Abstract
Aurora A kinase plays an important role in several aspects of cell division, including centrosome maturation and separation, a crucial step for the correct organization of the bipolar spindle. Although it has long been showed that this kinase accumulates at the centrosome throughout mitosis its precise contribution to centriole biogenesis and structure has until now not been reported. It is not surprising that so little is known, due to the small size of somatic centrioles, where only dramatic structural changes may be identified by careful electron microscopy analysis. Conversely, centrioles of Drosophila primary spermatocytes increase tenfold in length during the first prophase, thus making any change easily detectable. Therefore, we examined the consequence of the pharmacological inhibition of Aurora A by MLN8054 on centriole biogenesis during early Drosophila gametogenesis. Here, we show that depletion of this kinase results in longer centrioles, mainly during transition from prophase to prometaphase of the first meiosis. We also found abnormal ciliogenesis characterized by irregularly growing axonemal doublets. Our results represent the first documentation of a potential requirement of Aurora A in centriole integrity and elongation.
Keywords: Aurora A kinase, centriole elongation, Drosophila, male meiosis, MLN8054
Introduction
The proper separation of sister chromosomes during cell division and their faithful segregation at the opposite poles of the daughter cells require the correct organization of the meiotic and mitotic spindles. The assembly of these microtubule-based structures requires both centrosomal and acentrosomal routes. Although in some systems the centrosome function is redundant when the centrosomes are present they influence the shape and the orientation of the microtubule network. Moreover, besides its role in the organization of the microtubule network, the centrosome also represents an important coordination center of the cell because it contains cell-cycle regulatory, checkpoint and signaling proteins.1,2 Control of the centrosome number is essential during the cell life, since numerical alterations represent hallmarks of cell transformation and may contribute to genetic instability.3-5
Two orthogonally arranged centrioles, small cylindrical organelles with a beautiful and highly conserved 9 symmetry, are found at the heart of each centrosome.6 Because the centrioles mark the sites where the centrosomal material is recruited, the centrosome dynamics is closely correlated to the centriole duplication cycle.7 Therefore, the centriole duplication has to be accurately regulated so that centrioles and thus the centrosomes duplicate once and only once in even cell cycle, in concert with DNA replication, to avoid the formation of multiple centrosomes and thus multipolar spindles.8-10
Centrioles have a double life and also act as templates for cilia and flagella. During interphase or in quiescent cells the centriole pair migrates to the periphery and the mother one docks to the cell membrane and converts in a basal body that nucleate the ciliary axoneme. Since the primary cilium is implicated in sensing environmental cues and signal transduction pathway, it represents an essential organelle required for animal development and adult homeostasis. Although centrioles and basal bodies represent different functional aspects of the same structure, the basal bodies cannot assemble primary cilia and organize the centrosomal material at the same time. The presence of a primary cilium seems, indeed, to prevent cell division, 11-14 although some exceptions have been described.15,16 Thus, the reenter in mitosis of vertebrate cells needs an additional control, namely the disassembly of the ciliary axoneme. This process releases the basal body from the plasma membrane that migrates inwards and allows the centriole to organize the functional centrosomes that manage the assembly of the bipolar spindle. Some cell cycle regulatory proteins, and among them the serin-threonine kinase Aurora A, have been implicated in the process of ciliary resorption in proliferating cells.17,18
Aurora A was firstly described in Drosophila where the loss of function of this kinase leads to failure of centrosome separation and the formation of spindles with abnormally organized poles, including characteristic monopolar spindles.19 This characteristic phenotype has led to the widely accepted role of Aurora A in centrosome separation, even though the analysis of different model systems reported a range of apparent contradictory defects in the absence of Aurora A.20,21 Aurora A activity is also required to control centrosome maturation, mitotic entry, bipolar spindle assembly, chromosome congression, midzone formation at anaphase and cytokinesis.18,22 These pleiotropic functions of Aurora A depend on the interaction with different proteins that may modulate its activity.23,24
CALK, a distant Aurora A horthologue, has been reported to control the disassembly of the flagellar axoneme in Chlamydomonas after ionic stress, 25 suggesting that phosphorylation-based signaling may play a key role in the mechanism of axoneme microtubule depolymerisation. In mammalian cells, the resorption of the primary cilium depends by the interaction of Aurora A with HEF1, 26 Pitchfork, 27 and calmodulin, 28 to activate the histone deacetylase-6 (HDAC-6) and determine the depolymerisation of the axonemal microtubules. Aurora A also negatively regulates primary cilia assembly during mitosis by interacting with the keratin intermediate filament protein trichoplein that is localized in the subapical region of the centriole.29
Based on the importance of Aurora A in ciliary disassembly we ask whether the inhibition of this kinase activity could also lead to structural defects of the centriole that nucleate the ciliary axonemes. Although, Aurora A has been found at the centrosome in a variety of cells, 30,31 including Drosophila embryos, 32 no relationships with the centriole biogenesis and organization have been reported.
Since centrioles in somatic cells are very short, eventual defects in their organization may escape to conventional immunofluorescence observations and may be only detectable under careful EM analysis. To circumvent this issue we focused on the structure and dynamics of centrioles/CLRs complexes during Drosophila male gametogenesis. The centrioles of Drosophila mature primary spermatocytes are 10 times longer than somatic ones thus representing a good model to investigate structural modifications and length variations. In this study, we sought to examine the effects of Aurora A depletion on centriole/CLRs complexes during Drosophila spermatogenesis upon incubation in MLN8054, a small inhibitory molecule for this kinase.33 MLN8054 is a particularly useful biochemical tool in this context, as it has been demonstrated that the Aurora A inhibition occurs rapidly and is more than 150-fold selective for Aurora A over the family member Aurora B in cultured cells.33
Our data suggest that the Aurora A kinase may be involved in the control of centriole length during Drosophila male meiosis.
Results
Previous studies of aurora mutations in Drosophila have been restricted to the effects on the early mitoses in syncytial embryos obtained by homozygous females and in the neuroblasts divisions in third-instar larval brains. These studies revealed defects in centrosome separation leading to the formation of typical monopolar spindles.19 In the attempt to clarify the role of the aurora kinase during the meiotic divisions we analyzed male gametogenesis in the heterozygous aur209/aur287 pupae. However, we were unable to find a distinct phenotype except a slightly asynchrony of the germ cell divisions within the same cysts that did not affect the normal progression through meiosis. The ultrastructural analysis also failed to reveal specific abnormalities. Since the aurora mutants are weak hypomorphic alleles it is possible that a reduced amount of the protein could be sufficient to allow the proper meiotic progression. Thus, we decided to overcome this limitation by studying the pharmacological inhibition of the aurora kinase during male gametogenesis.
Germ cell line development in Drosophila males starts at the tip of the testis with the asymmetric division of the germ line stem cells that produce the primary spermatogonia. Spermatogonia undergo 4 round of mitosis to originate cysts of 16 primary spermatocytes that after 2 successive meiotic divisions form 64 round spermatids. Spermatids then differentiate in elongated sperm cells. Both cell division types rely on the proper organization of a spindle apparatus that supports the correct segregation of the sister chromosomes. Spermatogonia centrioles duplicate once during each cell cycle in concert with DNA replication, like centrioles of somatic tissues. Thus the early spermatocytes inherit at the end of fourth spermatogonial mitosis one centrosome with 2 orthogonally arranged centrioles that duplicate as the germ cells switch from the cell division program to the extended prophase. Each young primary spermatocyte has, therefore, at the beginning of prophase, 2 pairs of short centrioles that moved to the cell periphery to nucleate an axoneme that pushed against the plasma membrane to form a cilium-like region (CLR).
The product of the uncoordinated (unc) gene, previously identified as a protein involved in the process of centriole/basal body conversion, 34 represents a good marker for the centriole/CLR complex.16 Therefore, we examined the spermatogenesis in flies expressing an Unc-GFP fusion protein to look at the consequences on centriole and CLR organization upon treatment with MLN8054.
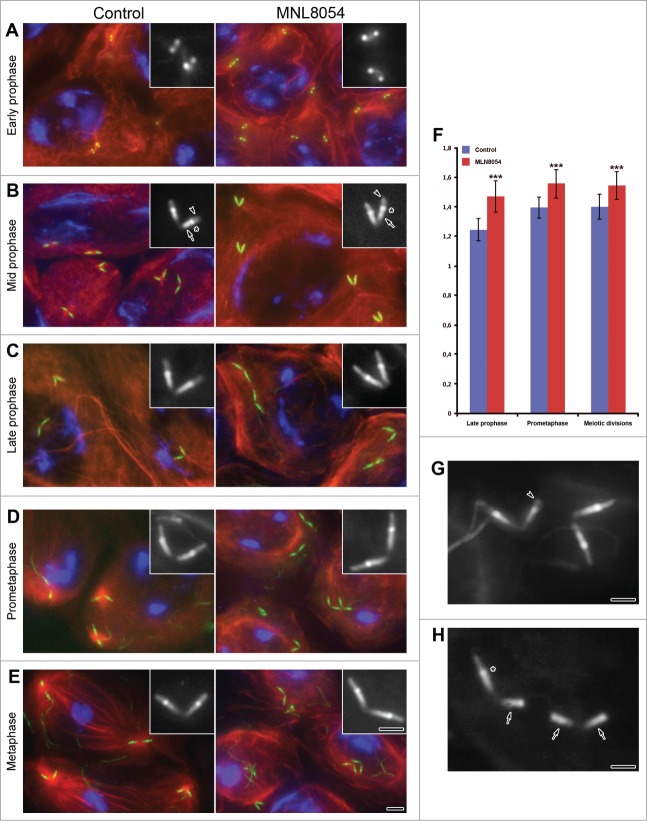
According to previous reports Unc-GFP labeling was not detected in asymmetric dividing germ line stem cells, during the spermatogonial mitoses and in early spermatocytes, but first appeared as 2 pairs of small close spots in apolar spermatocytes (Fig. 1A). As prophase progressed the centriole/CLR complexes elongated (Fig. 1B) and reached their full length in mature spermatocytes (Fig. 1C). The Unc labeling was localized in 3 distinct domains: the mid-apical region of the centriole, the CLR and an intermediate dot-like region (Fig. 1B). This distribution remained unchanged during transition from prometaphase (Fig. 1D) to metaphase (Fig. 1E) and persisted through the whole meiotic process.16
Figure 1.
(See previous page). Aurora-A dependent centriole elongation. Control (left panel) and treated (right panel) primary spermatocytes expressing Unc-GFP were counterstained for acetylated tubulin (red) and DNA (blue). The microtubule cytoskeleton does not show significant differences in control and treated primary prophase spermatocytes (A–C). By contrast, the organization of the cytoplasmic microtubules substantially diverges during prometaphase (D) and metaphase (E) in control and treated spermatocytes: the formers display large asters and bipolar spindles, whereas the others lack both these structures. Magnifications of the centriole/CLR complexes as recognized by Unc-GFP are shown in insets. During early prophase (A) Unc-GFP recognizes small spots in both control and treated spermatocytes. As prophase progressed (B, C) Unc-GFP shows 3 distinct localization: the centriole (arrows, B), the transition region (asterisks, B), the CLR (arrowheads, B). Although the centrioles and the CLRs concurrently elongate during prophase, the centrioles appear slightly longer in late prophase (C), prometaphase (D) and metaphase (E) treated spermatocytes. (F) Quantification of centriole length at different stages of spermatogenesis following 24 hr DMSO (control) or MLN8054 (MLN8054) incubation. Error bars represent SEM. p value from Student's t-test, ***P < 0.0001. Details of abnormally short CLRs (arrowhead, G) or centrioles lacking CLRs (arrows, H); note that the intermediate Unc-GFP dot (asterisk) is only present when the centriole nucleates the ciliary axoneme. Scale bars: (A–E) = 2.5 μm; (A–E) insets, (G, H) = 1 μm.
Therefore, the spermatocytes contain 4 ciliary processes, suggesting that all centrioles within the male germ cells have the same potential. Thus, the insect spermatocytes have 4 virtually mature centrioles that nucleate the ciliary axoneme. This is a remarkable condition since it is generally assumed that only the mother centriole is able to nucleate a ciliary axoneme. Vertebrate cells contain, indeed, one mature centriole, the mother centriole that is able to nucleate a cilium, and an immature centriole, the daughter one that will take full maturation 1.5 cell cycle later.35,36
The distribution of Unc-GFP on centrioles and CLRs of treated spermatocytes did not show significant differences (Fig. 1B–E). However, higher magnification revealed that the centrioles scored from late prophase to meiotic divisions appeared slightly elongated after incubation in MLN8054 (compare insets on left and right panels of Fig.1B-E; Fig. 1F).
The centriole/CLRs complexes were found in control prometaphase cells at the focus of large asters that will organize the forthcoming metaphase spindle. By contrast, distinct meiotic spindles never were found upon MLN8054 incubation and the cytoplasmic microtubules were loosely organized around the nuclear region. However, the chromatin condensed in 2–3 discrete masses as found in control prometaphase cells and compacted further in a single cluster, like that observed in metaphase or later control meiotic stages.
At the onset of the meiotic divisions the centriole/CLRs complexes retracted within a membrane pocket to become the poles of the meiotic spindles, 16,37,38 whereas in treated cells they remained at the cell periphery far from the nuclear region, suggesting a delay in their inward movement. The length of the CLRs slightly decreased in untreated cells during metaphase, whereas this reduction was usually less evident after incubation in MLN8054 (Fig. 1E).
Since, 1 μM concentration of MNL8054 also slightly affects Aurora B in vertebrate cells, 33 we cannot exclude off target activities of the drug against Aurora B. Therefore, the interpretation of the results involving Aurora A inhibition by MLN8054 may be complicated by low levels of Aurora B inhibition. However, the same phenotypes were also found upon incubation in 0.5 μM of MLN8054, a concentration that does not inhibit Aurora B activity.
The tripartite distribution of Unc-GFP was usually found in both control and treated cells. However, a low but significant amount of spermatocytes showed a changed pattern of Unc-GFP upon MLN8054 treatment. 8% of the centriole/CLR complexes examined (46, n = 576) displayed abnormally short CLRs (Fig. 1G) and maintained the intermediate dot-like Unc-GFP labeling. 10% of the centriole/CLR complexes (58, n = 576) lost a distinct CLR and the Unc-GFP was localized on the centriole only, lacking the typical intermediate dot (Fig. 1H).
Since the intermediate Unc-GFP has been correlated with the transition region between the centriole and the ciliary axoneme, 39 we would ascertain whether the abnormal CLRs observed with conventional immunofluorescence upon Aurora A depletion might reflect ultrastructural defects of the centriole/CLRs.
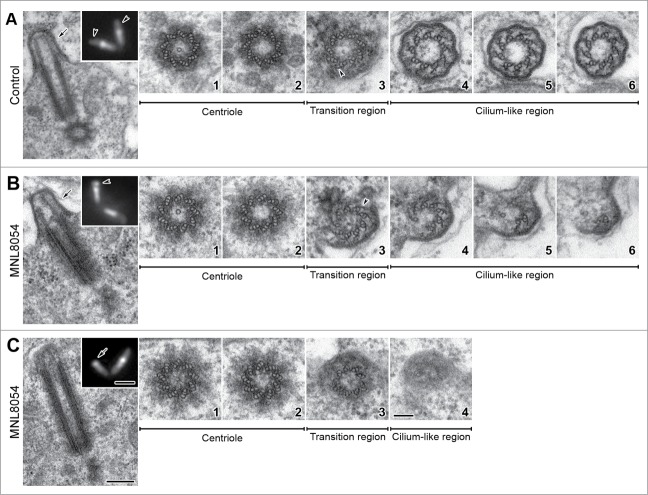
Both control and treated spermatogonia had one pair of short centrioles composed of 9 triplet microtubules and a central cartwheel (not shown). During spermatocyte grow the centriole pairs moved to the cell periphery and docked to the plasma membrane. The A-and B-tubules then elongate at the distal end of the centriole and pushed against the plasma membrane to form the ciliary axoneme.
As prophase proceeded both centrioles and CLRs elongated and during mid prophase the CLRs of untreated spermatocytes reached about half length of the centrioles (Fig. 2A). The basal region of the centriole still contained the cartwheel (Fig. 2A1), whereas the distal lumen was empty (Fig. 2A2). The hallmark of the axoneme formation in Drosophila primary spermatocytes was the reduction at the distal end of the centriole of the C-tubules to hook-like projections that represent cross-sectioned curved longitudinal blades (Fig. 2A3). This region was also marked by the emergence from the B-tubule of thin radial projections that persisted for all the ciliary length (Fig. 2A4–6). Distinct links usually connected the A and B tubules of adjacent doublets (Fig. 2A4–6), reminiscent of the nexin links found in motile cilia.
Figure 2.
Defects in CLR organization upon MLN8054 incubation. Longitudinal and cross sections of centriole/CLR complexes in control (A) and treated (B, C) mid prophase primary spermatocytes; insets represent Unc-GFP localization. Control spermatocytes display distinct CLRs evidenced by Unc-GFP (inset A, arrowheads) that protrude from the cell surface (A, arrow). Occasionally, treated spermatocytes display abnormal CLRs (inset B, arrowhead) with tubules of different length (arrow, B) or elongated centrioles that contact the plasma membrane (C), without nucleating the ciliary microtubules (inset C, arrow). The centrioles associated with normal (A1,2) and abnormal (B1,2, C1,2) CLRs display the same architecture; as usual the cartwheel is present in the basal region only. The transition regions display distinct C-blades (A3, B3, arrowheads) when normal (A4–6) or reduced (B4–6) axonemes are present; C-blades are missing (C3) when the axoneme is lacking (C4). Scale bars: (A–C) insets = 1 μm; (A–C) = 250 nm; A1–6, B1–6, C1–4 = 100 nm.
79% (n = 56) of the growing primary spermatocytes scored by electron microscopy (n = 71) showed the typical organization of the CLRs seen in controls and characterized by 9 microtubule doublets, lateral C-blades and radial links. However, in some cases (21%; n = 15) we found spermatocytes in which the centrioles reached a length comparable to controls but the CLRs appeared abnormal. The CLRs may be, indeed, present but highly disorganized with doublets incomplete (Fig. 2B) or absent (Fig. 2C). These features were consistent with the observations in immufluorescence of short (Fig. 1G, inset Fig. 2B) or lacking (Fig. 1H, inset Fig. 2C) CLRs.
The organization of the ciliary axoneme usually reflects the architecture of the centriole. However, while the ciliary axoneme was disrupted, the architecture of the centriole was unchanged in treated cells and a distinct cartwheel was always present (Fig. B1,2; Fig. C1,2). Thus, we asked whether the conversion of the centriole triplets to the axonemal doublets that occurs in the transition region might present eventual defects in Aurora A depleted spermatocytes.
Cross sections thought the transition regions of the abnormal centriole/CLRs complexes showed that these structures had an apparently normal organization compared to untreated controls and also consist of 9 doublets. The reduction of the C-tubules in hook-like projections was only found, however, when the ciliary axoneme was present (Fig. 2B3). Conversely, the hook-like projections lack (Fig. 2C3) when the transition region did not continue with the ciliary structures (Fig. 2C4). Only the few doublets that extended within the abnormal CLRs maintained, indeed, the remnants of the C-tubules in form of longitudinal blades (Fig. 2B4–6). The links between adjacent doublets were barely detectable only in the proximal region of the short axoneme (Fig. 2B4).
Centrioles and CLRs of both control and treated spermatocytes elongated further and reached their full size in late prophase (Fig. 3A). According to Unc-GFP localization (insets in Fig. 3A,B), EM analysis of late prophase (Fig. 3A) and prometaphase (Fig. 3B) spermatocytes confirmed that the centrioles were more elongated in treated than in control cells.
Figure 3.
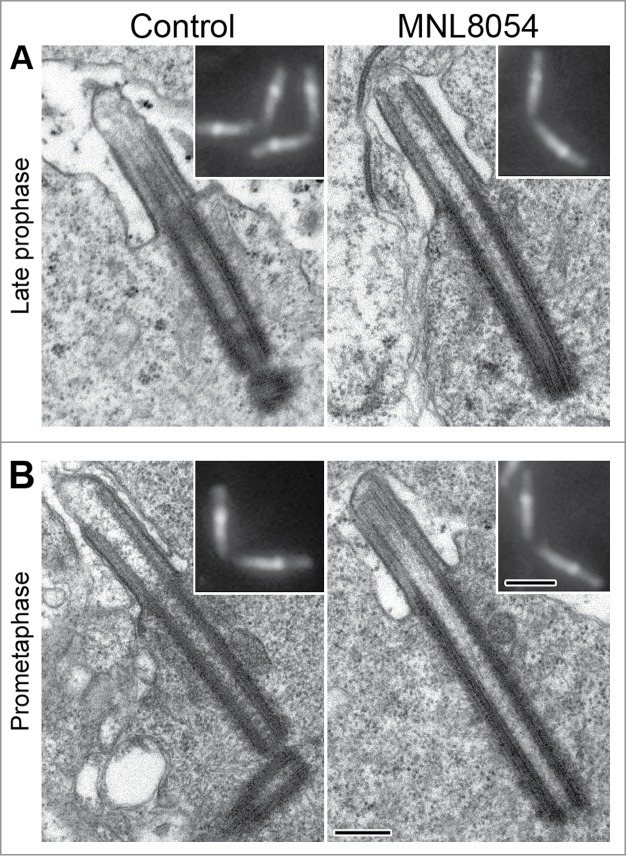
MLN8054 promotes centriole elongation at the onset of meiosis. Longitudinal sections of the centriole/CLR complexes in control (left panel) and treated (right panel) primary spermatocytes during late prophase (A) and prometaphase (B). Insets represent centriole/CLR complexes as recognized by Unc-GFP. Note that centrioles are longer in treated spermatocytes that in controls. Scale bars: insets = 1 μm; A-B = 250 nm.
Discussion
Our findings reveal that the inhibition of the Aurora A activity by the small molecule MNL8054 in Drosophila primary spermatocytes results in more elongated centrioles and, to minor extent, in abnormal CLRs. Therefore, the Aurora A kinase might play a novel role in regulating the centriole biogenesis. However, we find that male gametogenesis is normal in heterozygous aurora209/aurora287 pupae, whereas the brains of third-instar mutant larvae showed monopolar spindles focused on unseparated centrosomes. The normal meiotic phenotype contrasts with the defects of centrosome separations described in syncytial embryos obtained from homozygous females and in larval neuroblasts.19 Why should be easier to observe defects in centrosome dynamics during mitotic divisions? One possible explanation could be that there are different requirements upon aurora function in mitosis and meiosis. One hypothesis could be that a smaller amount of the kinase aurora is need for progression through male gametogenesis. This may be accentuated by the hypomorphic nature of the aurora mutations in which residual function may be sufficient to allow the correct meiotic divisions of the male germ cells. Treatment with MLN8054 might reduce the residual dose of the protein revealing a meiotic phenotype. However, we cannot appreciate feeble variations in centriole length during the early syncytial mitoses or in larval brains due to the reduced dimensions of the centrioles themselves.
A non-mitotic role of Aurora A kinase in controlling the dynamics of the primary cilia has been recently reported in mammalian cells. Aurora A promotes, indeed, the destabilization of the axonemal microtubules and the resorption of the primary cilium. Depletion of this kinase leads to the inactivation of the tubulin deacetylase HDAC6 resulting in stable primary cilia.26–28 However, the centriole/CLRs phenotype observed in Drosophila spermatocytes seems to be incompatible with this function. The MLN8054 treatment leaves unchanged the length of the CLRs during prophase-prometaphase, as it also occurs in control spermatocytes in which the Aurora A kinase activity is present. Rather, we observed abnormally reduced CLRs. This inconsistency is likely to stem from the fact that vertebrate primary cilia and Drosophila CLRs are similar but not homologous structures. Drosophila spermatocyte CLRs diverge, indeed, from conventional primary cilia by several structural aspects.40,41 Moreover, the CLRs persisted during the meiotic divisions until the onset of spermiogenesis when they give origin to the sperm axonemes.16,40 By contrast, true primary cilia disassemble when G0 growth-arrested vertebrate cells re-enter the cell cycle.42,43 This is a critical process needed to enter cell division.12,18,44–47 During cilia disassembly, indeed, the centrioles detach from the plasma membrane to organize the centrosomes that will make the mitotic spindle poles.11
How we can explain that the inhibition of Aurora A in the Drosophila primary spermatocytes results in more elongated centrioles? The axoneme of the mammalian primary cilia is nucleate by a basal body which does not change in length through ciliogenesis, when the ciliary microtubules elongate at their distal plus end by an IFT-mediated process. By contrast, both the centrioles and the associate CLRs grow in length concurrently in Drosophila primary spermatocytes in the absence of IFT. This raises the question of how the centrioles can elongate while at the same time they templates the assembly of the ciliary axoneme and points to novel dynamics at the distal end of these centrioles. The elongation of the A and B tubules and the conversion of the C-tubules in C-blades in the apical region of the centriole could build the proximal part of the growing axoneme. Since, the centrioles do not elongate in Unc mutant spermatocytes in which the aberrant CLRs lack distinct C-blades, 40 we speculate that the reverse transformation of the C-blades in complete C-tubules may lead to the elongation of the centriole. Such transformation would require a dynamic exchange between microtubule polymerization and depolymerisation. Alteration of this balance by microtubule stabilization may lead to longer centrioles. The CLRs of Drosophila spermatocyte are, indeed, directly affected by drugs that target microtubule polymerization. In the presence of nocodazole, a microtubule destabilizer, the axonemes of CLRs failed to assemble, whereas Taxol, a stabilizer, leads to unusually long centrioles and axonemes.48 These findings implicate the requirement of factors that control the dynamics of the transition from centriolar triplet microtubules to the axonemal doublets of the CLRs.
Given that Aurora A accumulates at the centrosomes in most of organisms, 21,40–53 including Drosophila, 54 it is tempting to speculate that this kinase might also regulate the dynamics of the centriole microtubules. Suppression of Aurora-A by small interfering RNA causes an incorrect separation of the centriole pairs in cultured cells, indicating that Aurora-A is essential for the proper execution of this process.55 This mechanism might require the phosphorylation of microtubule-related proteins including some involved in microtubule stabilization/depolymerisation.56–57 Most known Aurora A substrates are associated with centrosomes, 30 and the Aurora A depletion results in disconnection of centrosomes from mitotic spindle poles in Drosophila.59 Moreover, the Aurora A kinase activity is required to target factors involved in microtubule stabilization at the centrosome.60 Aurora A has been reported to play a main function in protecting centrosomal microtubules against depolymerisation by its interaction with TACC complexes in both vertebrate, 61 and Drosophila cells.32 Aurora A negatively regulates the mitotic centromere-associated kinesin (MCAK), 62 and supports the assembly of the central spindle at anaphase by promoting microtubule stabilization.63 Aurora A also regulates the Kinesin-13 microtubule depolymerase Kif2A at the spindle poles of mammalian cells during prometaphase.64
Several members of the microtubule-depolymerising kinesin-13 family have been shown to regulate the length of cilia in mammalian cells, 65 and flagella in protozoans.66–68 However, the only kinesin-13 demonstrated until now to play a role in centriole elongation is the Drosophila Klp10A.69 Drosophila mutant spermatocytes for Klp10A had elongated centrioles that organize irregular CLRs.40 Since the centriole phenotypes found in MLN8054 treated cells is strikingly similar to that seen in Klp10A mutant spermatocytes, it is conceivable that Klp10A may represent a logical target of the Aurora kinase to regulate the transition of centriolar triplet microtubules to doublets during the formation of the ciliary axoneme.
However, Aurora A promotes microtubule stabilization in mammalian cells by negatively regulating the activity of the microtubule depolymerases.20,57,62,70 Thus we should found shorter centrioles in Drosophila spermatocytes when the Klp10A activity is up-regulated in the absence of Aurora A, whereas we observed centrioles longer than usual. This discrepancy may be explained with a different function of Aurora A that during Drosophila spermatogenesis may activate rather than inhibit the Kinesin-13 microtubule depolymerase Klp10A. Alternatively, since many functions attributed to Aurora A are considered to be in part regulated by the interaction with multiple partner proteins, 23,52,71,72 additional actors may be work together to Aurora A in regulating the Klp10A activity at the transition between the centriole and the axoneme. This possibility may be consistent with the observation that some centrioles of treated spermatocytes lack the intermediate Unc-GFP labeling and are unable to nucleate axonemal microtubules. The dot-like Unc-GFP has been correlated, indeed, with the transition region. Mutations in chibby and dilatory that disrupt this region lead to defects in the organization of the ciliary axoneme.39,73
Further studies are, therefore, required to identify key substrates that may interact with Aurora A kinase to control and regulate the centriole dynamics during male meiosis of Drosophila.
Materials and Methods
Drosophila strains
The stock containing the Unc-GFP transgene was described previously.34 The aure209 and aur287 alleles were reported in Glover et al.19 Flies were raised on standard Drosophila medium at 24°C.
Antibodies and reagents
Mouse anti-acetylated tubulin (1:100) was from Sigma–Aldrich. Alexa Fluor 555 secondary antibody (1:800) was purchased from Invitrogen. The chemical inhibitor to Aurora A (MLN8054) was purchased by Selleck. Dimethyl sulfoxide (DMSO) and Sang M3 Insect Medium were purchased from Sigma-Aldrich. MLN8054 was dissolved in DMSO at stock concentration of 1000 μM and stored frozen at 20°C. The stock solution was diluted to the desired concentration in culture medium prior to incubation with testes.
Culture and drug treatment experiments
Testes were dissected from pupae between 5–7 d in M3 medium. To inhibit Aurora A, testes were incubated 24 hours in M3 medium containing 0.5 μM or 1 μM MLN8054 for 24 hours into a 24-well plate at 24°C.
Incubation of testes in M3 medium containing DMSO but lacking MLN8054 had no effect on the structure of the centrioles.
Indirect immunofluorescence staining
After incubation, the testes were washed in M3 medium for 10 minutes and then in phosphate buffered saline (PBS) for 5 minutes. Then testes were fixed as previously reported.41 To visualize microtubules testes were incubated with anti-acetylated tubulin antibody for 4–5 h at room temperature. After washing in PBS–BSA the samples were incubated for 1 h at room temperature with the appropriate secondary antibodies. DNA was visualized with incubation of 3–4 min in Hoechst. Samples mounted in small drops of 90% glycerol in PBS were observed by using an Axio Imager Z1 (Carl Zeiss) microscope equipped with an AxioCam HR cooled charge-coupled camera (Carl Zeiss). Grayscale digital images were collected separately and then pseudocolored and merged using Adobe Photoshop 7.0 software (Adobe Systems).
Transmission electron microscopy
Both drug-treated and control testes were carefully rinsed first in M3 medium and then in phosphate-buffered saline (PBS) for 5 minutes. Samples were pre-fixed in 2.5% glutaraldehyde in PBS overnight at 4°C. After washing in PBS, the testes were post-fixed in 1% osmium tetroxide in PBS for 1h at 4°C. Subsequently the material was rinsed again in PBS, dehydrated through a graded series of Ethanol, and embedded in a mixture of Epon-Araldite resin. Serial ultrathin sections (65–75 nm) were prepared with a Reichert ultramicrotome equipped with a diamond knife, collected with formvar-coated copper grids, and routinely stained with uranyl acetate and lead citrate. TEM observations were performed with a FEI Tecnai G2 Spirit transmission electron microscope operating at an accelerating voltage of 100 kV and equipped with a Morada CCD camera (Olympus).
Statistics
Centrioles from individual cysts of both control and treated spermatocytes were scored based on the Unc-GFP labeling. Only the longitudinal centrioles were measured. Centrioles scored were: control prophase (n = 103), control prometaphase (n = 85), control meiotic divisions (n = 84); treated prophase (n = 202); treated prometaphase (n = 138), treated meiotic divisions (n = 235). The error was measured as standard error of the means (SEM). Significance was measured using the Student's t test. For significance ranking values ***P < 0.0001.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The Authors would like to thank Timothy Megraw and Helene Rangone for generously providing the stocks and Laura Patrussi for help in statistical analysis.
Funding
This work was supported by a grant from PRIN2012 to GC.
References
- 1.Doxsey S, Zimmerman W, Mikule K. Centrosome Control of the Cell Cycle. Trends Cell Biol 2005; 15:303–11; PMID:15953548; http://dx.doi.org/ 10.1016/j.tcb.2005.04.008 [DOI] [PubMed] [Google Scholar]
- 2.Müller H, Schmidt D, Steinbrink S, Mirgorodskaya E, Lehmann V, Habermann K, Dreher F, Gustavsson N, Kessler T, Lehrach H. Proteomic and functional analysis of the mitotic Drosophila centrosome. EMBO J. 2010; 29:3344–57; PMID:20818332; http://dx.doi.org/ 10.1038/emboj.2010.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009; 460:278–82; PMID:19506557; http://dx.doi.org/ 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012; 482:53–8; PMID:22258507; http://dx.doi.org/ 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vitre BD, Cleveland DW. Centrosomes, chromosome instability (CIN) and aneuploidy. Curr Opin Cell Biol 2012; 24:809–15; PMID:23127609; http://dx.doi.org/ 10.1016/j.ceb.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson CG. Choosing sides – asymmetric centriole and basal body assembly. J Cell Sci 2014; 127:2803–10; PMID:24895399; http://dx.doi.org/ 10.1242/jcs.151761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sluder G, Rieder CL. Centriole number and the reproductive capacity of spindle poles. J Cell Biol 1985; 100:887–96; PMID:3972899; http://dx.doi.org/ 10.1083/jcb.100.3.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature 2006; 442:947–51; PMID:16862117; http://dx.doi.org/ 10.1038/nature04985 [DOI] [PubMed] [Google Scholar]
- 9.Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell 2009; 17:344–54; PMID:19758559; http://dx.doi.org/ 10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang WJ, Soni RK, Uryu K, Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J Cell Biol 2011; 193:727–39; PMID:21576395; http://dx.doi.org/ 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos N, Reiter JF. Building it up and taking it down: the regulation of vertebrate ciliogenesis. Dev Dyn 2008; 237:1972–81; PMID:18435467; http://dx.doi.org/ 10.1002/dvdy.21540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. J Cell Biol 2011; 193:435–44; PMID:21536747; http://dx.doi.org/ 10.1083/jcb.201101005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S, Tsiokas L. Cilia and cell cycle re-entry. Cell Cycle 2011; 10:2683–90; PMID:21814045; http://dx.doi.org/ 10.4161/cc.10.16.17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan J, Seeger-Nukpezah T, Golemis EA. The role of the cilium in normal and abnormal cell cycles: emphasis on renal cystic pathologies. Cell Mol Life Sci 2012; 70:1849–74; PMID:22782110; http://dx.doi.org/ 10.1007/s00018-012-1052-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bloodgood RA. From central to rudimentary to primary: the history of an under appreciated organelle whose time has come. The primary cilium. Methods Cell Biol 2009; 94:3–52; PMID:20362083 [DOI] [PubMed] [Google Scholar]
- 16.Riparbelli MG, Callaini G, Megraw TL. Assembly and persistence of primary cilia in dividing Drosophila spermatocytes. Dev Cell 2012; 23:425–32; PMID:22898783; http://dx.doi.org/ 10.1016/j.devcel.2012.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goto H, Inoko A, Inagaki M. Cell cycle progression by the repression of primary cilia formation in proliferating cells. Cell Mol Life Sci 2013; 70:3893–905; PMID:23475109; http://dx.doi.org/ 10.1007/s00018-013-1302-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ke YIN, Yang W. Primary cilium: an elaborate structure that blocks cell division? Gene 2014; 547:175–85; PMID:24971504; http://dx.doi.org/ 10.1016/j.gene.2014.06.050 [DOI] [PubMed] [Google Scholar]
- 19.Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 1995; 81:95–105; PMID:7720077; http://dx.doi.org/ 10.1016/0092-8674(95)90374-7 [DOI] [PubMed] [Google Scholar]
- 20.Hégarat N, Smith E, Nayak G, Takeda S, Eyers PA, Hochegger H. Aurora A and Aurora B jointly coordinate chromosome segregation and anaphase microtubule dynamics. J Cell Biol 2011; 195:1103–13; PMID:22184196; http://dx.doi.org/ 10.1083/jcb.201105058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochegger H, Hégarat N, Pereira-Leal JB. Aurora at the pole and equator: overlapping functions of Aurora kinases in the mitotic spindle. Open Biol 2013; 120185; PMID:23516109; http://dx.doi.org/ 10.1098/rsob.120185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL Jr, Golemis EA. Aurora A kinase (AURKA) in normal and pathological cell division. Cell Mol Life Sci 2013; 70:661–87; PMID:22864622; http://dx.doi.org/ 10.1007/s00018-012-1073-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol 2009; 21:796–805; PMID:19836940; http://dx.doi.org/ 10.1016/j.ceb.2009.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, Jiang Q, Zhang C. The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J Cell Sci 2014; 127:4111–22; PMID:25128564; http://dx.doi.org/ 10.1242/jcs.151753 [DOI] [PubMed] [Google Scholar]
- 25.Pan J, Wang Q, Snell WJ. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev Cell 2004; 6:445–51; PMID:15030766; http://dx.doi.org/ 10.1016/S1534-5807(04)00064-4 [DOI] [PubMed] [Google Scholar]
- 26.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 2007; 129:1351–63; PMID:17604723; http://dx.doi.org/ 10.1016/j.cell.2007.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinzel D, Boldt K, Davis EE, Burtscher I, Trumbach D, Diplas B, Attié-Bitach T, Wurst W, Katsanis N, Ueffing M, et al.. Pitchfork regulates primary cilia disassembly and left-right asymmetry. Dev Cell 2010; 19:66–77; PMID:20643351; http://dx.doi.org/ 10.1016/j.devcel.2010.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plotnikova OV, Nikonova AS, Loskutov YV, Kozyulina PY, Pugacheva EN, Golemis EA. Calmodulin activation of Aurora-A kinase (AURKA) is required during ciliary disassembly and in mitosis. Mol Biol Cell 2012; 23:2658–70; PMID:22621899; http://dx.doi.org/ 10.1091/mbc.E11-12-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoko A, Matsuyama M, Goto H, Ohmuro-Matsuyama Y, Hayashi Y, Enomoto M, Ibi M, Urano T, Yonemura S, Kiyono T, et al.. Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J Cell Biol 2012; 197:391–405; PMID:22529102; http://dx.doi.org/ 10.1083/jcb.201106101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barr AR, Gergely F. Aurora-A: the maker and breaker of spindle poles. J Cell Sci 2007; 120:2987–96; PMID:17715155; http://dx.doi.org/ 10.1242/jcs.013136 [DOI] [PubMed] [Google Scholar]
- 31.Joukov V, De Nicolo A, Rodriguez A, Walter JC, Livingston DM. Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc Natl Acad Sci USA 2010; 107:21022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, Glover DM. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol 2002; 156:437–451; PMID:11827981; http://dx.doi.org/ 10.1083/jcb.200108135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manfredi MG, Ecsedy JA, Meetze KA, Balani SK, Burenkova O, Chen W, Galvin KM, Hoar KM, Huck JJ, LeRoy PJ et al.. Antitumor activity of MLN8054, an orally active small-molecule inhibitor of Aurora A kinase. Proc Natl Acad Sci USA 2007; 104:4106–4111; PMID:17360485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker JD, Adhikarakunnathu S, Kernan MJ. Mechanosensory-defective, male-sterile unc mutants identify a novel basal body protein required for ciliogenesis in Drosophila. Development 2004; 131:3411–3422; PMID:15226257; http://dx.doi.org/ 10.1242/dev.01229 [DOI] [PubMed] [Google Scholar]
- 35.Vorobyev IA, Chentsov YS. Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol 1982; 98:938–949; PMID:7119006; http://dx.doi.org/ 10.1083/jcb.93.3.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azimzadeh J, Marshall WF: Building the centriole. Curr Biol 2010; 20:R816–825; PMID:20869612; http://dx.doi.org/ 10.1016/j.cub.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tates AD. Cytodifferentiation during spermatogenesis in Drosophila melanogaster: an electron microscope study. PhD thesis, Rijksuniversiteit, Leiden, Germany: 1971. [Google Scholar]
- 38.Fritz-Niggli H, Suda T. Bildung und bedeutung der zentriolen: eine studie und neuinterpretation der meiose von drosophila. Cytobiologie 1972; 5:12–41 [Google Scholar]
- 39.Enjolras C, Thomas J, Chhin B, Cortier E, Duteyrat JL, Soulavie F, Kernan MJ, Laurençon A, Durand B. Drosophila chibby is required for basal body formation and ciliogenesis but not for Wg signaling. J Cell Biol 2012;197:313–25; PMID:22508513; http://dx.doi.org/ 10.1083/jcb.201109148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gottardo M, Callaini G, Riparbelli MG. The cilium-like region of the Drosophila spermatocyte: an emerging flagellum? J Cell Sci 2013; 126:5441–52; PMID:24105264; http://dx.doi.org/ 10.1242/jcs.136523 [DOI] [PubMed] [Google Scholar]
- 41.Riparbelli MG, Gottardo M, Glover DM, Callaini G. Inhibition of Polo kinase by BI2536 affects centriole separation during Drosophila male meiosis. Cell Cycle 2014; 13:2064–72; PMID:24802643; http://dx.doi.org/ 10.4161/cc.29083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rieder CL, Jensen L, Jensen C. The resorption of primary cilia during mitosis in a vertebrate (PtK,) cell line. J Ultrastmct Res 1979; 68:173–85; PMID:480410; http://dx.doi.org/ 10.1016/S0022-5320(79)90152-7 [DOI] [PubMed] [Google Scholar]
- 43.Tucker RW, Pardee AB, Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell 1979; 17:527–35; PMID:476831; http://dx.doi.org/ 10.1016/0092-8674(79)90261-7 [DOI] [PubMed] [Google Scholar]
- 44.Kim S, Zaghloul NA, Bubenshchikova E, Oh EC, Rankin S, Katsanis N, Obara T, Tsiokas L. Nde1-mediated inhibition of ciliogenesis affects cell cycle re-entry. Nat Cell Biol 2011; 13:351–60; PMID:21394081; http://dx.doi.org/ 10.1038/ncb2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan J, Snell W. The primary cilium: keeper of the key to cell division. Cell 2007; 129:1255–7; PMID:17604715; http://dx.doi.org/ 10.1016/j.cell.2007.06.018 [DOI] [PubMed] [Google Scholar]
- 46.Quarmby LM, Parker JD. Cilia and the cell cycle? J Cell Biol 2005; 169:707–710; PMID:15928206; http://dx.doi.org/ 10.1083/jcb.200503053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seeley ES, Nachury MV. The perennial organelle: assembly and disassembly of the primary cilium. J Cell Sci 2010; 123:511–8; PMID:20144999; http://dx.doi.org/ 10.1242/jcs.061093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riparbelli MG, Cabrera OA, Callaini G, Megraw TL. Unique properties of Drosophila spermatocyte primary cilia. Biol Open 2013; 2:1137–47; PMID:24244850[http://dx.doi.org/ 10.1242/bio.20135355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva VC, Cassimeris L. Stathmin and microtubules regulate mitotic entry in HeLa cells by controlling activation of both Aurora kinase A and Plk1. Mol Biol Cell 2013; 24:3819–31; PMID:24152729; http://dx.doi.org/ 10.1091/mbc.E13-02-0108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toya M, Terasawa M, Nagata K, Iida Y, Sugimoto A. A kinase-independent role for Aurora A in the assembly of mitotic spindle microtubules in Caenorhabditis elegans embryos. Nat Cell Biol 2011; 13:710–6; http://dx.doi.org/ 10.1038/ncb2242 [DOI] [PubMed] [Google Scholar]
- 51.De Luca M, Brunetto L, Asteriti IA, Giubettini M, Lavia P, Guarguaglini G. Aurora-A and ch-TOG act in a common pathway in control of spindle pole integrity. Oncogene 2008; 27:6539–49; PMID:18663358; http://dx.doi.org/ 10.1038/onc.2008.252 [DOI] [PubMed] [Google Scholar]
- 52.Lukasiewicz KB, Lingle WL. Aurora A, centrosome structure, and the centrosome cycle. Environ Mol Mutagen 2009; 50:602–19; PMID:19774610; http://dx.doi.org/ 10.1002/em.20533 [DOI] [PubMed] [Google Scholar]
- 53.Mahen R, Venkitaraman AR. Pattern formation in centrosome assembly. Curr Opin Cell Biol 2012; 24:14–23; PMID:22245706; http://dx.doi.org/ 10.1016/j.ceb.2011.12.012 [DOI] [PubMed] [Google Scholar]
- 54.Terada Y, Uetake Y, Kuriyama R. Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J Cell Biol 2003; 162:757–763; PMID:12939255; http://dx.doi.org/ 10.1083/jcb.200305048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, Saya H. Aurora A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem 2003; 278:51786–95; PMID:14523000; http://dx.doi.org/ 10.1074/jbc.M306275200 [DOI] [PubMed] [Google Scholar]
- 56.Jang C-Y, Fang G. DDA3 associates with MCAK and controls chromosome congression. Biochem Biophys Res Commun. 2011; 407:610–4; PMID:21426902; http://dx.doi.org/ 10.1016/j.bbrc.2011.03.081 [DOI] [PubMed] [Google Scholar]
- 57.Zhang X, Ems-McClung SC, Walczak CE. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol Biol Cell 2008; 19:2752–65; PMID:18434591; http://dx.doi.org/ 10.1091/mbc.E08-02-0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Asteriti IA, Giubettini M, Lavia P, Guarguaglini G. Aurora-A inactivation causes mitotic spindle pole fragmentation by unbalancing microtubule-generated forces. Mol Cancer 2011; 10:131; PMID:22011530; http://dx.doi.org/ 10.1186/1476-4598-10-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romé P, Montembault E, Franck N, Pascal A, Glover DM, Giet R. Aurora A contributes to p150(glued) phosphorylation and function during mitosis. J Cell Biol 2010; 189:651–9; PMID:20479466; http://dx.doi.org/ 10.1083/jcb.201001144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sardon T, Peset I, Petrova B, Vernos I. Dissecting the role of Aurora A during spindle assembly. EMBO J. 2008; 27:2567–79; PMID:18756265; http://dx.doi.org/ 10.1038/emboj.2008.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barros TP, Kinoshita K, Hyman AA, Raff JW. Aurora A activates D-TACC-Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules. J Cell Biol 2005; 170:1039–46; PMID:16186253; http://dx.doi.org/ 10.1083/jcb.200504097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. A Complex of Kif18b and MCAK Promotes Microtubule Depolymerization and Is Negatively Regulated by Aurora Kinases. Curr Biol 2011; 21:1356–65; PMID:21820309; http://dx.doi.org/ 10.1016/j.cub.2011.07.017 [DOI] [PubMed] [Google Scholar]
- 63.Lioutas A, Vernos I. Aurora A kinase and its substrate TACC3 are required for central spindle assembly. EMBO Rep 2013; 14:829–36; PMID:23887685; http://dx.doi.org/ 10.1038/embor.2013.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jang CY, Coppinger JA, Seki A, Yates JR, Fang G. Plk1 and Aurora A regulate the depolymerase activity and the cellular localization of Kif2a. J Cell Sci 2009; 122:1334–41; PMID:19351716; http://dx.doi.org/ 10.1242/jcs.044321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell 2011; 145:914–25; PMID:21620453; http://dx.doi.org/ 10.1016/j.cell.2011.04.028 [DOI] [PubMed] [Google Scholar]
- 66.Blaineau C, Tessier M, Dubessay P, Tasse L, Crobu L, Pagès M, Bastien P. A novel microtubule-depolymerizing kinesin involved in length control of a eukaryotic flagellum. Curr Biol 2007; 17:778–82; PMID:17433682; http://dx.doi.org/ 10.1016/j.cub.2007.03.048 [DOI] [PubMed] [Google Scholar]
- 67.Dawson SC, Sagolia MS, Mancuso JJ, Woessner DJ, House SA, Fritz-Laylin L, Cande WZ, et al.. Kinesin-13 regulates flagellar, interphase, mitotic microtubule dynamics in Giardia intestinalis. Eukaryot Cell 2007; 6:2354–64; PMID:17766466; http://dx.doi.org/ 10.1128/EC.00128-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Piao T, Luo MW, Wang L, Guo Y, Li D, Li P, Snell WJ, Pan J. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc Nat Acad Sci USA 2009; 106:4713–8; PMID:19264963; http://dx.doi.org/ 10.1073/pnas.0808671106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Delgehyr N, Rangone H, Fu J, Mao G, Tom B, Riparbelli MG, Callaini G, Glover DM. Klp10A, a microtubule-depolymerizing kinesin-13, cooperates with CP110 to control Drosophila centriole length. Curr Biol 2012; 22:502–9; PMID:22365849; http://dx.doi.org/ 10.1016/j.cub.2012.01.046 [DOI] [PubMed] [Google Scholar]
- 70.Knowlton AL, Vorozhko VV, Lan W, Gorbsky GJ, Stukenberg PT. ICIS and Aurora B coregulate the microtubule depolymerase Kif2a. Curr Biol 2009; 19:758–63; PMID:19327998; http://dx.doi.org/ 10.1016/j.cub.2009.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta 2008; 1786:60–72; PMID:18662747 [DOI] [PubMed] [Google Scholar]
- 72.Reboutier D, Troadec MB, Cremet JY, Chauvin L, Guen V, Salaun P, Prigent C. Aurora A is involved in central spindle assembly through phosphorylation of Ser 19 in P150Glued. J Cell Biol 2013; 201: 65–79; PMID:23547029; http://dx.doi.org/ 10.1083/jcb.201210060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ma L, Jarman AP. Dilatory is a Drosophila protein related to AZI1 (CEP131) that is located at the ciliary base and required for cilium formation. J Cell Sci 2011; 124:2622–30; PMID:21750193; http://dx.doi.org/ 10.1242/jcs.084798 [DOI] [PMC free article] [PubMed] [Google Scholar]