from patients with primary biliary cirrhosis show an increased LC3 expression and p62 positive aggregates in biliary epithelial cells of small bile ducts.116,117 Moreover, inclusion bodies or MDBs have been found in patients with primary biliary cirrhosis and primary sclerosing cholangitis respectively.118-120 Elevated levels of cytotoxic BAs play a critical role in MDB formation. Both cholic acid feeding and common bile duct ligation result in the formation of MDBs in drug-primed mice, suggesting that cytotoxic BAs may drive the formation of MDBs in cholestatic liver disease.118,121,122 Indeed, liver MDBs in patients with primary biliary cirrhosis are found in acinar zone 1, where bile acids levels are highest.119 Autophagy as reflected by LC3/p62 involves the pathogenesis of cholestasis.123 MDB formation may be resulted from a blockage of autophagic flux that is inhibited by cytotoxic BAs. In addition, the nutrient sensing TOR pathway regulates translation and protein synthesis, which modulates autophagy and aging processes.124 Therefore, rapamycin and other inhibitors of mTOR may be beneficial to the prevention of liver injury. Autophagy in response to rapamycin or nutrient limitation can be attenuated in multiple cell lines subsequent to the treatment of resveratrol, a plant-derived polyphenol. The mechanism by which resveratrol suppresses autophagy is due to inhibition of p70 S6 kinase (S6K1) activity.125 An interaction between resveratrol and S6K1 may be new targets to develop therapeutic drugs for liver disease.
Figure 4.
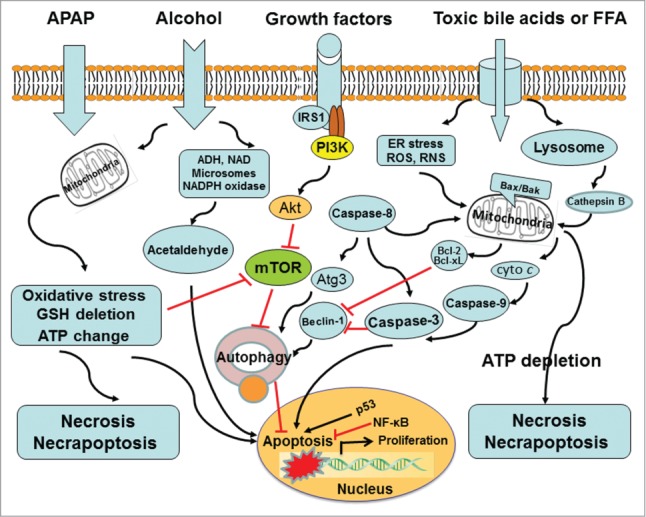
Modulation of autophagy affects the progression of liver injury. Autophagy promotes cell survival via a series of response to damaged organelles, ER stress, DNA stability, or loss of nutrient and growth factor signaling pathways. Of note, autophagy can enable and/or antagonize apoptosis. The coordinate regulation of these opposite effects of autophagy relies upon a complex network of signal transducers, most of which also participate in non-autophagic signaling cascades.
Dual-Effect of Hepatic Autophagy
Dual role of autophagy is obviously observed during carcinogenesis. Autophagy can protect against cancer by removing damaged organelles, increasing protein catabolism, promoting cell differentiation, and even inducing cell death of cancerous cells.125 However, autophagy can also contribute to the growth of cancer through nutrient recycle and cellular energy production.126 Dual effects of autophagy in both tumor suppression and tumor cell survival can cause completely different outcomes. When autophagic mechanism is used to design anticarcinogenic therapeutics, different strategies should be carried out. The first strategy is to induce autophagy and enhance its tumor suppression attributes. The second strategy is to inhibit autophagy and then induce apoptosis.127 The functionality of autophagy is closely related to liver pathophysiology as well. A fast development of steatosis detected in mouse liver with defective autophagy strongly supports the contribution of autophagy in the pathogenesis of fatty liver disease.110 A compromise of hepatic autophagy is the basis for the accumulation of lipid droplets (LD) upon exposure to high concentration of ethanol.114 Furthermore, pharmacological up-regulation of autophagy reduces hepatotoxicity and steatosis in alcohol-treated mouse livers.113,114 The liver responds to stressful conditions through global activation of autophagy, selectively to eliminate damaged mitochondria and accumulated lipid droplets. Especially, upregulation of hepatic autophagy may be specific for lipophagy in some instances. The autophagy activation as a protective mechanism can target lipid droplets, which is a primary defense against alcohol-induced hepatotoxicity and steatosis.114 Autophagy activation is thus considered as a generalized treatment against steatosis, but the up-regulation of autophagy in hepatic stellate cells favors their activation and consequently initiate liver fibrosis128 (Fig. 5). On the contrary, an inhibition of autophagy can lead to the accumulation of lipid droplets in hepatic stellate cells, resulting in apoptosis of hepatic stellate cells.129 Future efforts should focus on how selectively to modulate cell-specific autophagy for therapeutic purpose. Interestingly, an ability of autophagy-mobilized hepatic lipids is also utilized by viruses to favor their replication. Usually, autophagy is an efficient mechanism in the defense against the most viral infection, but upregulation of autophagy can favor replication of some viruses, such as dengue virus. Autophagy-activated by dengue virus mediates LD breakdown and release of FFA necessary to maintain the high levels of intracellular ATPs that are required for the replication of dengue viruses.130 Further studies are needed to determine which subset of hepatotropic virus makes use of lipophagy for their replication and whether the blockage of autophagic response can be a selective way to prevent virogenesis, but not hinder normal metabolism of hepatocytes. Autophagy promotes viral RNA replication of hepatitis C virus (HCV). There is the transient interaction of Atg5 with NS4B and NS5B. HCV-induced autophagic membrane can be used as a compartment for the replication of viral RNA. NS5A, NS5B, and nascent viral RNA are co-localized with the autophagosome. The autophagy machinery is required to initiate HCV replication. Autophagy proteins (i.e., Beclin-1, Atg4B, Atg5, and Atg12) are proviral factors for translation of incoming HCV RNA. In infected hepatocytes HCV infection transcriptionally up-regulates Beclin-1, which forms complex with Vps34, the class III PI3-kinase, to trigger autophagic response. HCV infection enhances phospho-mTOR expression and its downstream target 4EBP1 activation, which in turn may promote hepatocyte growth.131 HCV infection up-regulates expression of Drp1 and its mitochondrial receptor. HCV induces the phosphorylation of Drp1 (Ser616) and causes its subsequent translocation to the mitochondria. HCV boosts Parkin-mediated elimination of damaged mitochondria. Silencing Drp1 or Parkin significantly amplifies apoptotic signaling as demonstrated by cytochrome c release, caspase 3 activity, and cleavage of poly(ADP-ribose) polymerase. HCV-induced mitochondrial fission and mitophagy serve to attenuate apoptosis and are potential mechanisms for persistent HCV infection.132 Autophagy regulates the assembly of infectious virions. HCV exploits autophagic machinery in favor of virus growth and survival in host cells.133,134 In addition, HBV- or HBx-induced autophagosome formation is accompanied by decreased degradation of LC3 and SQSTM1/p62. HBx impairs lysosomal acidification leading to a diminishment of lysosomal degradative capacity. Moreover, there is an upregulation of SQSTM1 and immature lysosomal hydrolase CTSD in human liver with HBV-associated liver cancer. HBx protein inhibits autophagic degradation by impairing lysosomal maturation, and this may be crucial to the development of HBV-induced HCC.135 Autophagy suppresses tumorigenesis of HBV-associated HCC through degradation of microRNA-224 as well.136 Autophagy-deficient mice derived from genetic modification develop multiple liver tumors. The mosaic deletion of Atg5 and liver-specific Atg7(−)/(−) genes causes a high rate of benign adenomas in mouse liver. These tumor cells originate autophagy-deficient hepatocytes, showing mitochondrial swelling, p62 accumulation, oxidative stress and genomic damage responses. Autophagy through a cell-intrinsic mechanism is an important mediator for the suppression of tumorigenesis.49 Furthermore, autophagy defect is associated with a poor prognosis of HCC. The expression of autophagic Beclin-1 in HCC tissue is down-regulated as compared with adjacent non-tumor tissues. There is a significant correlation between Beclin-1 expression and tumor differentiation in Bcl-xL(+) but not in Bcl-xL(−) HCC patients. The expression of Beclin-1 is also positively correlated with disease-free survival and overall survival in the Bcl-xL(+) group.48 Autophagy has roles in various cellular functions, but sometimes modulates them in 2 opposite directions. An enhancement of autophagy promotes hepatocyte survival during liver injury, whereas the activating effect of autophagy in hepatic stellate cells favors fibrogenesis. Autophagy participates in hepatitis virus infection and replication, yet its role still is inconsistent between HCV and HBV. Although defects or suppression of autophagy is identified in HCC, a dual effect of autophagy for cancer cells has to be considered while novel therapeutic targets are chosen.
Figure 5.
Dual-effect of hepatic autophagy. The dual roles of autophagy are obviously observed in cancer development and liver fibrosis. Autophagy can serve either to promote cell/tumor survival at certain stages, or to stimulate its elimination at other stages. Autophagy activation is beneficial for hepatocyte proliferation and liver repair, but the upregulation of autophagy in hepatic stellate cells favors their activation and consequently initiates fibrogenesis.
Summary and Future Study
Autophagy as a self-digestive program is a lysosomal degradation process that regulates organelle and protein homeostasis and serves as a cell survival mechanism under a variety of stress conditions. It provides an essential means of refreshing and remodeling cells. As such, it is required for normal development, metabolic homeostasis, and prevention of degenerative disease. Once stressful stimuli occur, autophagy may contribute to relevant resiliency. Thus, both activation and inhibition of autophagy hold promise for the improvement of related clinical diseases. Autophagy modulates the progression of liver injury. Cross-talks between autophagy and apoptosis play an important role during pathogenesis of liver disease. According to the known evidence, a regulation of autophagic pathway can be an effective approach to deal with liver disease. However, there are a number of standing questions that deserve immediate attention. How to target the autophagic machinery selectively? How to overcome dual-effect of hepatic autophagy? What determines the threshold of autophagy to switch from a survival mechanism to autophagic cell death? In a general context, is up-regulation of autophagy protective against all types of liver injury? These questions need time to get answers. There is ample evidence that liver-specific targeting of autophagic process can be a novel therapeutic intervention against liver disease.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med 2013; 368(7):651-62; Epub 2013/02/15; PMID:23406030; http://dx.doi.org/ 10.1056/NEJMra1205406 [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132(1):27-42; Epub 2008/01/15; PMID:18191218; http://dx.doi.org/ 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Revi Drug Discov 2012; 11(9):709-30; Epub 2012/09/01; PMID:22935804; http://dx.doi.org/ 10.1038/nrd3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K. Molecular mechanisms of hepatic apoptosis. Cell Death Dis 2014; 5:e996; Epub 2014/01/18; PMID:24434519; http://dx.doi.org/ 10.1038/cddis.2013.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabinowitz JD, White E. Autophagy and metabolism. Science 2010; 330(6009):1344-8; Epub 2010/12/04; PMID:21127245; http://dx.doi.org/ 10.1126/science.1193497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu JJ, Quijano C, Wang J, Finkel T. Metabolism meets autophagy. Cell Cycle 2010; 9(24):4780-1; Epub 2010/12/15; PMID:21150322; http://dx.doi.org/ 10.4161/cc.9.24.14273 [DOI] [PubMed] [Google Scholar]
- 7.Bortoluci KR, Medzhitov R. Control of infection by pyroptosis and autophagy: role of TLR and NLR. Cell Mol Life Sci 2010; 67(10):1643-51; Epub 2010/03/17; PMID:20229126; http://dx.doi.org/ 10.1007/s00018-010-0335-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni HM, Bockus A, Boggess N, Jaeschke H, Ding WX. Activation of autophagy protects against acetaminophen-induced hepatotoxicity. Hepatology 2012; 55(1):222-32; Epub 2011/09/21; PMID:21932416; http://dx.doi.org/ 10.1002/hep.24690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen ZH, Lam HC, Jin Y, Kim HP, Cao J, Lee SJ, Ifedigbo E, Parameswaran H, Ryter SW, Choi AM. Autophagy protein microtubule-associated protein 1 light chain-3B (LC3B) activates extrinsic apoptosis during cigarette smoke-induced emphysema. Proc Natl Acad Sci U S A 2010; 107(44):18880-5; Epub 2010/10/20; PMID:20956295; http://dx.doi.org/ 10.1073/pnas.1005574107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB, Dhir R, Landreneau RJ, Schuchert MJ, Yousem SA, et al.. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PloS One 2008; 3(10):e3316; Epub 2008/10/03; PMID:18830406; http://dx.doi.org/ 10.1371/journal.pone.0003316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta 2013; 1833(12):3448-59; Epub 2013/06/19; PMID:23770045; http://dx.doi.org/ 10.1016/j.bbamcr.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 12.Shi S, Wang Q, Xu J, Jang JH, Padilla MT, Nyunoya T, Xing C, Zhang L, Lin Y. Synergistic anticancer effect of cisplatin and Chal-24 combination through IAP and c-FLIPL degradation, Ripoptosome formation and autophagy-mediated apoptosis. Oncotarget 2015; 6(3):1640-51; Epub 2015/02/16; PMID:25682199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bursch W. The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ 2001; 8(6):569-81; Epub 2001/09/06; PMID:11536007; http://dx.doi.org/ 10.1038/sj.cdd.4400852 [DOI] [PubMed] [Google Scholar]
- 14.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 2004; 6(12):1221-8; Epub 2004/11/24; PMID:15558033; http://dx.doi.org/ 10.1038/ncb1192 [DOI] [PubMed] [Google Scholar]
- 15.Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 2004; 304(5676):1500-2; Epub 2004/05/08; PMID:15131264; http://dx.doi.org/ 10.1126/science.1096645 [DOI] [PubMed] [Google Scholar]
- 16.Wang WJ, Wang Y, Chen HZ, Xing YZ, Li FW, Zhang Q, Zhou B, Zhang HK, Zhang J, Bian XL, et al.. Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway. Nat Chem Biol 2014; 10(2):133-40; Epub 2013/12/10; PMID:24316735; http://dx.doi.org/ 10.1038/nchembio.1406 [DOI] [PubMed] [Google Scholar]
- 17.Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS, Cheng AL, Chen PJ, Chen KF. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis 2013; 4:e485; Epub 2013/02/09; PMID:23392173; http://dx.doi.org/ 10.1038/cddis.2013.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung KH, Noh JH, Kim JK, Eun JW, Bae HJ, Chang YG, Kim MG, Park WS, Lee JY, Lee SY, et al.. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology 2012; 56(2):644-57; Epub 2012/03/07; PMID:22392728; http://dx.doi.org/ 10.1002/hep.25699 [DOI] [PubMed] [Google Scholar]
- 19.Schwartz LM, Smith SW, Jones ME, Osborne BA. Do all programmed cell deaths occur via apoptosis? Proc Natl Acad Sci U S A 1993; 90(3):980-4; Epub 1993/02/01; PMID:8430112; http://dx.doi.org/ 10.1073/pnas.90.3.980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsujimoto Y, Shimizu S. Another way to die: autophagic programmed cell death. Cell Death Differ 2005; 12(Suppl 2):1528-34; Epub 2005/10/26; PMID:16247500; http://dx.doi.org/ 10.1038/sj.cdd.4401777 [DOI] [PubMed] [Google Scholar]
- 21.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22(2):140-9; Epub 2010/01/26; PMID:20097051; http://dx.doi.org/ 10.1016/j.ceb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19(12):5360-72; Epub 2008/10/10; PMID:18843052; http://dx.doi.org/ 10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robert G, Gastaldi C, Puissant A, Hamouda A, Jacquel A, Dufies M, Belhacene N, Colosetti P, Reed JC, Auberger P, et al.. The anti-apoptotic Bcl-B protein inhibits BECN1-dependent autophagic cell death. Autophagy 2012; 8(4):637-49; Epub 2012/04/14; PMID:22498477; http://dx.doi.org/ 10.4161/auto.19084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y, Massen S, Terenzio M, Lang V, Chen-Lindner S, Eils R, Novak I, Dikic I, Hamacher-Brady A, Brady NR. Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. J Biol Chem 2013; 288(2):1099-113; Epub 2012/12/05; PMID:23209295; http://dx.doi.org/ 10.1074/jbc.M112.399345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ 2010; 17(2):268-77; Epub 2009/08/29; PMID:19713971; http://dx.doi.org/ 10.1038/cdd.2009.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo W, Yan L, Yang L, Liu X, E Q, Gao P, Ye X, Liu W, Zuo J. Targeting GRP75 improves HSP90 inhibitor efficacy by enhancing p53-mediated apoptosis in hepatocellular carcinoma. PloS One 2014; 9(1):e85766; Epub 2014/01/28; PMID:24465691; http://dx.doi.org/ 10.1371/journal.pone.0085766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng Z, Zhang H, Levine AJ, Jin S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc Natl Acad Sci U S A 2005; 102(23):8204-9; Epub 2005/06/02; PMID:15928081; http://dx.doi.org/ 10.1073/pnas.0502857102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006; 126(1):121-34; Epub 2006/07/15; PMID:16839881; http://dx.doi.org/ 10.1016/j.cell.2006.05.034 [DOI] [PubMed] [Google Scholar]
- 29.Patschan S, Chen J, Polotskaia A, Mendelev N, Cheng J, Patschan D, Goligorsky MS. Lipid mediators of autophagy in stress-induced premature senescence of endothelial cells. Am J Physiol Heart Circu Physiol 2008; 294(3):H1119-29; Epub 2008/01/22; PMID:18203850; http://dx.doi.org/ 10.1152/ajpheart.00713.2007 [DOI] [PubMed] [Google Scholar]
- 30.Galluzzi L, Vicencio JM, Kepp O, Tasdemir E, Maiuri MC, Kroemer G. To die or not to die: that is the autophagic question. Curr Mol Med 2008; 8(2):78-91; Epub 2008/03/14; PMID:18336289; http://dx.doi.org/ 10.2174/156652408783769616 [DOI] [PubMed] [Google Scholar]
- 31.Bejarano E, Yuste A, Patel B, Stout RF Jr., Spray DC, Cuervo AM. Connexins modulate autophagosome biogenesis. Nat Cell Biol 2014; 16(5):401-14; Epub 2014/04/08; PMID:24705551; http://dx.doi.org/ 10.1038/ncb2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klionsky DJ, Eskelinen EL, Deretic V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes… wait, I'm confused. Autophagy 2014; 10(4):549-51; Epub 2014/03/25; PMID:24657946; http://dx.doi.org/ 10.4161/auto.28448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reimers K, Choi CY, Bucan V, Vogt PM. The Bax Inhibitor-1 (BI-1) family in apoptosis and tumorigenesis. Curr Mol Med 2008; 8(2):148-56; Epub 2008/03/14; PMID:18336295; http://dx.doi.org/ 10.2174/156652408783769562 [DOI] [PubMed] [Google Scholar]
- 34.Takahashi M, Kakudo Y, Takahashi S, Sakamoto Y, Kato S, Ishioka C. Overexpression of DRAM enhances p53-dependent apoptosis. Cancer Med 2013; 2(1):1-10; Epub 2013/10/18; PMID:24133622; http://dx.doi.org/ 10.1002/cam4.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghavami S, Mutawe MM, Sharma P, Yeganeh B, McNeill KD, Klonisch T, Unruh H, Kashani HH, Schaafsma D, Los M, et al.. Mevalonate cascade regulation of airway mesenchymal cell autophagy and apoptosis: a dual role for p53. PloS One 2011; 6(1):e16523; Epub 2011/02/10; PMID:21304979; http://dx.doi.org/ 10.1371/journal.pone.0016523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, Chang H, Zhou FC, Gao SJ, Liang C, et al.. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol 2009; 11(11):1355-62; Epub 2009/10/20; PMID:19838173; http://dx.doi.org/ 10.1038/ncb1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu K, Lou J, Wen T, Yin J, Xu B, Ding W, Wang A, Liu D, Zhang C, Chen D, et al.. Depending on the stage of hepatosteatosis, p53 causes apoptosis primarily through either DRAM-induced autophagy or BAX. Liver Int 2013; 33(10):1566-74; Epub 2013/07/24; PMID:23875779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cha HH, Hwang JR, Kim HY, Choi SJ, Oh SY, Roh CR. Autophagy induced by tumor necrosis factor alpha mediates intrinsic apoptosis in trophoblastic cells. Reprod Sci 2014; 21(5):612-22; Epub 2013/11/08; PMID:24198074; http://dx.doi.org/ 10.1177/1933719113508816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pliyev BK, Menshikov M. Differential effects of the autophagy inhibitors 3-methyladenine and chloroquine on spontaneous and TNF-alpha-induced neutrophil apoptosis. Apoptosis 2012; 17(10):1050-65; Epub 2012/05/29; PMID:22638980; http://dx.doi.org/ 10.1007/s10495-012-0738-x [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Singh R, Massey AC, Kane SS, Kaushik S, Grant T, Xiang Y, Cuervo AM, Czaja MJ. Loss of macroautophagy promotes or prevents fibroblast apoptosis depending on the death stimulus. J Biol Chem 2008; 283(8):4766-77; Epub 2007/12/13; PMID:18073215; http://dx.doi.org/ 10.1074/jbc.M706666200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, Roelandt R, De Rycke R, Verspurten J, Declercq W, et al.. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 2010; 1:e18; Epub 2010/01/01; PMID:21364619; http://dx.doi.org/ 10.1038/cddis.2009.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 2007; 462(2):245-53; Epub 2007/05/04; PMID:17475204; http://dx.doi.org/ 10.1016/j.abb.2007.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Nartiss Y, Steipe B, McQuibban GA, Kim PK. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy 2012; 8(10):1462-76; Epub 2012/08/15; PMID:22889933; http://dx.doi.org/ 10.4161/auto.21211 [DOI] [PubMed] [Google Scholar]
- 44.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 2007; 21(13):1621-35; Epub 2007/07/04; PMID:17606641; http://dx.doi.org/ 10.1101/gad.1565707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie R, Wang F, McKeehan WL, Liu L. Autophagy enhanced by microtubule- and mitochondrion-associated MAP1S suppresses genome instability and hepatocarcinogenesis. Cancer Res 2011; 71(24):7537-46; Epub 2011/11/01; PMID:22037873; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaachouay H, Ohneseit P, Toulany M, Kehlbach R, Multhoff G, Rodemann HP. Autophagy contributes to resistance of tumor cells to ionizing radiation. Radiother Oncol 2011; 99(3):287-92; Epub 2011/07/05; PMID:21722986; http://dx.doi.org/ 10.1016/j.radonc.2011.06.002 [DOI] [PubMed] [Google Scholar]
- 47.Zhang H, Kong X, Kang J, Su J, Li Y, Zhong J, Sun L. Oxidative stress induces parallel autophagy and mitochondria dysfunction in human glioma U251 cells. Toxicol Sci 2009; 110(2):376-88; Epub 2009/05/20; PMID:19451193; http://dx.doi.org/ 10.1093/toxsci/kfp101 [DOI] [PubMed] [Google Scholar]
- 48.Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z, Shi GM, Wang XY, Ke AW, Wu B, et al.. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res 2008; 68(22):9167-75; Epub 2008/11/18; PMID:19010888; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-1573 [DOI] [PubMed] [Google Scholar]
- 49.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 2011; 25(8):795-800; Epub 2011/04/19; PMID:21498569; http://dx.doi.org/ 10.1101/gad.2016211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Métivier D, Meley D, Souquere S, Yoshimori T, et al.. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 2005; 25(3):1025-40; Epub 2005/01/20; PMID:15657430; http://dx.doi.org/ 10.1128/MCB.25.3.1025-1040.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al.. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005; 169(3):425-34; Epub 2005/05/04; PMID:15866887; http://dx.doi.org/ 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature 2004; 432(7020):1032-6; Epub 2004/11/05; PMID:15525940; http://dx.doi.org/ 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- 53.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem 2007; 282(7):4702-10; Epub 2006/12/01; PMID:17135238; http://dx.doi.org/ 10.1074/jbc.M609267200 [DOI] [PubMed] [Google Scholar]
- 54.Oh SH, Lim SC. Endoplasmic reticulum stress-mediated autophagy/apoptosis induced by capsaicin (8-methyl-N-vanillyl-6-nonenamide) and dihydrocapsaicin is regulated by the extent of c-Jun NH2-terminal kinase/extracellular signal-regulated kinase activation in WI38 lung epithelial fibroblast cells. J Pharmacol Exp Ther 2009; 329(1):112-22; Epub 2009/01/14; PMID:19139269; http://dx.doi.org/ 10.1124/jpet.108.144113 [DOI] [PubMed] [Google Scholar]
- 55.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ 2007; 14(2):230-9; Epub 2006/06/24; PMID:16794605; http://dx.doi.org/ 10.1038/sj.cdd.4401984 [DOI] [PubMed] [Google Scholar]
- 56.Qiu W, Zhang J, Dekker MJ, Wang H, Huang J, Brumell JH, Adeli K. Hepatic autophagy mediates endoplasmic reticulum stress-induced degradation of misfolded apolipoprotein B. Hepatology 2011; 53(5):1515-25; Epub 2011/03/02; PMID:21360721; http://dx.doi.org/ 10.1002/hep.24269 [DOI] [PubMed] [Google Scholar]
- 57.Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ 2008; 15(9):1460-71; Epub 2008/06/14; PMID:18551133; http://dx.doi.org/ 10.1038/cdd.2008.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ullman E, Fan Y, Stawowczyk M, Chen HM, Yue Z, Zong WX. Autophagy promotes necrosis in apoptosis-deficient cells in response to ER stress. Cell Death Differ 2008; 15(2):422-5; Epub 2007/10/06; PMID:17917679; http://dx.doi.org/ 10.1038/sj.cdd.4402234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Janssen K, Horn S, Niemann MT, Daniel PT, Schulze-Osthoff K, Fischer U. Inhibition of the ER Ca2+ pump forces multidrug-resistant cells deficient in Bak and Bax into necrosis. J Cell Sci 2009; 122(Pt 24):4481-91; Epub 2009/11/19; PMID:19920074; http://dx.doi.org/ 10.1242/jcs.055772 [DOI] [PubMed] [Google Scholar]
- 60.Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, Kalkanis SN, Mikkelsen T, Brodie C. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer 2009; 125(3):717-22; Epub 2009/05/12; PMID:19431142; http://dx.doi.org/ 10.1002/ijc.24402 [DOI] [PubMed] [Google Scholar]
- 61.Chen YJ, Huang WP, Yang YC, Lin CP, Chen SH, Hsu ML, Tseng YJ, Shieh HR, Chen YY, Lee JJ. Platonin induces autophagy-associated cell death in human leukemia cells. Autophagy 2009; 5(2):173-83; Epub 2008/12/11; PMID:19066447; http://dx.doi.org/ 10.4161/auto.5.2.7360 [DOI] [PubMed] [Google Scholar]
- 62.Zhao X, Gao S, Ren H, Huang H, Ji W, Hao J. Inhibition of autophagy strengthens celastrol-induced apoptosis in human pancreatic cancer in vitro and in vivo models. Curr Mol Med 2014; 14(4):555-63; Epub 2014/04/16; PMID:24730520; http://dx.doi.org/ 10.2174/1566524014666140414211223 [DOI] [PubMed] [Google Scholar]
- 63.Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood 2007; 110(1):313-22; Epub 2007/03/17; PMID:17363733; http://dx.doi.org/ 10.1182/blood-2006-10-050260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen TM, Subramanian IV, Kelekar A, Ramakrishnan S. Kringle 5 of human plasminogen, an angiogenesis inhibitor, induces both autophagy and apoptotic death in endothelial cells. Blood 2007; 109(11):4793-802; Epub 2007/02/03; PMID:17272502; http://dx.doi.org/ 10.1182/blood-2006-11-059352 [DOI] [PubMed] [Google Scholar]
- 65.Debnath J. Detachment-induced autophagy during anoikis and lumen formation in epithelial acini. Autophagy 2008; 4(3):351-3; Epub 2008/01/17; PMID:18196957; http://dx.doi.org/ 10.4161/auto.5523 [DOI] [PubMed] [Google Scholar]
- 66.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Mol Biology Cell 2008; 19(3):797-806; Epub 2007/12/21; PMID:18094039; http://dx.doi.org/ 10.1091/mbc.E07-10-1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 2007; 128(5):931-46; Epub 2007/03/14; PMID:17350577; http://dx.doi.org/ 10.1016/j.cell.2006.12.044 [DOI] [PubMed] [Google Scholar]
- 68.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014; 15(2):81-94; Epub 2014/01/10; PMID:24401948; http://dx.doi.org/ 10.1038/nrm3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gordy C, He YW. The crosstalk between autophagy and apoptosis: where does this lead? Protein Cell. 2012;3(1):17-27; Epub 2012/02/09; PMID:22314807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy 2010; 6(7):891-900; Epub 2010/08/21; PMID:20724831; http://dx.doi.org/ 10.4161/auto.6.7.13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han J, Hou W, Goldstein LA, Lu C, Stolz DB, Yin XM, Rabinowich H. Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J Biol Chem 2008; 283(28):19665-77; Epub 2008/04/01; PMID:18375389; http://dx.doi.org/ 10.1074/jbc.M710169200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S, Gozuacik D. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis 2012; 17(8):810-20; Epub 2012/05/31; PMID:22644571; http://dx.doi.org/ 10.1007/s10495-012-0735-0 [DOI] [PubMed] [Google Scholar]
- 73.Zhang J, Ma K, Qi T, Wei X, Zhang Q, Li G, Chiu JF. P62 regulates resveratrol-mediated Fas/Cav-1 complex formation and transition from autophagy to apoptosis. Oncotarget 2015; 6(2):789-801; Epub 2015/01/19; PMID:25596736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci 2009; 122(Pt 14):2554-66; Epub 2009/06/25; PMID:19549685; http://dx.doi.org/ 10.1242/jcs.046250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003; 100(25):15077-82; Epub 2003/12/06; PMID:14657337; http://dx.doi.org/ 10.1073/pnas.2436255100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al.. Ambra1 regulates autophagy and development of the nervous system. Nature 2007; 447(7148):1121-5; Epub 2007/06/26; PMID:17589504 [DOI] [PubMed] [Google Scholar]
- 77.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 2006; 8(7):688-99; Epub 2006/06/27; PMID:16799551; http://dx.doi.org/ 10.1038/ncb1426 [DOI] [PubMed] [Google Scholar]
- 78.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep 2001; 2(4):330-5; Epub 2001/04/18; PMID:11306555; http://dx.doi.org/ 10.1093/embo-reports/kve061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 2005; 1(1):46-52; Epub 2006/07/29; PMID:16874027; http://dx.doi.org/ 10.4161/auto.1.1.1542 [DOI] [PubMed] [Google Scholar]
- 80.Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A 2008; 105(49):19211-6; Epub 2008/12/04; PMID:19050071; http://dx.doi.org/ 10.1073/pnas.0810452105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol 1998; 72(11):8586-96; Epub 1998/10/10; PMID:9765397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, et al.. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 2007; 26(10):2527-39; Epub 2007/04/21; PMID:17446862; http://dx.doi.org/ 10.1038/sj.emboj.7601689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122(6):927-39; Epub 2005/09/24; PMID:16179260; http://dx.doi.org/ 10.1016/j.cell.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 84.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, Cho DH, Choi B, Lee H, Kim JH, et al.. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem 2005; 280(21):20722-9; Epub 2005/03/22; PMID:15778222; http://dx.doi.org/ 10.1074/jbc.M413934200 [DOI] [PubMed] [Google Scholar]
- 85.Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol Cell 2011; 44(5):698-709; Epub 2011/12/14; PMID:22152474; http://dx.doi.org/ 10.1016/j.molcel.2011.10.014 [DOI] [PubMed] [Google Scholar]
- 86.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 2006; 8(10):1124-32; Epub 2006/09/26; PMID:16998475; http://dx.doi.org/ 10.1038/ncb1482 [DOI] [PubMed] [Google Scholar]
- 87.Lepine S, Allegood JC, Edmonds Y, Milstien S, Spiegel S. Autophagy induced by deficiency of sphingosine-1-phosphate phosphohydrolase 1 is switched to apoptosis by calpain-mediated autophagy-related gene 5 (Atg5) cleavage. J Biol Chem 2011; 286(52):44380-90; Epub 2011/11/05; PMID:22052905; http://dx.doi.org/ 10.1074/jbc.M111.257519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu L, Xing D, Chen WR, Chen T, Pei Y, Gao X. Calpain-mediated pathway dominates cisplatin-induced apoptosis in human lung adenocarcinoma cells as determined by real-time single cell analysis. Int J Cancer 2008; 122(10):2210-22; Epub 2008/01/25; PMID:18214855; http://dx.doi.org/ 10.1002/ijc.23378 [DOI] [PubMed] [Google Scholar]
- 89.He MX, He YW. A role for c-FLIP(L) in the regulation of apoptosis, autophagy, and necroptosis in T lymphocytes. Cell Death Differ 2013; 20(2):188-97; Epub 2012/11/24; PMID:23175183; http://dx.doi.org/ 10.1038/cdd.2012.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, et al.. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 2008; 10(6):676-87; Epub 2008/05/06; PMID:18454141; http://dx.doi.org/ 10.1038/ncb1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Alberti A, Michelet X, Djeddi A, Legouis R. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy 2010; 6(5):622-33; Epub 2010/06/05; PMID:20523114; http://dx.doi.org/ 10.4161/auto.6.5.12252 [DOI] [PubMed] [Google Scholar]
- 92.Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat Cell Biol 2010; 12(9):842-6; Epub 2010/09/03; PMID:20811357; http://dx.doi.org/ 10.1038/ncb0910-842 [DOI] [PubMed] [Google Scholar]
- 93.Camougrand N, Kissova I, Velours G, Manon S. Uth1p: a yeast mitochondrial protein at the crossroads of stress, degradation and cell death. FEMS Yeast Res 2004; 5(2):133-40; Epub 2004/10/19; PMID:15489196; http://dx.doi.org/ 10.1016/j.femsyr.2004.05.001 [DOI] [PubMed] [Google Scholar]
- 94.Wang JH, Ahn IS, Fischer TD, Byeon JI, Dunn WA Jr., Behrns KE, Leeuwenburgh C, Kim JS. Autophagy suppresses age-dependent ischemia and reperfusion injury in livers of mice. Gastroenterology 2011; 141(6):2188-99.e6; Epub 2011/08/23; PMID:21854730; http://dx.doi.org/ 10.1053/j.gastro.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med 2008; 14(9):959-65; Epub 2008/08/12; PMID:18690243; http://dx.doi.org/ 10.1038/nm.1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hubbard VM, Valdor R, Macian F, Cuervo AM. Selective autophagy in the maintenance of cellular homeostasis in aging organisms. Biogerontology 2012; 13(1):21-35; Epub 2011/04/05; PMID:21461872; http://dx.doi.org/ 10.1007/s10522-011-9331-x [DOI] [PubMed] [Google Scholar]
- 97.Kondomerkos DJ, Kalamidas SA, Kotoulas OB. An electron microscopic and biochemical study of the effects of glucagon on glycogen autophagy in the liver and heart of newborn rats. Microsc Res Tech 2004; 63(2):87-93; Epub 2004/01/15; PMID:14722905; http://dx.doi.org/ 10.1002/jemt.20000 [DOI] [PubMed] [Google Scholar]
- 98.Donati A, Cavallini G, Paradiso C, Vittorini S, Pollera M, Gori Z, Bergamini E. Age-related changes in the autophagic proteolysis of rat isolated liver cells: effects of antiaging dietary restrictions. J Gerontol A Biol Sci Med Sci 2001; 56(9):B375-83; Epub 2001/08/29; PMID:11524438; http://dx.doi.org/ 10.1093/gerona/56.9.B375 [DOI] [PubMed] [Google Scholar]
- 99.Chan SH, Kikkawa U, Matsuzaki H, Chen JH, Chang WC. Insulin receptor substrate-1 prevents autophagy-dependent cell death caused by oxidative stress in mouse NIH/3T3 cells. J Biomed Sci 2012; 19:64; Epub 2012/07/14; PMID:22788551; http://dx.doi.org/ 10.1186/1423-0127-19-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gohla A, Klement K, Nurnberg B. The heterotrimeric G protein G(i3) regulates hepatic autophagy downstream of the insulin receptor. Autophagy 2007; 3(4):393-5; Epub 2007/04/26; PMID:17457037; http://dx.doi.org/ 10.4161/auto.4256 [DOI] [PubMed] [Google Scholar]
- 101.Jung HS, Lee MS. Role of autophagy in diabetes and mitochondria. Ann N Y Acad Sci 2010; 1201:79-83; Epub 2010/07/24; PMID:20649543; http://dx.doi.org/ 10.1111/j.1749-6632.2010.05614.x [DOI] [PubMed] [Google Scholar]
- 102.Cahill A, Stabley GJ, Wang X, Hoek JB. Chronic ethanol consumption causes alterations in the structural integrity of mitochondrial DNA in aged rats. Hepatology 1999; 30(4):881-8; Epub 1999/09/25; PMID:10498638; http://dx.doi.org/ 10.1002/hep.510300434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chistiakov DA, Sobenin IA, Revin VV, Orekhov AN, Bobryshev YV. Mitochondrial aging and age-related dysfunction of mitochondria. Biomed Res Int 2014; 2014:238463; Epub 2014/05/13; PMID:24818134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Laos M, Anttonen T, Kirjavainen A, af Hallstrom T, Laiho M, Pirvola U. DNA damage signaling regulates age-dependent proliferative capacity of quiescent inner ear supporting cells. Aging 2014; 6(6):496-510; Epub 2014/07/27; PMID:25063730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Settembre C, Fraldi A, Jahreiss L, Spampanato C, Venturi C, Medina D, de Pablo R, Tacchetti C, Rubinsztein DC, Ballabio A. A block of autophagy in lysosomal storage disorders. Hum Mol Genet 2008; 17(1):119-29; Epub 2007/10/05; PMID:17913701; http://dx.doi.org/ 10.1093/hmg/ddm289 [DOI] [PubMed] [Google Scholar]
- 106.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta 2009; 1793(4):664-73; Epub 2008/08/19; PMID:18706940; http://dx.doi.org/ 10.1016/j.bbamcr.2008.07.014 [DOI] [PubMed] [Google Scholar]
- 107.Ng'oma E, Reichwald K, Dorn A, Wittig M, Balschun T, Franke A, Platzer M, Cellerino A. The age related markers lipofuscin and apoptosis show different genetic architecture by QTL mapping in short-lived Nothobranchius fish. Aging 2014; 6(6):468-80; Epub 2014/08/06; PMID:25093339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab 2014; 20(3):417-32; Epub 2014/07/22; PMID:25043815; http://dx.doi.org/ 10.1016/j.cmet.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Apostolova N, Gomez-Sucerquia LJ, Gortat A, Blas-Garcia A, Esplugues JV. Compromising mitochondrial function with the antiretroviral drug efavirenz induces cell survival-promoting autophagy. Hepatology 2011; 54(3):1009-19; Epub 2011/06/01; PMID:21626526; http://dx.doi.org/ 10.1002/hep.24459 [DOI] [PubMed] [Google Scholar]
- 110.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature 2009; 458(7242):1131-5; Epub 2009/04/03; PMID:19339967; http://dx.doi.org/ 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mei S, Ni HM, Manley S, Bockus A, Kassel KM, Luyendyk JP, Copple BL, Ding WX. Differential roles of unsaturated and saturated fatty acids on autophagy and apoptosis in hepatocytes. J Pharmacol Exp Ther 2011; 339(2):487-98; Epub 2011/08/23; PMID:21856859; http://dx.doi.org/ 10.1124/jpet.111.184341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science 2004; 306(5698):990-5; Epub 2004/11/06; PMID:15528435; http://dx.doi.org/ 10.1126/science.1099993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin CW, Zhang H, Li M, Xiong X, Chen X, Dong XC, Yin XM. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol 2013; 58(5):993-9; Epub 2013/01/24; PMID:23339953; http://dx.doi.org/ 10.1016/j.jhep.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, Lu B, Stolz DB, Clemens DL, Yin XM. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology 2010; 139(5):1740-52; Epub 2010/07/28; PMID:20659474; http://dx.doi.org/ 10.1053/j.gastro.2010.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Manley S, Ni HM, Kong B, Apte U, Guo G, Ding WX. Suppression of autophagic flux by bile acids in hepatocytes. Toxicol Sci 2014; 137(2):478-90; Epub 2013/11/06; PMID:24189133; http://dx.doi.org/ 10.1093/toxsci/kft246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sasaki M, Miyakoshi M, Sato Y, Nakanuma Y. A possible involvement of p62/sequestosome-1 in the process of biliary epithelial autophagy and senescence in primary biliary cirrhosis. Liver Int 2012; 32(3):487-99; Epub 2011/11/22; PMID:22098537 [DOI] [PubMed] [Google Scholar]
- 117.Sasaki M, Miyakoshi M, Sato Y, Nakanuma Y. Autophagy may precede cellular senescence of bile ductular cells in ductular reaction in primary biliary cirrhosis. Dig Dis Sci 2012; 57(3):660-6; Epub 2011/10/13; PMID:21989821; http://dx.doi.org/ 10.1007/s10620-011-1929-y [DOI] [PubMed] [Google Scholar]
- 118.Fickert P, Trauner M, Fuchsbichler A, Stumptner C, Zatloukal K, Denk H. Bile acid-induced Mallory body formation in drug-primed mouse liver. Am J Pathol 2002; 161(6):2019-26; Epub 2002/12/06; PMID:12466118; http://dx.doi.org/ 10.1016/S0002-9440(10)64480-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gerber MA, Orr W, Denk H, Schaffner F, Popper H. Hepatocellular hyalin in cholestasis and cirrhosis: its diagnostic significance. Gastroenterology 1973; 64(1):89-98; Epub 1973/01/01; PMID:4119149 [PubMed] [Google Scholar]
- 120.Strnad P, Zatloukal K, Stumptner C, Kulaksiz H, Denk H. Mallory-Denk-bodies: lessons from keratin-containing hepatic inclusion bodies. Biochim Biophys Acta 2008; 1782(12):764-74; Epub 2008/09/23; PMID:18805482; http://dx.doi.org/ 10.1016/j.bbadis.2008.08.008 [DOI] [PubMed] [Google Scholar]
- 121.Yokoo H, Craig RM, Harwood TR, Cochrane C. Griseofulvin-induced cholestasis in Swiss albino mice. Gastroenterology 1979; 77(5):1082-7; Epub 1979/11/01; PMID:488635 [PubMed] [Google Scholar]
- 122.Meerman L, Koopen NR, Bloks V, Van Goor H, Havinga R, Wolthers BG, Kramer W, Stengelin S, Müller M, Kuipers F, et al.. Biliary fibrosis associated with altered bile composition in a mouse model of erythropoietic protoporphyria. Gastroenterology 1999; 117(3):696-705; Epub 1999/08/28; PMID:10464147; http://dx.doi.org/ 10.1016/S0016-5085(99)70464-6 [DOI] [PubMed] [Google Scholar]
- 123.Manley S, Williams JA, Ding WX. Role of p62/SQSTM1 in liver physiology and pathogenesis. Exp Biol Med (Maywood) 2013; 238(5):525-38; Epub 2013/07/17; PMID:23856904; http://dx.doi.org/ 10.1177/1535370213489446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hands SL, Proud CG, Wyttenbach A. mTOR's role in ageing: protein synthesis or autophagy? Aging 2009; 1(7):586-97; Epub 2010/02/17; PMID:20157541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al.. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137(6):1062-75; Epub 2009/06/16; PMID:19524509; http://dx.doi.org/ 10.1016/j.cell.2009.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy 2007; 3(1):28-31; Epub 2006/09/14; PMID:16969128; http://dx.doi.org/ 10.4161/auto.3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 2011; 10(9):1533-41; Epub 2011/09/01; PMID:21878654; http://dx.doi.org/ 10.1158/1535-7163.MCT-11-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thoen LF, Guimaraes EL, Dolle L, Mannaerts I, Najimi M, Sokal E, van Grunsven LA. A role for autophagy during hepatic stellate cell activation. J Hepatol 2011; 55(6):1353-60; Epub 2011/08/02; PMID:21803012; http://dx.doi.org/ 10.1016/j.jhep.2011.07.010 [DOI] [PubMed] [Google Scholar]
- 129.Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, Czaja MJ, Friedman SL. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology 2012; 142(4):938-46; Epub 2012/01/14; PMID:22240484; http://dx.doi.org/ 10.1053/j.gastro.2011.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 2010; 8(5):422-32; Epub 2010/11/16; PMID:21075353; http://dx.doi.org/ 10.1016/j.chom.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shrivastava S, Bhanja Chowdhury J, Steele R, Ray R, Ray RB. Hepatitis C virus upregulates Beclin1 for induction of autophagy and activates mTOR signaling. J Virol 2012; 86(16):8705-12; Epub 2012/06/08; PMID:22674982; http://dx.doi.org/ 10.1128/JVI.00616-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kim SJ, Syed GH, Khan M, Chiu WW, Sohail MA, Gish RG, Siddiqui A. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natil Acad Sci U S A 2014; 111(17):6413-8; Epub 2014/04/16; PMID:24733894; http://dx.doi.org/ 10.1073/pnas.1321114111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shrivastava S, Raychoudhuri A, Steele R, Ray R, Ray RB. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology 2011; 53(2):406-14; Epub 2011/01/29; PMID:21274862; http://dx.doi.org/ 10.1002/hep.24073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ke PY, Chen SS. Autophagy in hepatitis C virus-host interactions: potential roles and therapeutic targets for liver-associated diseases. World J Gastroenterol 2014; 20(19):5773-93; Epub 2014/06/11; PMID:24914338; http://dx.doi.org/ 10.3748/wjg.v20.i19.5773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu B, Fang M, Hu Y, Huang B, Li N, Chang C, Huang R, Xu X, Yang Z, Chen Z, et al.. Hepatitis B virus X protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy 2014; 10(3):416-30; Epub 2014/01/10; PMID:24401568; http://dx.doi.org/ 10.4161/auto.27286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology 2014; 59(2):505-17; Epub 2013/08/06; PMID:23913306; http://dx.doi.org/ 10.1002/hep.26659 [DOI] [PMC free article] [PubMed] [Google Scholar]


