Abstract
Background
The para-Bombay phenotype results from a variety of mutations in the α-(1,2)-fucosyltransferase gene (FUT1). We investigated samples from seven Chinese probands serologically typed as having the para-Bombay phenotype.
Materials and methods
The para-Bombay phenotype was identified by standard serological methods. Genetic mutations of FUT1 and FUT2 genes were analysed by DNA sequencing. Heterozygous mutations of FUT1 were identified by TOPO cloning sequencing. Blood samples from 331 randomly-selected Chinese donors were analysed with the SNaPshot system to distinguish five known mutations (Se C357T, A385T, G428A, G716A and FUT1 880delTT) in the FUT1 and FUT2 genes. The genetic characteristics of all para-Bombay probands identified in the Fujian Blood Centre, including those in the present study, were also summarised.
Results
Three FUT1 genotypes, h1/h1 (5 individuals), h1/h6 (1 individual) and h3/h2 (1 individual), and three FUT2 genotypes, Se357/Se357 (5 individuals), Se357/Se357, 385 (1 individual) and Se357/Se357, 716 (1 individual) were observed in seven para-Bombay probands. Among 331 donors, only one individual carried the G716A and 880delTT mutations in heterozygosity; this subjects FUT1 and FUT2 genotypes were H/h2 and Se357/Se357, 716, respectively.
Conclusion
The review of all para-Bombay probands identified in the Fujian Blood Centre showed that h1 and h2 are the predominant non-functional FUT1 alleles in Fujian para-Bombay individuals. Our data confirm the hypothesis that the h2 allele is linked to Se357, 716, and the concurrence of unique FUT1 and FUT2 mutations is geographically specific.
Keywords: para-Bombay phenotype, FUT1 gene, FUT2 gene, α-(1, 2)-fucosyltransferase
Introduction
It is well known that the ABH blood group antigens are expressed on both erythrocyte membranes and in secretions such as saliva1. The expression of the ABH antigens on erythrocyte membranes depends on the expression of human FUT1 gene (or H gene) that encodes α-(1,2)-fucosyltransferase (H enzyme), while the expression of the secreted ABH antigens depends on the human FUT2 gene (or Se gene) that encodes α-(1,2)-fucosyltransferase (Se enzyme)1.
The H antigen is a precursor for both A and B antigens. The H enzyme is also essential for the formation of A and B antigens2, since it catalyses the formation of the H antigen by transferring fucose from an α-(1,2) linkage to the terminal galactose of a precursor molecule.
Most people express H antigen on the surface of their red blood cells, but this antigen is absent from the erythrocytes of a few individuals. Individuals with a deficiency in H antigen are said to have an H-deficient phenotype. Bombay phenotypes are H-deficient non-secretors, while para-Bombay phenotypes are H-deficient secretors1. Progress in molecular genetic analysis of H-deficient phenotypes showed that the absence of H antigen on the erythrocytes of individuals with Bombay phenotype results from silenced mutations in FUT1 (h/h) and FUT2 (se/se), whereas such an absence in individuals with para-Bombay phenotype results either from a silenced FUT1 (h/h) gene present with active FUT2 (Se/Se or Se/se) gene or from a mutated FUT1 (h/h) gene present with or without an active FUT2 gene1,3–7.
To date, more than 40 FUT1 alleles associated with either the Bombay or para-Bombay phenotype have been described in the red blood cell database of the National Center for Biotechnology Information (NCBI)8. These FUT1 alleles include nucleotide substitutions, and insertions and deletions of short DNA strands. In our previous studies9,10, we screened blood donors from Fujian province (on the south-eastern coast of China) and reported on 30 para-Bombay individuals. In this study, we analysed the FUT1 and FUT2 genes in seven newly-identified para-Bombay probands, characterised the genes of 37 para-Bombay individuals in our blood centre, and calculated the frequency of five known mutations (Se C357T, Se A385T, Se G428A, Se G716A and FUT1 880delTT) in another 331 randomly selected Chinese donators by SNaPshot.
Material and methods
Sample collection and immunohaematology
Venous blood samples from seven unrelated Han Chinese with para-Bombay phenotype were collected in Fujian Blood Centre. A total of 331 additional Han Chinese donors with normal ABO blood groups were recruited to analyse the frequency of five known mutations in the FUT1 and FUT2 genes.
ABO serology was performed with standard serological techniques. Absorption and elution testing were conducted to detect the trace amount of ABH antigens on the surface of red blood cells. A standard serological haemagglutination inhibition test or prediction based on the Lewis blood group typing was used to analyse secreted ABH antigens in the saliva. The study was approved by the Ethics Committee of Fujian Blood Centre.
DNA preparation
Genomic DNA from whole blood was prepared with the QIAamp Blood Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions and then kept at −30 °C prior to analysis.
ABO genotyping
The ABO genotyping of para-Bombay individuals were performed with a sequence-specific priming kit (Ready Gene ABO SSP kit, G&T, Rockville, USA) according to the manufacturer’s instructions.
Molecular cloning and sequence analysis of the FUT1 and FUT2 genes
To clone the gene for sequencing, polymerase chain reactions (PCR) were performed using the primer targeting FUT1 and FUT2 as reported by Yip et al.11. The PCR products were sequenced by an ABI 3730XL sequencer (Applied Biosystems, Foster City, CA, USA) as described previously9,10. Heterozygous mutations were confirmed by direct sequencing after TOPO cloning (TaKaRa, Dalian, China). The FUT1 and FUT2 genotypes of 37 para-Bombay probands (including those in the present study) identified in Fujian Blood Centre were summarised.
SNaPshot for the screening of Se C357T, A385T, G428A, G716A and FUT1 880delTT mutations
To analyse the polymorphisms of FUT1 and FUT2, genomic DNA regions surrounding five known mutations (Se C357T, A385T, G428A, G716A and FUT1 880delTT) were amplified with a multiplex PCR method (Table I). In the PCR, 100 ng of genomic DNA were amplified by 2.5 U of Taq DNA polymerase (Fermentas, Ontario, Canada) in a 25 μL reaction mixture containing 2.5 mmol/L of MgCl2, 0.2 mmol/L of dNTP, PCR buffer and primer sets (see Table I for final concentrations). PCR amplification was performed on a thermal cycler (EDC-810, Eastwin Life Sciences Inc., Beijing, China). The conditions for the multiplex PCR included an initial denaturation cycle at 95 °C for 3 minutes; 11 cycles of denaturation at 94 °C for 15 seconds, annealing at 60 °C for 15 seconds and extension at 72 °C for 30 seconds; another 24 cycles of denaturation at 94 °C for 15 seconds, annealing at 54 °C for 15 seconds and extension at 72 °C for 30 seconds; followed by a final extension of 7 minutes at 72 °C. After amplification, PCR products were purified to remove unincorporated dNTP and primers. During purification, 10 U of exonuclease I and 3 U of shrimp alkaline phosphatase (Fermentas, Ontario, Canada) were used for 60 minutes at 37 °C, followed by enzyme inactivation at 80 °C for 15 minutes.
Table I.
The primer targeting the FUT1 and FUT2 genes in the multiplex PCR assay.
| Single nucleotide polymorphisms | Forward (5′→3′) | Reverse (5′→3′) | Product size (bp) | Concentration (nmol/L) |
|---|---|---|---|---|
| FUT2 C357T/A385T/G428A/G716A | CCATCTTCAGAATCACCCTG | CAATGGTCATGATGGTGTGG | 556 | 800 |
| FUT1 880del TT | GTCACCAGCAACGGCATGGAG | AGATCTTCAGGAACTCAGAGT | 238 | 500 |
Next, single-base primer extension was performed using the SNaPshot kit (Applied Biosystems) with reference to t he manufacturer’s protocol. The final reaction volume was 6 μL, comprising 2 μL of the above purified amplicons, 3 μL of SNaPshot ready reaction premix, and extension primers (see Table II for final concentrations). The extension reaction was performed under stringent conditions (25 cycles at 96 °C for 10 seconds, 52 °C for 5 seconds, 60 °C for 30 seconds). After treatment with shrimp alkaline phosphatase, 0.5 μL of SNaPshot primer extension product were mixed with formamide before analysis on a capillary sequencer (ABI 3730XL, Applied Biosystems) using POP 7 polymer according to the manufacturer’s instructions. Results were analysed using appropriate software (Peak Scanner™ v 1.0., Applied Biosystems).
Table II.
The probe primers used for the multiplex assay for five single nucleotide polymorphisms (SNP).
| Allele | SNP | Polymorphisms | Extension nucleotides | Forward/reverse | 5′→3′ extension primer | Size (bp) | Concentration (nmol/L) |
|---|---|---|---|---|---|---|---|
| FUT1 | 880del TT | TT/- | A/G | Reverse | GTTGCACTGTGTGAGCAGGGCA | 22 | 50 |
| FUT2 | C357T | C/T | C/T | Forward | T(3)CCTGGCAGAACTACCACCTGAA | 25 | 50 |
| FUT2 | G716A | A/G | A/G | Forward | T(7)AGTAATGGCATGGCCTGGTGTC | 29 | 200 |
| FUT2 | G428A | A/G | C/T | Reverse | T(11)GCGGAGGTGGTGGTAGAAGGTC | 33 | 80 |
| FUT2 | A385T | A/T | A/T | Reverse | T(16)AAGCGGACGTACTCCCCCGGGA | 37 | 150 |
Results
Serological results
We failed to detect the presence of ABH antigens on seven para-Bombay probands using ordinary methods of haemagglutination. However, by using a more sensitive assay, named the adsorption-elution test, we found that three individuals were para-Bombay A, one was Para-Bombay B, and three were para-Bombay O. The seven probands were demonstrated to be secretors either by their Le (a−b+) blood group or the ABH substances present in the saliva. With the exception of the serum from one para-Bombay B individual, the sera from all probands contained anti-H. The ABO genotypes were consistent with the serological phenotypes.
FUT1 and FUT2 gene analysis in para-Bombay individuals
To identify the genetic mutations of FUT1, we sequenced the full coding region of FUT1 in the seven para-Bombay probands and found four previously reported defective FUT1 alleles, h1 (547delAG), h2 (880del TT), h3 (C658T), h6 (C522A). In total, three FUT1 genotypes, h1/h1 (5 individuals), h1/h6 (1 individual) and h3/h2 (1 individual), were detected.
Sequencing analysis of the FUT2 coding region revealed three alleles, Se357, Se357, 716 and Se357, 385, in the seven para-Bombay probands. Three FUT2 genotypes, Se357/Se357 (5 individuals), Se357/Se357, 385 (1 individual) and Se357/Se357, 716 (1 individual), were observed.
Estimation of the frequency of alleles (h2, Se357, Se428, Se357, 385 and Se357, 716) with the SNaPshot system
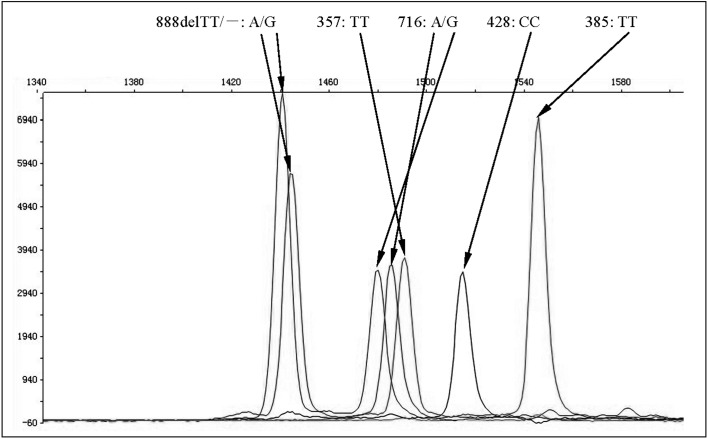
Since h1 and h2 alleles have been observed in conjunction with Se357 and Se357, 716 respectively, we further analysed the distribution of five single nucleotide polymorphisms (Se C357T, A385T, G428A, G716A and FUT1 880delTT) in the Fujian population with SNaPshot. Among the 662 alleles considered, the frequency of C and T alleles in the C357T polymorphism were 12.09% and 87.91%, respectively, while the frequency of A and T alleles in the A385T polymorphism were 58.16% and 41.84%, respectively. No G428A mutation was found in the Fujian population. In addition, only one donor carried a G716A mutation, and this donor also had the 880delTT mutation in the FUT1 allele. Sequencing confirmed that the FUT1 and FUT2 mutations of the donor were H/h2 and Se357/Se357, 716, respectively (Figure 1). Together, five FUT2 alleles were detected in the normal population: Se357, Se357, 385, Se, Se385and Se357, 716, and their frequencies were 46.37%, 41.39%, 11.63%, 0.45% and 0.15%, respectively (Table III).
Figure 1.
SNPs identified in FUT1 and FUT2 genes.
Table III.
The distribution of partial FUT1 and FUT2 alleles in the Fujian population (n=331).
| Alleles | Number | Percentage (%) |
|---|---|---|
| Se357 | 307 | 46.37 |
| se357, 385 | 274 | 41.39 |
| Se | 77 | 11.63 |
| se385 | 3 | 0.45 |
| Se357, 716 | 1 | 0.15 |
| se428 | 0 | 0 |
| h2 | 1 | 0.15 |
Discussion
The para-Bombay phenotype is a rare blood phenotype worldwide, while it is relatively frequently observed in the Fujian population (1:8,500)9. A total of 43 unrelated para-Bombay individuals have been identified in our laboratory. Among the 37 para-Bombay individuals in this study, six different h alleles were observed: h1 (547delAG, Arg183Argfs*86), h2 (880delTT, Phe297Cysfs*40), h3 (C658T, Arg220Cys), h6 (C522A, Phe174Leu), h9 (C424T, Arg142Trp), and h328A (G328A, Ala110Thr) (Table IV). More than 90% of the h alleles associated with para-Bombay were h1 and h2, indicating that the h1 and h2 alleles were non-functional FUT1 alleles which are prevalent in the Fujian population. This was consistent with non-functional FUT1 alleles in para-Bombay individuals found in Taiwan12.
Table IV.
Genotypes and phenotypes of 37 Chinese para-Bombay individuals.
| N. | Phenotypes | Genotypes | N. | Phenotypes | Genotypes | ||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| FUT 1 | FUT 2 | FUT 1 | FUT 2 | ||||
| 1 | O(h) | h1/h1 | Se357/Se357 | 2 | A(h) | h1/h1 | Se357/Se357 |
| 3 | A(h) | h1/h2 | Se357/Se357,716 | 4 | A(h) | h1/h1 | Se357/Se357 |
| 5 | B(h) | h1/h1 | Se357/Se357 | 6 | A(h) | h1/h2 | Se357/Se357, 716 |
| 7 | O(h) | h1/h1 | Se357/Se357 | 8 | A(h) | h1/h2 | Se357/Se357, 716 |
| 9 | B(h) | h328/h3 | Se357/Se357 | 10 | AB(h) | h1/h2 | Se357/Se357, 716 |
| 11 | A(h) | h1/h1 | Se357/Se357 | 12 | B(h) | h1/h2 | NT |
| 13 | AB(h) | h1/h2 | Se357/Se357, 716 | 14 | A(h) | h1/h2 | NT |
| 15 | O(h) | h1/h2 | Se357/Se357, 716 | 16 | A(h) | h1/h1 | Se357/Se357 |
| 17 | O(h) | h2/h2 | Se357, 716/Se357, 716 | 18 | B(h) | h1/h9 | Se357/Se357 |
| 19 | A(h) | h1/h1 | Se357/Se357 | 20 | A(h) | h3/h2 | Se357/Se357, 716 |
| 21 | O(h) | h1/h2 | Se357/Se357, 716 | 22 | A(h) | h1/h1 | Se357/Se357 |
| 23 | A(h) | h1/h2 | Se357/Se357, 716 | 24 | A(h) | h1/h1 | Se357/Se357 |
| 25 | AB(h) | h1/h2 | Se357/Se357, 716 | 26 | O(h) | h1/h1 | Se357/Se357 |
| 27 | A(h) | h1/h1 | Se357/Se357 | 28 | A(h) | h1/h1 | Se357/Se357 |
| 29 | A(h) | h1/h1 | Se357/Se357 | 30 | O(h) | h1/h1 | Se357/Se357 |
| 31 | O(h) | h1/h1 | Se357/Se357 | 32 | A(h) | h1/h1 | Se357/Se357 |
| 33 | A(h) | h1/h6 | Se357/Se357,385 | 34 | A(h) | h3/h2 | Se357/Se357, 716 |
| 35 | O(h) | h1/h1 | Se357/Se357 | 36 | B(h) | h1/h1 | Se357/Se357 |
| 37 | O(h) | h1/h1 | Se357/Se357 | ||||
Among the 37 para-Bombay individuals who underwent FUT1 genotyping, FUT2 genotyping analysis was only conducted in 35 probands including those in the present study. Three FUT2 alleles (Se357, Se357, 716 and Se357, 385) were observed, and their frequencies were 81.43%, 17.14% and 1.43%, respectively. The frequencies of Se357, Se357, 716 and Se357, 385 in the Fujian population were 46.37%, 0.15% and 41.39%, respectively (Table III). The neutral mutation C357T is commonly found in the Chinese population (87.91%), whereas the frequency of the missense mutation G716A is relatively low (0.15%).
Our study also showed that 51 h1 alleles were associated with Se357, 13 h2 alleles were associated with Se357, 716, and the remaining 4 h alleles (i.e., h3, h6, h9 and h328A) had various sporadic mutations and were nun-functional alleles (Table IV). These results indicate that the h1 and h2 alleles form h1-Se357and h2-Se357, 716 haplotypes, respectively. Although the C357T mutation is commonly observed, the G716A mutation is rarely re ported in the Chinese population. In our studies, G716A was only found in the para-Bombay individuals in association with the h2 gene. Since h2-Se357, 716 was also observed in other Chinese para-Bombay individuals13,14, we further analysed the frequency of the G716A mutation and 880delTT in the 331 randomly selected Chinese with SNaPshot. Only one donor carried the G716A and 880delTT mutations in heterozygosity (1/662); this donor’s FUT1 and FUT2 genotypes were H/h2 and Se357/Se357, 716, respectively. This further supports the hypothesis that the h2 allele is linked to Se357, 716. In addition, different haplotype linkages were also observed in populations screened in other regions. The h695 allele is frequently found linking to sew385 in the Japanese population15. A majority of FUT1 349T inactivating mutations is associated with the se428 mutation in the population of Reunion Island16. FUT1 725G is associated with the FUT2 deletion mutation in the Indian population17.
The para-Bombay phenotype is characterised by a deficiency of ABH antigens on red blood cells, and the underlying molecular basis of this phenotype is mutations in the FUT1 gene. The h2 is a null FUT1 gene that has a deletion of two bases at nucleotides 880–882 and results in frame shift and inactivation of the H enzyme5. Se357, 716, which carries a C357T neutral mutation and G716A missense mutation in the FUT2 gene, does not influence the Se enzyme14. As discussed previously, both h2 and Se357, 716 are prevalent alleles in Chinese para-Bombay individuals, and have never been reported in any other ethnic population. The present study indicates that G716A and 880delTT are rarely present in the normal population. The haplotype h2-Se357, 716 may be present in both Chinese para-Bombay individuals and the normal population. These results indicate that the h2 allele is linked to Se357, 716 and that the linkage is geographically specific.
The FUT1 and FUT2 genes are located at the proximal end of chromosome 19q13.3 and are translated in the same direction 35 kb apart (Cent-FUT2-FUT1-Ter). Both loci encode homologous α-(1,2)-fucosyltransferases (H and Se enzyme) with subtle discrepancies in substrate specificity2,18, suggesting linkage disequilibrium between the two genes. Our previous family analysis9 of para-Bombay individuals confirmed that the altered FUT1 and FUT2 genes also occurred in Mendelian inheritance. The pedigree indicated that the two haplotypes (h1-Se357and h2-Se357, 716) were inherited according to the Mendelian generations.
The frequency of se385 and se428 alleles in the Chinese population was also included (Table III). The nonsense mutation G428A is a common non-secretor allele and responsible for the non-secretor phenotype in Caucasian and African populations18. The missense mutation A385T is responsible for a partial secretor phenotype and has only been found in populations of Asia and the Pacific islands19,20. In this study, we analysed the distribution of the two major non-functional alleles in Fujian population and found that the distribution of the two major null alleles in the Fujian population was obviously different from that of Europeans. We did not observe the G428A mutation, although this was present at a frequency of 0.7% in another Chinese population20; in contrast to this rare mutation, A385T is a frequently observed mutation (41.84%). These results further confirmed previous reports that FUT2 mutations are geographically distributed or have race specificity.
Conclusion
Our study suggesed two prevalent non-functional FUT1 alleles (h1, h2) in Fujian para-Bombay individuals and these h alleles were observed in conjunction with Se357 and Se357, 716 respectively (i.e. h1-Se357and h2-Se357, 716). The joint occurrence of unique sets of mutations in FUT1 and FUT2 was geographically specific.
Acknowledgements
We thank all blood donors, especially those individuals with the para-Bombay blood phenotype. We would also thank technicians and faculties in the laboratory for their technical assistance in the study and Ms Jenny Cai for the English editing of the manuscript.
Footnotes
Funding
This work was supported by grants from the Youth Foundation of Fujian Provincial Public Health Board Bureau (2012-2-31) and the Fujian Natural Science Foundation (2014J01295).
Authorship contributions
AZ and QC designed the research, analysed the data and prepared the manuscript; BR performed the serological studies.
The Authors declare that they have no conflicts of interest.
References
- 1.Daniels G. Human Blood Groups. 2th ed. Oxford: Blackwell Science Ltd; 2002. [Google Scholar]
- 2.Larsen RD, Ernst LK, Nair RP, Lowe JB. Molecular cloning, sequence, and expression of a human GDP-L-fucose: beta-D-galactoside 2-αlpha-L-fucosyltransferase cDNA that can form the H blood group antigen. Proc Natl Acad Sci USA. 1990;87:6674–8. doi: 10.1073/pnas.87.17.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storry JR, Johannesson JS, Poole J, et al. Identification of six new alleles at FUT1 and FUT2 loci in ethnically diverse individuals with Bombay and para-Bombay phenotypes. Transfusion. 2006;46:2149–55. doi: 10.1111/j.1537-2995.2006.01045.x. [DOI] [PubMed] [Google Scholar]
- 4.Yan L, Zhu F, Xu X, et al. Molecular basis for para-Bombay phenotypes in Chinese persons, including a novel nonfunctional FUT1 allele. Transfusion. 2005;45:725–30. doi: 10.1111/j.1537-2995.2005.04305.x. [DOI] [PubMed] [Google Scholar]
- 5.Yu LC, Yang YH, Broadberry RE, et al. Heterogeneity of the human H blood group alpha(1,2)fucosyltransferase gene among para-Bombay individuals. Vox Sang. 1997;72:36–40. doi: 10.1046/j.1423-0410.1997.00036.x. [DOI] [PubMed] [Google Scholar]
- 6.Luo G, Wei L, Wang Z, et al. The summary of FUT1 and FUT2 genotyping analysis in Chinese para-Bombay individuals including additional nine probands from Guangzhou in China. Transfusion. 2013;53:3224–9. doi: 10.1111/trf.12183. [DOI] [PubMed] [Google Scholar]
- 7.Matzhold EM, Helmberg W, Wagner T, et al. Identification of 14 new alleles at the fucosyltransferase 1, 2, and 3 loci in Styrian blood donors, Austria. Transfusion. 2009;49:2097–108. doi: 10.1111/j.1537-2995.2009.02293.x. [DOI] [PubMed] [Google Scholar]
- 8.Blumenfeld OO, Patnaik SK. Allelic genes of blood group antigens: a source of human mutations and cSNPs documented in the Blood Group Antigen Gene Mutation Database. Human Mutation. 2004;23:8–16. doi: 10.1002/humu.10296. [DOI] [PubMed] [Google Scholar]
- 9.Chi Q, Tang W, Wang CQ, et al. Molecular genetics analysis and frequency survey of H deficient phenotype. Chin J Blood Transfusion. 2006;19:446–8. [In Chinese] [Google Scholar]
- 10.Chi Q, Zhang A, Ren BC. Analysis of FUT1 and FUT2 genes in 16 para- Bombay individuals and identification of a novel null FUT allele. Chin J Blood Transfusion. 2012;25:1152–4. [In Chinese] [Google Scholar]
- 11.Yip SP, Chee KY, Chan PY, et al. Molecular genetic analysis of para-Bombay phenotypes in Chinese: a novel non-functional FUT1 allele is identified. Vox Sang. 2002;83:258–62. doi: 10.1046/j.1423-0410.2002.00184.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen DP, Tseng CP, Wang WT, et al. Two prevalent h alleles in para-Bombay haplotypes among 250,000 Taiwanese. Ann Clin Lab Sci. 2004;34:314–8. [PubMed] [Google Scholar]
- 13.Cai XH, Jin S, Liu X, et al. Molecular genetic analysis for the para-Bombay blood group revealing two novel alleles in the FUT1 gene. Blood Transfus. 2011;9:466–8. doi: 10.2450/2011.0115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo ZH, Xiang D, Zhu ZY, et al. Analysis on FUT1 and FUT2 gene of 10 para-Bombay individuals in China. Chin J Med Genet. 2004;21:417–21. [In Chinese] [PubMed] [Google Scholar]
- 15.Kaneko M, Nishihara S, Shinya N, et al. Wide variety of point mutations in the H gene of Bombay and para-Bombay individuals that inactivate H enzyme. Blood. 1997;90:839–49. [PubMed] [Google Scholar]
- 16.Fernandez MP, Cailleau A, Henry S, et al. Point mutations and deletion responsible for the Bombay H null and the Reunion H weak blood groups. Vox Sang. 1998;75:37–46. [PubMed] [Google Scholar]
- 17.Koda Y, Soejima M, Johnson PH, et al. Missense mutation of FUT1 and deletion of FUT2 are responsible for Indian Bombay phenotype of ABO blood group system. Biochem Biophys Res Commun. 1997;238:21–5. doi: 10.1006/bbrc.1997.7232. [DOI] [PubMed] [Google Scholar]
- 18.Kelly RJ, Rouquier S, Giorgi D, et al. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2) fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the nonsecretor phenotype. J Biol Chem. 1995;270:4640–9. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- 19.Yip SP, Lai SK, Wong ML. Systematic sequence analysis of the human fucosyltransferase 2 (FUT2) gene identifies novel sequence variations and alleles. Transfusion. 2007;47:1369–80. doi: 10.1111/j.1537-2995.2007.01280.x. [DOI] [PubMed] [Google Scholar]
- 20.Liu YH, Koda Y, Soejima M, et al. Extensive polymorphism of the FUT2 gene in an African (Xhosa) population of South Africa. Hum Genet. 1998;103:204–10. doi: 10.1007/s004390050808. [DOI] [PubMed] [Google Scholar]