Abstract
Poly(ADP-ribosyl)ation is an unique posttranslational modification and required for spindle assembly and function during mitosis. However, the molecular mechanism of poly(ADP-ribose) (PAR) in mitosis remains elusive. Here, we show the evidence that PAR is recognized by ECT2, a key guanine nucleotide exchange factor in mitosis. The BRCT domain of ECT2 directly binds to PAR both in vitro and in vivo. We further found that α-tubulin is PARylated during mitosis. PARylation of α-tubulin is recognized by ECT2 and recruits ECT2 to mitotic spindle for completing mitosis. Taken together, our study reveals a novel mechanism by which PAR regulates mitosis.
Keywords: BRCT domain, ECT2, mitosis, PAR, PAR binding
Abbreviations:
- PAR
poly(ADP-ribose)
- PARylation
poly(ADP-ribosyl)ation
- ECT2
epithelial cell transforming sequence 2 oncogene
- BRCA1
Breast Cancer Gene 1
- BARD1
BRCA1-associated RING domain protein 1
- BRCT
BRCA1 C Terminus
- CHFR
checkpoint with forkhead-associated and ring finger
- DH
Dbl homology
- PH
pleckstrin homology
- PBC
polybasic cluster
- EDTA
Ethylenediaminetetraacetic acid
- GST
glutatione S-transferase
- PARG
poly(ADP-ribose) glycohydrolase gene
- PARP-1
Poly(ADP-ribose) polymerase 1
- RhoGEF
Rho guanine nucleotide exchange factor
- TBST
Tris-Buffered Saline and Tween 20.
Introduction
Poly(ADP-ribosyl)ation (PARylation) is an unique posttranslational modification that plays an important role in chromatin remodeling during DNA damage response.1-4 Using nicotinamide adenine dinucleotide (NAD) as the donor, a group of enzymes, namely poly (ADP-ribose) polymerases (PARPs), catalyze this posttranslational modification by covalently linking ADP-ribose to protein targets and building the linear and branched chains of poly (ADP-ribose) (PAR).5-7 Interestingly, recent studies suggest that PARylation occurs at the spindle apparatus during mitosis,8-10 implying a novel function of PAR during mitosis. However, the molecular mechanism of PARylation in mitosis is not clear.
Recently, we found that the tandem BRCT motifs of BARD1, a functional partner of BRCA1, is a PAR-binding domain that targets the BRCA1/BARD1 complex to DNA lesions for the BRCA1-dependent DNA damage repair.3 Interestingly, ECT2, a guanine nucleotide exchange factor (GEF) that plays a key role during mitosis, contains tandem BRCT motifs.11-13 ECT2 activates Rho family small GTPases during mitosis, especially at cytokinesis, the last step of mitosis, by recruiting and activating RhoA at central spindle complex.12-13 Moreover, lacking ECT2 impairs not only cytokinesis but also the chromosomal segregation,11 suggesting that ECT2 may also be an important regulator at multiple stages during mitosis via the activation of different Rho GTPases. Consistent with its role in mitosis, ECT2 is a biomarker for cell proliferation and tumorigenesis.14-15
It has been reported that the BRCT domain of ECT2 plays a key role to regulate the function of ECT during mitosis.12 However, the underlying molecular mechanism remains elusive. Here, we found that the BRCT domain of ECT2 is PAR-binding domain and recognizes PARylated α-tubulin during mitosis. The interaction between the BRCT domain of ECT2 and PARylated α-tubulin is required for the function of ECT2 in mitosis.
Results
The ECT2 BRCT domain recognizes poly(ADP-ribose) in vitro
Our previous study suggests that the BRCT domain is a PAR-binding domain.3-4 Since ECT2 has the tandem BRCT motifs at the N-terminus (Fig. 1A), we asked whether the BRCT domain of ECT2 recognizes PAR during mitosis. We first expressed and purified the recombinant GST-ECT2 BRCT domain in vitro and performed in vitro binding assay by incubating recombinant GST-ECT2 BRCT with PAR. Since CHFR directly interacts with PAR via its C-terminal PBZ motif,16-17 we used GST and recombinant CHFR as the negative and positive control in this binding assay, respectively. Purified recombinant proteins were dot-blotted on the nitrocellulose membrane after incubation with PAR. We found that, like CHFR, the GST-ECT2 BRCT domain could easily pull down PAR (Fig. 1B). Moreover, biotinylated PAR could also pull down the GST-ECT2 BRCT domain (Fig. 1C), suggesting a potential direct interaction between the ECT2 BRCT domain and PAR. To demonstrate the direct interaction and measure the affinity between the recombinant ECT2 BRCT domain and PAR, we performed isothermal titration calorimetry (ITC) assay, and measured the dissociation constant of the interaction with a KD at around 4.85 μM (Fig. 1D). Since PAR is mixed length of branched polymer of ADP-ribose, the affinity between the PAR and ECT2 BRCTs might not be measured very accurately. Therefore, we also measured the affinity between the ECT2 BRCT domain and ADP-ribose, the basic unit of PAR using the ITC assays. The KD between the ECT2 BRCT domain and ADP-ribose is around 8.70 μM (Fig. 1D).
Figure 1.
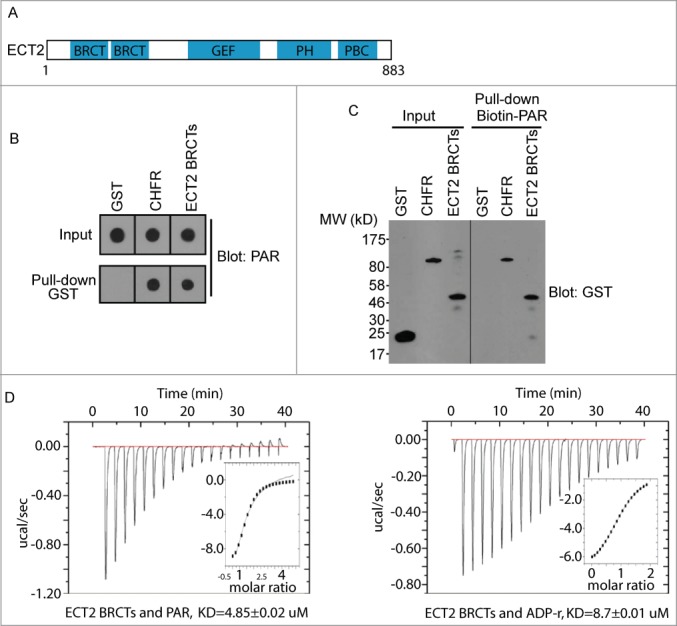
ECT2 BRCT is a PAR binding domain. (A) Schematic structure of ECT2, containing BRCT repeats, a DH-type RhoGEF domain, a pleckstrin homology (PH) domain and a polybasic cluster (PBC). (B) The interaction between PAR and GST (negative control), GST-CHFR (positive control), or GST-ECT2 BRCTs was examined by GST pull-down and dot blot with anti-PAR antibody. PAR was blotted and shown as the input. (C) The interaction between biotin-PAR and the recombinant proteins in (A) was examined by the reciprocal pull-down with streptavidin beads and blotted with anti-GST antibody. Recombinant proteins were blotted and shown as the input. (D) The affinity between PAR and GST-ECT2 BRCTs was measured by ITC (left); the affinity between ADP-ribose and GST-ECT2 BRCTs was measured by ITC (right). Titration of PAR or ADP-ribose was injected into a solution containing the purified protein. The inset shows the fit of the data to an equilibrium binding isotherm. The fit provides an equilibrium dissociation constant (KD) for the binding of PAR or ADP-ribose to the protein.
Mapping the PAR-binding site in the ECT2 BRCT domain
Next, we examined the PAR-binding site in the ECT2 BRCT domain. The BRCT domain, first identified in BRCA1, is known as a phospho-Ser binding domain.18-19 Although the homology of the primary sequence of different BRCT domains is very low, BRCT domains retain several key residues for the similar secondary folding. In particular, a set of BRCT domains containing tandem BRCT motifs have a putative phospho-protein binding pocket formed by the conserved residues within these BRCT domains.20-23 However, the phospho-protein binding partners of these BRCT domains are largely unknown. Recently, we found that the BRCT domain of BARD1 is an ADP-ribose-binding domain, and the ADP-ribose binding site is coincided with the putative phospho-protein binding pocket in the BARD1 BRCT domain.3 These results suggest that the BARD BRCT domain may not interact with the phospho-Ser residue. Instead, it is likely to interact with a phosphate group in ADP-ribose. Like the BARD1 BRCT domain, the ECT2 BRCT domain comprises 2 BRCT motifs and has a putative phospho-group binding pocket. Thus, we asked if the putative phosphate group binding pocket in the ECT2 BRCT domain could recognize ADP-ribose.24 From the secondary structure prediction, the K195 of ECT2 is likely to be the conserved residue in the binding pocket (Fig. 2A). Thus, we mutated K195 into alanine. The K195A mutant of ECT2 failed to interact with PAR in pull down assays (Fig. 2B and 2C). Moreover, the KD between the recombinant K195A mutant and ADP-ribose is higher than 100 μM (Fig. 2D), suggesting that the K195A mutation abolishes the interaction between the ECT2 BRCT domain and ADP-ribose. Thus, it is likely that the conserved phosphate group binding pocket of the ECT2 BRCT domain recognizes ADP-ribose.
Figure 2.
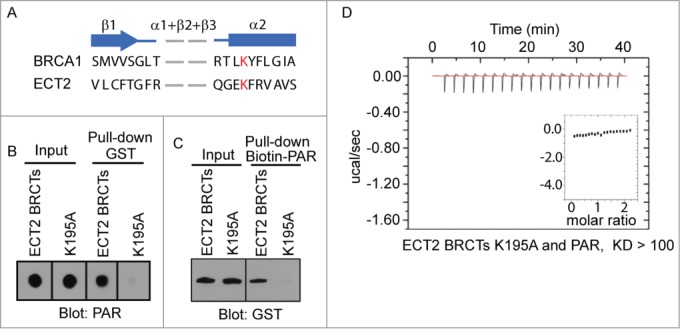
PAR binding site in ECT2 BRCTs. (A) Secondary structure comparison between BRCA1 BRCTs and ECT2 BRCTs shows the conserved binding pocket for phosphate group. β strand is shown as arrow and α helix is shown as box. Conserved residues are marked in red. (B) The interaction between PAR and GST-ECT2 BRCTs (positive control), or GST-ECT2 BRCTs K195A was examined by GST pull-down and dot blotted with anti-PAR antibody. PAR was blotted and shown as the input. (C) The interaction between biotin-PAR and the recombinant proteins in (A) was examined by the reciprocal pull-down with Streptavidin beads and blotted with anti-GST antibody. Recombinant proteins were blotted and shown as the input. (D) The affinity between PAR and GST-ECT2 BRCTs K195A was measured by ITC. Titration of PAR was injected into a solution containing the purified protein. The inset shows the fit of the data to an equilibrium binding isotherm. The fit provides an equilibrium dissociation constant (KD) for the binding of PAR to the protein.
The ECT2 BRCT domain binds PAR during mitosis
Next, we examined the interaction between PAR and ECT2 in vivo. It has been shown that PARylation occurs during mitosis,9-10 which is regulated by Tankyrase 1.10 Consistently, PAR is remarkably synthesized in the mitotic cells induced by nocodazole treatment (Fig. 3A). We then examined the interaction between PAR and ECT2 in the nocodazole-treated cells using co-immunoprecipitation (co-IP) assays. As shown in Fig. 3B, PAR could co-IP with ECT2 in the nocodazole-treated cells. Moreover, we used siRNA to specifically down-regulate the expression of Tankyrase 1 (Fig. 3C). When cells were pretreated with Tankyrase 1 siRNA to suppress PAR synthesis, cells were arrested in mitosis and the interaction between PAR and ECT2 was abolished (Fig. 3B). Moreover, compared with wild type ECT2, the K519A mutant failed to interact with PAR (Fig. 3D). Collectively, these data suggest that ECT2 binds to PAR in vivo.
Figure 3.
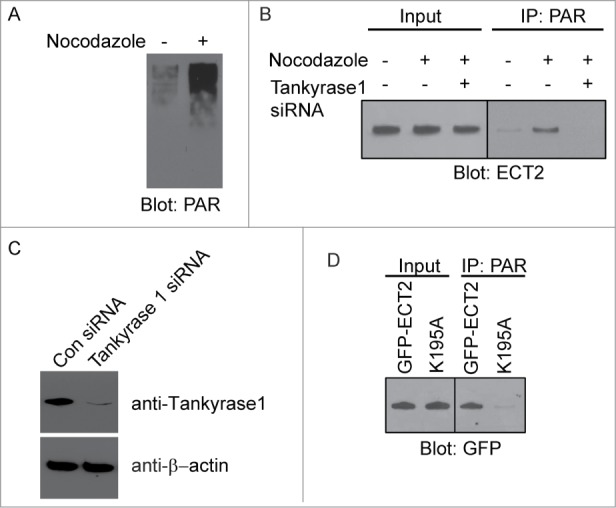
The ECT2 BRCT domain binds PAR during mitosis. (A) PAR is remarkably synthesized in vivo with the nocodazole treatment. (B) The in vivo interaction between PAR and ECT2 with or without the treatment of Tankyrase 1 siRNAs was measured by co-IP with the indicated antibodies. Whole cell lysates were blotted and shown as the input. (C) Tankyrase 1 protein was significantly decreased in Tankyrase 1 siRNA treated HeLa cells. β-actin was used as loading control. (D) The K519A mutant failed to interact with PAR in vivo. Wild type ECT2 was used as control. Whole cell lysates were blotted and shown as the input.
α-tubulin is PARylated by tankyrase 1 during mitosis
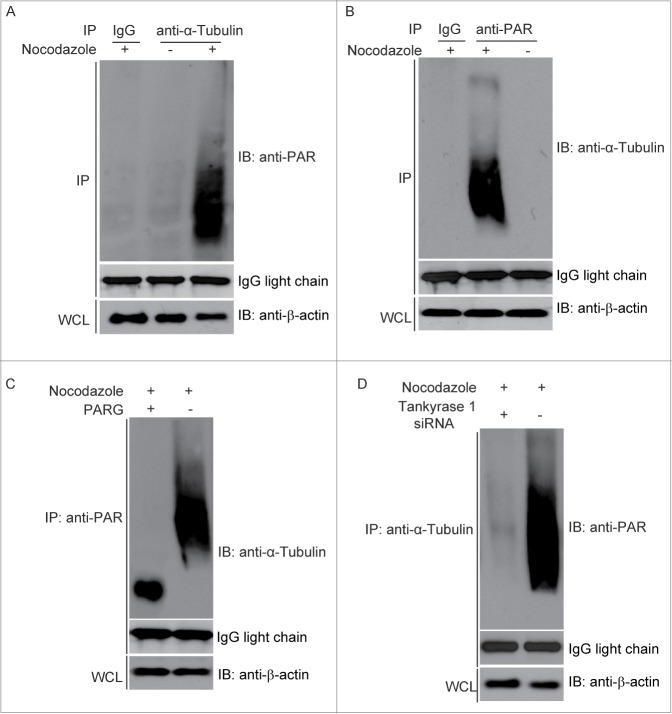
During DNA damage response, the most PARylated proteins are histones and PARP1, the enzyme itself.25-27 However, it is not clear which protein is PARylated during mitosis and recognized by ECT2. From PARylation proteomics, α-tubulin, a major scaffold protein in mitotic spindles, is one of most abundant PARylated protein in vivo.28 To examine whether PARylation of α-tubulin occurs during mitosis, we treated cells with Nocodazole and arrest cells in mitosis. Compared with mock treated cells, α-tubulin is significantly PARylated (Fig. 4A). The results were further confirmed by the reciprocal co-IP assay (Fig. 4B). Due to that PARylation brings huge amount of phosphate moieties, PARylated α-tubulin slowly migrated into the SDS-PAGE. Since PARG is a major enzyme to digest PAR from its targets, we treated precipitated α-tubulin with PARG to remove the covalently bound PAR from α-tubulin. Compared with the mock treated precipitates, PARylation of α-tubulin was significantly reduced in the presence of PARG (Fig. 4C). Collectively, these results suggest that α-tubulin is PARylated during mitosis.
Figure 4.
PARylation of α-tubulin during cytokinesis. (A) α-tubulin is PARylated during mitosis. Co-IP assay was performed by using Nocodazole-treated HeLa cells. The α-tubulin antibody was used for IP and PAR antibody used for IB, and IgG used as negative control. β-actin and IgG light chain were used as loading control for IP and input, respectively. (B) PARylation of α-tubulin during mitosis was examined by reverse Co-IP. Co-IP assay was performed by using Nocodazole-treated HeLa cells. The PAR antibody was used for IP and α-tubulin antibody used for IB and IgG used as negative control. β-actin and IgG light chain were used as loading control for IP and input, respectively. (C) PARG induced the PAR degradation for PARylated α-tubulin. Co-IP assay was performed by using Nocodazole-treated HeLa cells. The PAR antibody was used for IP. After IP, the sepharose was treated with PARG protein. Then α-tubulin antibody was used for IB. β-actin and IgG light chain were used as loading control for IP and input, respectively. (D) PARylation of α-tubulin is dependent on Tankyrase 1. PARylation of α-tubulin was significantly reduced in Tankyrase 1 siRNA treated HeLa cells. β-actin and IgG light chain were used as loading control for IP and input, respectively.
Since mitotic protein PARylation is controlled by Tankyrase 1,10 we further examined if α-tubulin is PARylated by Tankyrase 1. We found that loss of Tankyrase 1 by Tankyrase 1 siRNA significantly suppressed the PARylation of α-tubulin during mitosis (Fig. 4D).
ECT2 recognizes PARylated α-tubulin, which regulates cytokinesis
Previous studies have shown that ECT2 is recruited to mitotic spindle during the metaphase and plays a role in chromosome segregation.29 Moreover, PARylation during mitosis is mainly occurred on mitotic spindles.9-10 Since we and others found that α-tubulin, a major scaffold protein in mitotic spindles, is PARylated by Tankyrase 1, we asked whether ECT2 recognizes PARylated α-tubulin. We arrested cell cycle at M phase by Nocodazole treatment and found that α-tubulin interacted with ECT2 only in the M phase cells (Fig. 5A). Reciprocal co-IP shows that ECT2 only interacted with PARylated α-tubulin (Fig. 5B). Moreover, treating precipitated α-tubulin with PARG to remove covalently bound PAR abolished the interaction between α-tubulin and ECT2 (Fig. 5C). Collectively, these results suggest that ECT recognizes PARylated α-tubulin but not unmodified α-tubulin during mitosis. Since K195 in the BRCT domain of ECT2 plays a key role for the interaction with PAR, we mutated K195 into alanine and expressed the K195A mutant of ECT2. Compared with wild type ECT2, the K195A mutant failed to bind PARylated α-tubulin (Fig. 5D). Consistently, the mutant K195A, failed to be recruited to the mitotic spindle during the metaphase (Fig. S1).
Figure 5.
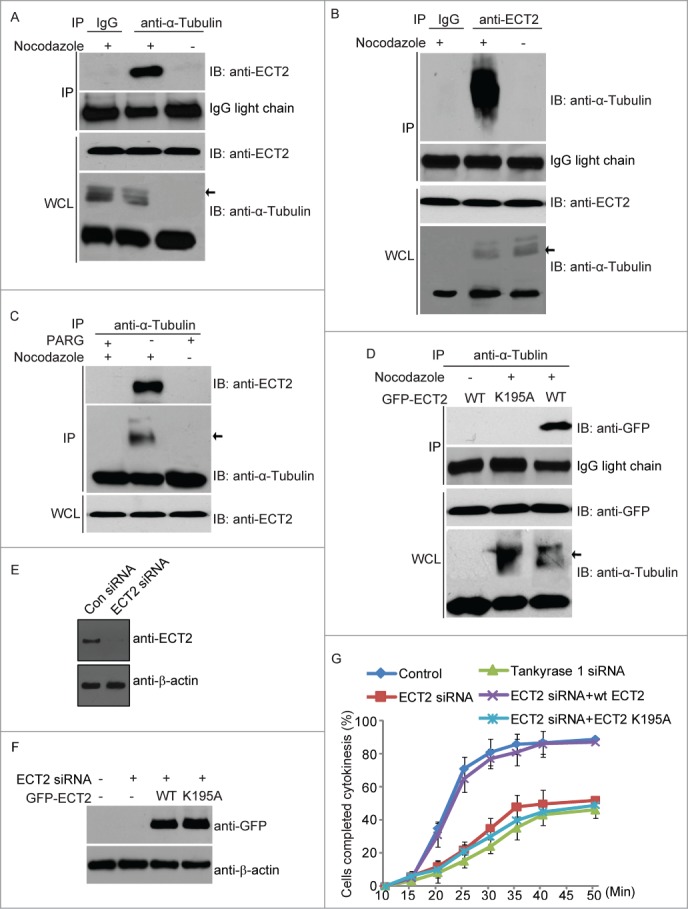
The interaction between PARylated α-tubulin and ECT2. (A) ECT2 interacts with PARylated α-tubulin but not unmodified α-tubulin. Co-IP was performed in Nocodazole treated HeLa cells. The α-tubulin antibody was used for IP and ECT2 antibody used for IB. The whole cell lysates (WCL) of HeLa cells were used as the input. An irrelevant IgG was used as the IP control. IgG light chain was used as IP loading control. Arrow shows the PARylated α-tubulin. (B) Interaction between PARylated α-tubulin and ECT2 was further examined by reverse Co-IP in Nocodazole treated HeLa cells. The ECT2 antibody was used for IP and α-tubulin antibody used for IB. The whole cell lysates (WCL) of HeLa cells were used as the input. An irrelevant IgG was used as the IP control. IgG light chain was used as IP loading control. Arrow shows the PARylated α-tubulin. (C) The interaction between PARylated α-tubulin and ECT2 was abolished by PARG treatment. Co-IP assay was performed by using Nocodazole-treated HeLa cells. The α-tubulin antibody was used for IP. After IP, the sepharose was treated with PARG protein. Then ECT2 and α-tubulin antibody was used for IB. The whole cell lysates (WCL) of HeLa cells were used as the ECT2 input. (D) PARylated α-tubulin interacts with wild type ECT2, but not ECT2 mutant (K195A). GFP-tagged ECT2 and ECT2 mutant (K195A) were transfected into HeLa cells respectively. Co-IP was performed by using the α-tubulin antibody for IP and Myc antibody for IB. The whole cell lysates (WCL) of HeLa cells were used as the input. IgG light chain was used as IP loading control. Arrow shows the PARylated α-tubulin. (E) ECT2 protein was significantly decreased in ECT2 siRNA treated HeLa cells. β-actin was used as loading control. (F) The siRNA-resistant GFP-tagged WT and mutant ECT2 (k195) were expressed in ECT2 siRNA treated HeLa cells. β-actin was used as loading control. (G) The effects of ECT2 and Tankyrase 1 down-regulation on the processes of cytokinesis. Live cells were stained with Hoechst 33342 were counted for cytokinesis in a time course. The effect of ECT2 knockdown on cytokinesis could be rescued by WT ECT2, but not ECT2 K195A. The percentages from at least 3 repeated experiments were expressed as mean ± SD.
The initial recruitment of ECT2 to the mitotic spindle indicates its potential role in chromosome segregation. We found that, compared with wild type ECT2, K195A mutant of ECT2 induced slight delay of chromosome segregation (Figure S2). But chromosome segregation is completed in most cells expressing the K195A mutant. The major function of ECT2 is to facilitate cytokinesis, which is likely through the activation of Rho family GTPase, such as RhoA.11-13,30,31 To study the functional interaction between ECT2 and PARylated α-tubulin during mitosis, we examined ECT2-dependent cytokinesis. In agreement with previous reports, loss of ECT2 by siRNA suppressed cytokinesis (Fig. 5G). However, when cell expressing siRNA-resistant ECT2, cytokinesis is restored. Compared with wild type ECT2, the K195A mutant, abolished the interaction with PARylated α-tubulin, failed to rescue cytokinesis (Fig. 5E–G). Moreover, we also depleted Tankyrase 1 by siRNA and found that loss of Tankyrase also arrested cytokinesis (Fig. 5G). Taken together, these results suggest that the interaction between ECT2 and PARylated α-tubulin is important for cytokinesis.
Discussion
In this study, we found that the BRCT domain of ECT2 is a PAR-binding domain and recognizes PARylated α-tubulin during mitosis, which plays a key role for the completion of cytokinesis. It has been shown that ECT2 and PAR are colocalized on the mitotic spindle together with α-tubulin.9-11 Our study provides the molecular basis of this colocalization phenomenon. The initial recruitment of ECT2 to the mitotic spindle indicates its role in chromosome segregation. However, major of cellular phenotype of loss of ECT2 is cytokinesis failure. Our study shows that recruitment of ECT2 to mitotic spindle is prerequisite for its role in cytokinesis. It is likely that ECT2 is recruited by PARylated α-tubulin to mitotic spindle, where ECT2 meets other partners as ECT2 has several other protein-protein binding domains besides the BRCT domain. Along with the extension of mitotic spindle, ECT2 arrives and is enriched at the middle body and participate in cytokinesis (Fig. 6).
Figure 6.
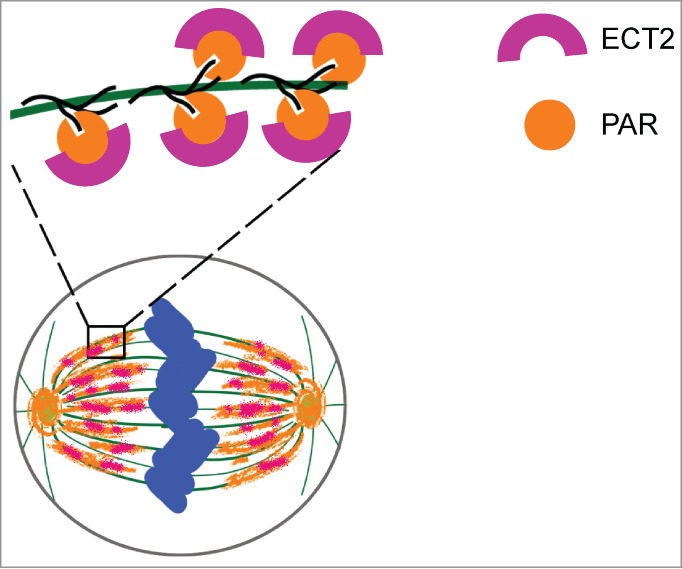
The role of PAR during cytokinesis. ECT2 is recruited by PAR to the spindle during metaphase via binding to PAR by its BRCT domains.
Although PARylation is known to play a key role in mitosis,9-10 the molecular mechanism is still unclear. Here, in our study, we demonstrate ECT2 as the first PAR-binding effector during mitosis. Besides ECT2, it is very likely that many other PAR-binding effectors recognize PARylated substrates during mitosis. Further exploring PAR-binding partners will reveal novel function of PARylation in mitosis.
Materials and Methods
Plasmids, siRNAs and antibodies
For the constructs used in immunofluorescence, ECT2 cDNA was cloned into pEGFP-C1 vector to generate plasmids encoding GFP-ECT2. For GST fusion protein, ECT2 BRCTs (amino acids 1-332) was cloned into the pGEX-4T1 vector. The mutations in the full length of ECT2 and the ECT2 BRCTs were generated using the QuikChange site-directed mutagenesis kit (Stratagene, #200519). The siRNA duplexes were purchased from Dharmacon Research (Lafayette, CO). The sequence of ECT2 siRNA and Tankyrase 1 siRNA were: 5’- UGA AAU GCA AGG AGG UAA AdTdT -3’ and5’ CAU GAA GAG UUG AAA GAA AdTdT 3’, respectively. siRNA was transfected into cells using Oligofectamine (Invitrogen, #12252-011) according to the manufacturer's instructions. Anti-PAR monoclonal antibody was purchased from Trevigen. Anti-GST monoclonal antibody was purchased from Millipore (#05-311). Anti-GFP polyclonal antibody was purchased from Novus (NB600-308). Anti-Tankyrase 1 polyclonal antibody was purchased from R & D (AF7116). Anti-ECT2 polyclonal antibody was purchased from Abcam (ab4863). Anti-α-tubulin monoclonal antibody was purchased from Sigma (T9026).
Immunoprecipitation and western blot
HeLa cells were lysed with NETN-100 buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40) on ice. To protect PAR against PARG degradation, PARG inhibitor, Gallotannin (Enzo Life Science, ALX-270-418), was added to the lysis buffer with the final concentration of 100 μM. Soluble fractions were subjected to Immunoprecipitation and Western blot, and probed with antibodies as indicated.
Generation and purification of PAR
PAR (or biotin labeled PAR) was synthesized and purified in vitro according to the previous work with some modifications.32 Briefly, PAR was synthesized in a 15 ml incubation mixture comprising 100 mM Tris-HCl pH 7.8, 10 mM MgCl2, 1 mM NAD+ (or plus 5 μM biotin-NAD+), 10 mM DTT, 60 mg/ml histone H1, 60 mg/ml histone type IIa, 50 mg/ml octameric oligonucleotide GGAATTCC and 150 nM human PARP-1. The reaction was stopped after 60 min by addition of 20 ml ice-cold 20% TCA. Following precipitation the pellet was washed with ice-cold 99.8% ethanol. Polymer was detached using 0.5 M KOH/50 mM EDTA. After extraction by phenol-chloroform-isoamyl alcohol, PAR was precipitated by isopropanol followed by centrifugation. PAR was then dried in air and stored at -20°C. For the ITC assay, PAR was diluted to the indicated concentrations by the buffer containing 10 mM Na2HPO4 (pH 7.5), 100 mM NaCl.
GST fusion protein expression and dot blot
GST fusion proteins were expressed in Escherichia coli or using the Bac-to-Bac Baculovirus expression system (Invitrogen, #10359-016) for GST-CHFR and purified under standard procedures. Purified GST fusion proteins (10 pmol) were conjugated to the Glutathione beads and incubated with PAR (100 pmol, calculated as the ADP-ribose unit) for 2 hours at 4°C. The beads were washed with NETN-100 buffer four times. GST-fusion proteins were eluted from beads by glutathione and spotted onto a nitrocellulose membrane. The membrane was blocked with TBST buffer supplemented with 5% milk, extensively washed with TBST. After drying in the air, the membrane was examined by anti-PAR antibody.
Pull-down assay
Purified GST fusion proteins (1 pmol) were incubated with biotin-PAR (5 pmol) and streptavidin beads for 2 hours at 4°C. The beads were collected by centrifugation at 600 g for 5 minutes and the supernatant removed. After washing with NETN-100 buffer 4 times, the samples were boiled in the SDS-sample buffer. The eluates were analyzed by Western blot with anti-GST antibody.
Isothermal titration calorimetry (ITC)
ITC was carried out at 16°C with an ITC 200 Microcalorimeter (GE Healthcare). Proteins were dialyzed extensively into the buffer containing 10 mM Na2HPO4 (pH 7.5), 100 mM NaCl at the final concentrations of 20∼60 μM. Ligands (PAR, ADP-ribose) in the injection syringe were also diluted by the same buffer at the final concentration of 150∼750 μM (the concentration of PAR was calculated as the ADP-ribose unit). A typical titration consisted of 19 consecutive 2-μl injections ligands following a pre-injection of 0.4 μl into the protein solution at time intervals of 120 s while stirring at 1000 rpm. Binding isotherms were integrated and analyzed using the software Origin 7.0 (OriginLab) provided by the manufacturer.
Immunofluorescence staining
Cells grown on glass-bottomed culture dishes were fixed with 3% paraformaldehyde for 20 minutes and permeabilized with 0.5% Triton X-100 in PBS for 10 minutes at room temperature. Samples were blocked with 5% goat serum and then incubated with primary antibody for 60 minutes. Samples were washed three times and incubated with secondary antibody for 30 minutes. After further washing, samples were stained with Hoechst 33342 and visualized by a fluorescence microscope. HeLa cells stably expressing RFP-histone H2B were used to observe the chromosome segregation.
Statistical analyses
All experiments were performed in triplicates unless indicated otherwise. Data were presented as mean ± SD.
Funding Statement
This work was supported by the National Institute of Health (CA132755 and CA130899 to XY), and the University of Michigan Cancer Center and GI Peptide Research Center. XY is a recipient of the Era of Hope Scholar Award from the Department of Defense.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Kalisch T, Ame JC, Dantzer F, Schreiber V. New readers and interpretations of poly(ADP-ribosyl)ation. Trends Biochem Sci 2012; 37:381-90; PMID:22766145; http://dx.doi.org/ 10.1016/j.tibs.2012.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang F, Chen Y, Li M, Yu X. The oligonucleotide/oligosaccharide-binding fold motif is a poly(ADP-ribose)-binding domain that mediates DNA damage response. Proc Natl Acad Sci U S A 2014; 111:7278-83; PMID:24799691; http://dx.doi.org/ 10.1073/pnas.1318367111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell 2013; 23:693-704; PMID:23680151; http://dx.doi.org/ 10.1016/j.ccr.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li M, Lu LY, Yang CY, Wang S, Yu X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev 2013; 27:1752-68; PMID:23964092; http://dx.doi.org/ 10.1101/gad.226357.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 2012. 13:411-24; PMID:22713970; http://dx.doi.org/ 10.1038/nrm3376 [DOI] [PubMed] [Google Scholar]
- 6. Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 2006; 7:517-28; PMID:16829982; http://dx.doi.org/ 10.1038/nrm1963 [DOI] [PubMed] [Google Scholar]
- 7. Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev 2005; 19:1951-67; PMID:16140981; http://dx.doi.org/ 10.1101/gad.1331805 [DOI] [PubMed] [Google Scholar]
- 8. Compton DA. Mitosis: PARty time in the spindle. Curr Biol 2005; 15:R178-9; PMID:15753032; http://dx.doi.org/ 10.1016/j.cub.2005.02.047 [DOI] [PubMed] [Google Scholar]
- 9. Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature 2004; 432:645-9; PMID:15577915; http://dx.doi.org/ 10.1038/nature03061 [DOI] [PubMed] [Google Scholar]
- 10. Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol 2005; 7:1133-9; PMID:16244666; http://dx.doi.org/ 10.1038/ncb1322 [DOI] [PubMed] [Google Scholar]
- 11. Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol 1999; 147:921-8; PMID:10579713; http://dx.doi.org/ 10.1083/jcb.147.5.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim JE, Billadeau DD, Chen J. The tandem BRCT domains of Ect2 are required for both negative and positive regulation of Ect2 in cytokinesis. J Biol Chem 2005; 280:5733-9; PMID:15545273; http://dx.doi.org/ 10.1074/jbc.M409298200 [DOI] [PubMed] [Google Scholar]
- 13. Kimura K, Tsuji T, Takada Y, Miki T, Narumiya S. Accumulation of GTP-bound RhoA during cytokinesis and a critical role of ECT2 in this accumulation. J Biol Chem 2000; 275:17233-6; PMID:10837491; http://dx.doi.org/ 10.1074/jbc.C000212200 [DOI] [PubMed] [Google Scholar]
- 14. Iyoda M, Kasamatsu A, Ishigami T, Nakashima D, Endo-Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K. Epithelial cell transforming sequence 2 in human oral cancer. PLoS One 2010; 5:e14082; PMID:21124766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murata Y, Minami Y, Iwakawa R, Yokota J, Usui S, Tsuta K, Shiraishi K, Sakashita S, Satomi K, Iijima T, et al. . ECT2 amplification and overexpression as a new prognostic biomarker for early-stage lung adenocarcinoma. Cancer Sci 2014; 105:490-7; PMID:24484057; http://dx.doi.org/ 10.1111/cas.12363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 2008; 451:81-5; PMID:18172500; http://dx.doi.org/ 10.1038/nature06420 [DOI] [PubMed] [Google Scholar]
- 17. Oberoi J, Richards MW, Crumpler S, Brown N, Blagg J, Bayliss R. Structural basis of poly(ADP-ribose) recognition by the multizinc binding domain of checkpoint with forkhead-associated and RING Domains (CHFR). J Biol Chem 2010; 285:39348-58; PMID:20880844; http://dx.doi.org/ 10.1074/jbc.M110.159855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science 2003; 302:639-42; PMID:14576433; http://dx.doi.org/ 10.1126/science.1088753 [DOI] [PubMed] [Google Scholar]
- 19. Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 2003; 302:636-9; PMID:14576432; http://dx.doi.org/ 10.1126/science.1088877 [DOI] [PubMed] [Google Scholar]
- 20. Shiozaki EN, Gu L, Yan N, Shi Y. Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol Cell 2004; 14:405-12; PMID:15125843; http://dx.doi.org/ 10.1016/S1097-2765(04)00238-2 [DOI] [PubMed] [Google Scholar]
- 21. Williams RS, Lee MS, Hau DD, Glover JN. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat Struct Mol Biol 2004; 11:519-25; PMID:15133503; http://dx.doi.org/ 10.1038/nsmb776 [DOI] [PubMed] [Google Scholar]
- 22. Clapperton JA, Manke IA, Lowery DM, Ho T, Haire LF, Yaffe MB, Smerdon SJ. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nat Struct Mol Biol 2004; 11:512-8; PMID:15133502; http://dx.doi.org/ 10.1038/nsmb775 [DOI] [PubMed] [Google Scholar]
- 23. Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 2005; 123:1213-26; PMID:16377563; http://dx.doi.org/ 10.1016/j.cell.2005.09.038 [DOI] [PubMed] [Google Scholar]
- 24. Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev 2012; 26:417-32; PMID: 22391446; http://dx.doi.org/ 10.1101/gad.183509.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 1999; 342 (Pt 2):249-68; PMID:10455009; http://dx.doi.org/ 10.1042/0264-6021:3420249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huletsky A, de Murcia G, Muller S, Hengartner M, Menard L, Lamarre D, Poirier GG. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem 1989; 264:8878-6; PMID:2498319 [PubMed] [Google Scholar]
- 27. Ogata N, Ueda K, Kawaichi M, Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J Biol Chem 1981; 256:4135-7; PMID:6260786 [PubMed] [Google Scholar]
- 28. Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods 2013; 10:981-4; PMID: 23955771; http://dx.doi.org/ 10.1038/nmeth.2603 [DOI] [PubMed] [Google Scholar]
- 29. Oceguera-Yanez F, Kimura K, Yasuda S, Higashida C, Kitamura T, Hiraoka Y, Haraguchi T, Narumiya S. Ect2 and MgcRacGAP regulate the activation and function of Cdc42 in mitosis. J Cell Biol 2005; 168:221-32; PMID:15642749; http://dx.doi.org/ 10.1083/jcb.200408085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niiya F, Tatsumoto T, Lee KS, Miki T. Phosphorylation of the cytokinesis regulator ECT2 at G2/M phase stimulates association of the mitotic kinase Plk1 and accumulation of GTP-bound RhoA. Oncogene 2006; 25:827-37; PMID:16247472; http://dx.doi.org/ 10.1038/sj.onc.1209124 [DOI] [PubMed] [Google Scholar]
- 31. Su KC, Takaki T, Petronczki M. Targeting of the RhoGEF Ect2 to the equatorial membrane controls cleavage furrow formation during cytokinesis. Dev Cell 2011; 21:1104-15; PMID:22172673; http://dx.doi.org/ 10.1016/j.devcel.2011.11.003 [DOI] [PubMed] [Google Scholar]
- 32. Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res 2007; 35:e143; PMID:17991682; http://dx.doi.org/ 10.1093/nar/gkm944 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.