Abstract
The high-affinity binding of the Tus protein to specific 21-bp sequences, called Ter, causes site-specific, and polar, DNA replication fork arrest in E coli. The Tus-Ter complex serves to coordinate DNA replication with chromosome segregation in this organism. A number of recent and ongoing studies have demonstrated that Tus-Ter can be used as a heterologous tool to generate site-specific perturbation of DNA replication when reconstituted in eukaryotes. Here, we review these recent findings and explore the molecular mechanism by which Tus-Ter mediates replication fork (RF) arrest in the budding yeast, S. cerevisiae. We propose that Tus-Ter is a versatile, genetically tractable, and regulatable RF blocking system that can be utilized for disrupting DNA replication in a diverse range of host cells.
Keywords: DnaB helicase, replication fork, homologous recombination repair, RecQ helicase, MCM helicase
Introduction
Replication fork (RF) stalling can occur when the DNA replication machinery encounters DNA adducts, secondary structures, topological constraints, or DNA-bound proteins. Failure to adequately resolve or counteract these obstacles may lead to unresolved DNA structures persisting into mitosis1 and/or the induction of pathological genome rearrangements.2 In some cases, these RF arrests are programmed replication pauses, caused by the binding of specific RF-arresting proteins to specific loci to coordinate DNA replication with important physiological processes.3 A number of these well-characterized RF-arresting systems have subsequently been exploited to further understand how stalled RFs are processed in vivo. By placing these systems at ectopic sites, they permit the detailed and controlled analysis of molecular events occurring at single perturbed RF at a unique genomic locus.4-6 This also circumvents the requirement for DNA damaging agents that cause multiple types of lesions at various sites throughout the genome, and subsequent activation of either a cell cycle checkpoint-mediated growth delay or loss of viability.
A system that shows exceptional promise as a tool to induce site-specific DNA replication perturbation tool is the E coli Tus-Ter system. This system exploits the high-affinity binding of the E coli terminator protein, Tus, to specific 21-bp DNA sequences called Ter.7 This system is utilized in certain species of bacteria to ensure that polar, site-specific, DNA replication termination occurs diametrically opposite to the single replication origin, oriC.8 Three groups have independently demonstrated recently that Tus-Ter can function as an RF barrier when reconstituted in yeast,9 mouse,10 or human cells (S. N. Powell, unpublished observation). The heterologous nature of Tus-Ter when introduced into non-bacterial organisms is an important aspect, because endogenous (programmed) RF pauses are established and dealt with differently than are 'alien' RF impediments.9,11 The Tus-Ter system therefore most likely induces cellular responses similar to those that occur when the replisome encounters certain types of DNA damage.9,10
Reconstitution of the Tus-Ter system in eukaryotes has already been used successfully to glean important information about the molecular events occurring at stalled RFs. Willis et al. integrated 6xTer sequences and an associated homologous recombination (HR) reporter into the genome of mouse cells to provide direct evidence that Brca1, Brca2 and RAD51 regulate HR at stalled RFs.10 In the absence of Brca1, Brca2, or RAD51, an altered outcome of (increased 'long-tract') HR products was detected at the HR reporter harboring Tus-Ter. Importantly, this result was not observed when HR was induced by an I-Sce-I-mediated site-specific double strand DNA break (DSB). Therefore, these data suggest that HR is regulated differently at stalled RFs, as compared to (DNA replication-independent) DSBs. This important study validates the Tus-Ter system as a tool to further understand the biological functions of the important tumor suppressor proteins, Brca1 and Brca2. For example, this system will serve as a novel tool to examine BRCA1 and BRCA2 “variants of uncertain significance” in the human population.12 These are alleles that have yet to be fully ascertained as high or low-risk BRCA mutations. By testing if any of these variants promote aberrant HR at stalled RFs, the Tus-Ter system could classify high-risk mutations that are likely to contribute to cancer predisposition. Other ongoing studies are aimed at testing the contribution other key proteins (as well as variants of these) that have also been implicated in regulating stalled RFs.
In yeast, we demonstrated recently that a tandem array of 3× or 7× Tus-Ter barriers causes transient RF arrest in S. cerevisiae.9 A major advantage of yeast for these studies is that DNA replication intermediates can easily be observed in the genome using the well-established 2D gel electrophoresis technique.13 Importantly, we demonstrated that Tus-Ter in yeast retains its intrinsic polarity for arresting RFs. This finding was somewhat unexpected, given the important role played by putative interactions between Tus and the replicative helicase in E. coli, DnaB.14,15 However, this intrinsic polarity can now be exploited in several ways in yeast. For example, Ter arrays arranged in the non-blocking (permissive) orientation provide an ideal control for the analysis of RF blocking by Ter sites arranged in the restrictive orientation.
Using the Tus-Ter system in yeast, we demonstrated that RF pausing at restrictive Tus-Ter barriers elicits the formation of unprocessed HR intermediates (that are detectable as X-shaped DNA molecules on 2D gels) in sgs1 mutants.9 Sgs1 is an evolutionarily conserved RecQ helicase that is required for the dissolution of Holliday junction-containing HR intermediates.16 These unprocessed HR intermediates are similar to those observed (genome-wide) in sgs1 mutants treated with DNA adduct-generating agents such as MMS and 4NQO.9,17,18, Importantly, although the HR machinery is clearly active at RFs stalled at Tus-Ter, it is not required for RF resumption at Tus-Ter barriers in yeast. This contrasts with the well characterized (and programmed) RTS1 barrier in S pombe.6,19 Therefore, we propose that the unprocessed HR intermediates formed at Tus-Ter in sgs1 mutants probably arise due to defects in completing the post-replicative gap filling of ssDNA gaps by HR. Indeed, post-replicative ssDNA gaps have been detected in yeast cells by electron microscopy following MMS or UV exposure.20,21 The Tus-Ter system will serve, therefore, as an important tool to analyze the role of Sgs1 (and, by inference, of its human ortholog, BLM) at the resolution level of a single stalled RF, without the need to perturb genome-wide DNA replication using DNA-damaging agents.
Despite their seemingly common ability to trigger aberrant HR events at stalled RFs, the Tus-Ter systems show a number of key differences in yeast and mouse cells. Most notably, 6xTus-Ter is proposed to cause bidirectional RF stalling and a subsequent 2-ended break in mouse cells,10 whereas 7xTus-Ter causes transient (and one-sided) RF stalling, without any detectable breaks, in yeast.9 Therefore, it is probable that RF-stalling at Tus-Ter barriers has different consequences in yeast and mouse cells. One way these seemingly disparate differences may be reconciled, however, is that cellular responses to Tus-Ter barriers probably depends on a number of key parameters, including: a) organism- or cell type-specific responses to DNA replication impediments, b) the precise mechanisms of Tus-Ter arrest (discussed further below), and c) the location of the Tus-Ter barriers in the host genome. Importantly, we have demonstrated that increasing the number of Ter sites correlates with an increased blocking efficiency in yeast.9 Given the small size (21-bp) of individual Ter sites, it will be possible to multimerize these and create stronger RF barriers in intrinsically difficult-to-replicate regions of the genome. Our ongoing studies aim to analyze stalled RFs and their biological consequences in these contexts.
Two, non-mutually exclusive, mechanisms have been proposed to explain how Tus-Ter functions as a polar RF barrier in E coli. One proposal is that Tus-Ter has intrinsic and polar RF-arresting capabilities, forming a tight 'lock' that is mediated through the capture of a 'flipped' C6 residue in Ter that is revealed upon dsDNA unwinding.22 However, this so called 'molecular mousetrap' model has evolved to specifically arrest the 5′→3′ DnaB helicase, and should theoretically not function in non-bacterial organisms with 3′→5′ replicative helicases (which would sequester the C6 residue within the central chamber of the helicase). The second mechanistic proposal for Tus-Ter function is that specific interactions between Tus and the E coli replicative helicase, DnaB, are required to elicit polar RF arrest.14,15 However, as there are no DnaB orthologs in yeast23 and, as discussed above, the yeast MCM helicase has the opposite strand polarity to that of DnaB,24 it is difficult to envisage how Tus-Ter could elicit polar RF arrest through specific protein-protein interactions in yeast. One way that these 2 models of polar RF arrest at Tus-Ter can be reconciled, however, is that an intrinsic RF-arresting activity in Tus-Ter initiates the RF barrier, but this is then reinforced through specific protein-protein interactions. This latter mechanism would only contribute to RF fork blocking in E coli, and could explain why the Tus-Ter RF barrier is apparently ∼15-fold less efficient at holding stalled RFs in yeast (that lack this putative reinforcement step) than in E coli.9
To further explore the mechanism of RF arrest elicited by Tus-Ter in yeast, we compared the in vivo RF-arresting ability of 3 previously validated Tus amino acid substitutions: E47Q, E49K and F140A. The E47Q substitution causes enhanced Ter interaction,25 and functions as an effective block to E coli DnaB in in vitro assays.14 However, this mutation reduces Tus-Ter RF arrest in vivo,25 suggesting that Glu47 is critical to reinforce RF arrest in E coli. The E49K mutation does not affect binding to Ter, but it fails to support RF arrest in any of the in vivo or in vitro assays tested so far.14 Finally, the F140A mutation has been proposed to abolish the locking-C mechanism of Tus-Ter. This mutation causes a 10-fold increase in Tus-Ter interaction, but a corresponding 18-fold reduction in the half-life of the locked configuration.22
Results
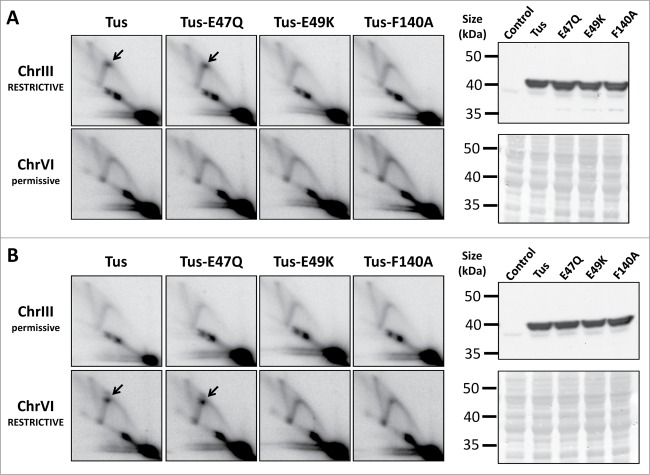
To directly compare the mutated versus non-mutated Tus alleles, we used previously validated strains that harbor either restrictive or permissive configurations of 3xTer arrays positioned to the right of ARS305 and ARS607, respectively.9 ChrIIIRESTRICTIVE/ChrVIpermissive and ChrIIIpermissive/ChrVIRESTRICTIVE strains harboring a low-copy GAL1-regulated plasmid containing HA-tagged wild-type Tus, or HA-Tus with E47Q, E49K, or F140A substitutions, were synchronized in G1 with α-factor pheromone. Expression of Tus was induced during the cell synchronization step. Similar levels of protein expression were confirmed for all 4 of the Tus constructs by Western blotting (Fig. 1). Cells were then released from G1-arrest and DNA replication intermediates were analyzed at ChrIII and ChrVI by 2D gel electrophoresis13. We confirmed that expression of HA-Tus caused detectable RF pausing at ChrIIIRESTRICTIVE, but not ChrVIpermissive (Fig. 1A). As shown previously, the reciprocal effect was observed in the ChrIIIpermissive/ChrVIRESTRICTIVE strain (Fig. 1B), consistent with Tus-Ter being a polar RF-arresting complex that operates at several different loci when reconstituted in the yeast genome.9 Direct comparison of wild type Tus with the E47Q, E49K and F140A mutants revealed that only Tus-E47Q was proficient at arresting RFs (Fig. 1). Furthermore, neither Tus, nor any of the mutated Tus constructs examined here, could elicit RF-pausing when Ter sites were arranged in the permissive orientation (Fig. 1). Therefore, our data demonstrate that Glu47 is not required for Tus to arrest RFs in yeast, despite its critical in vivo role in E coli.25 The Glu49 and Phe140 residues of Tus, however, are required to support efficient RF-arresting in yeast. The mechanistic interpretation of these findings is discussed further below.
Figure 1.
Mutational analysis of the Tus-Ter complex when reconstituted in S. cerevisiae. Yeast strains engineered with either restrictive (blocking) or permissive (non-blocking) 3xTerB modules adjacent to ARS305 (on ChrIII) and ARS607 (on ChrVI) were transformed with a low-copy GAL1-regulated plasmid containing HA-tagged wild-type Tus, or HA-Tus with E47Q, E49K, or F140A substitutions. The 3xTerB modules examined here were arranged in either (A) the ChrIII RESTRICTIVE / ChrVI permissive orientation, or (B) the reciprocal ChrIII permissive / ChrVI RESTRICTIVE configuration. At both of these genomic loci, replication forks emanating from either ARS305 or ARS607 replicate these 3xTerB modules from left to right. Cultures were synchronized in G1 with α-factor pheromone, and expression of Tus was induced for 2.5 hours during the cell synchronization step. Following a 35 min release from G1-arrest, cells were harvested for 2D gel analysis (left panels), or Western blotting for HA-Tus (right panels). The 2D gel images show DNA replication intermediates detectable at (a BamHI-HindIII fragment of) ChrIII and (a HindIII-HindIII fragment of) ChrVI restriction fragments. Paused RFs at Tus-Ter are indicated by the black arrow. Western blotting confirmed equivalent levels of wild type and mutated HA-Tus proteins at this time point. A control (uninduced HA-Tus) sample was also included as a negative control for the Western blot analysis. Membranes were stained with Ponceau S to confirm equivalent protein loading (lower right panels).
Discussion
Of the 3 mutated Tus alleles (E47Q, E49K and F140A) tested here, only Tus-E47Q was proficient at arresting RFs at Tus-Ter barriers in yeast. The Glu47 and Glu49 residues of Tus lie in the so-called “L1 loop,” which comprises a highly charged domain that is located on the restrictive face of Tus that causes arrest of the DnaB helicase. The E47Q and E49K substitutions have previously been demonstrated to cause a reduced ability to arrest DNA replication in E coli, without negatively affecting the affinity of Tus for Ter sites.14,25 Furthermore, the E47Q and E49K substitutions cause significant loss of Tus-DnaB interactions, indicating that these interactions are required for robust RF arrest in E coli.14,15 However, it was also demonstrated previously that Tus-E47Q, but not Tus-E49K, could support RF arrest in vitro.14 Our data in yeast therefore appear consistent with the in vitro, rather than the in vivo, data for Tus-Ter. Taken together, we propose that the E47K substitution does not abolish the intrinsic RF-arresting activity of Tus-Ter, but that the Glu47 residue is probably required to reinforce or sustain the Tus-Ter block once it forms in E coli.25 The absence of this reinforcement/stabilization mechanism in yeast probably explains why Tus-Ter is ∼15-fold less efficient at holding RFs in yeast than in E coli.9 If true, the failure of the Tus-E49K mutant to support RF arrest in any Tus-Ter assays suggests that this particular mutation may disrupt the intrinsic RF-arresting activity of Tus-Ter, in addition to abolishing the specific Tus-DnaB interactions.14
The F140A mutation was also unable to support RF arrest at Tus-Ter in yeast. Because this mutation increases the affinity of Tus for Ter, but reduces the half-life of the locked-C configuration,22 this further suggests that the locking mechanism is required for Tus-Ter to function in yeast. Curiously, Tus-F140A causes RF arrest at 6xTer in mouse cells and also induces higher levels of HR than wild-type Tus.10 As discussed above, this suggests that the Tus-Ter blocking mechanism is different in yeast and mouse cells. We propose, therefore, that the bidirectional RF arrest elicited by Tus (or Tus-F140A) in mouse cells arises due to the high affinity Tus-Ter interaction, without the need to form the locked-C6 intermediate per se. If true, this suggests that either the basal Tus-Ter interaction is enhanced or prolonged in mouse cells as compared to yeast, or that the mouse replication machinery or stress response is intrinsically more sensitive to DNA replication impediments. With regards to the latter possibility, it is worth noting that mammalian cells have a much more complex Fanconi anemia pathway for dealing with RF stress than that proposed to exist in yeast.26,27
Although our mutational analyses are consistent with the locking-C6 model22 for polar RF-stalling at Tus-Ter (Fig. 1), it still remains unclear how such a mechanism could occur when the yeast replisome machinery encounters Tus-Ter. Because the strand polarity of the MCM helicase is opposite to that of DnaB,24 the C6 residue of Ter is predicted to be sequestered within the central channel of the MCM helicase and therefore unable to interact with Tus. Further experiments will be required to fully understand how this process works in yeast, and should provide a novel insight into how Tus-Ter and/or the yeast replisome operates in vivo. One possibility is that DNA unwinding of Ter does not occur solely within the central chamber of the MCM helicase. In this scenario, we speculate that Tus-Ter could form a tightly bound protein-DNA complex that is highly responsive to the positive supercoiling generated by the approaching helicase. As the replisome approaches, a topologically-induced conformation change in the Tus-Ter complex could perhaps promote some localized/transient melting within Ter and trigger the subsequent locking of the C6 residue within the Phe140 pocket. Possible evidence in support of this theory comes from the observation that topA mutations (with increased levels of negative supercoiling) interfere with the ability of Tus-Ter to arrest RFs.28 Another possible scenario is that an as-yet-unidentified DNA helicase is recruited to the replisome (perhaps in response to Tus-Ter-induced positive supercoiling), and the subsequent DNA unwinding catalyzed by this enzyme can then unwittingly induce the Tus-Ter “mousetrap.” Although the Rrm3 helicase was a possible candidate for this,11,29 we observed that loss of Rrm3 had no obvious effects on the establishment, or resolution, of 3xTus-Ter barriers in yeast.9 Future studies should be aimed therefore at identifying which proteins are recruited to RFs stalled at Tus-Ter. This will provide an insight both into how the Tus-Ter barrier is elicited, and how it is resolved.
Recent advances in genome editing capabilities in mammalian cells will greatly extend the capabilities of Tus-Ter to understand site-specific RF stalling. Coupled with its genetic tractability, we propose that the use of Tus-Ter as a novel tool to induce site-specific DNA replication perturbation in any genomic locus or cell type has the potential to contribute greatly to the DNA replication field in the way that site-specific endonucleases such as I-Sce-I and HO-endonuclease subsequently revolutionized the DNA DSB repair field.30,31
Funding Statement
Work on yeast in the authors' laboratory is supported by the European Research Council, The Danish Science Research Council (FNU) and The Novo Nordisk Foundation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ralph Scully and Nicholas Dixon for helpful comments, Deepak Bastia for advice on Tus mutations to analyze, and Simon Powell for sharing unpublished data.
References
- 1. Mankouri HW, Huttner D, Hickson ID. How unfinished business from S-phase affects mitosis and beyond. EMBO J 2013; 32:2661-71; PMID:24065128; http://dx.doi.org/ 10.1038/emboj.2013.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lambert S, Carr AM. Impediments to replication fork movement: stabilisation, reactivation and genome instability. Chromosoma 2013; 122:33-45; PMID:23446515; http://dx.doi.org/ 10.1007/s00412-013-0398-9 [DOI] [PubMed] [Google Scholar]
- 3. Bastia D, Zaman S. Mechanism and physiological significance of programmed replication termination. Semin Cell Dev Biol 2014; 30:165-73; PMID:24811316; http://dx.doi.org/ 10.1016/j.semcdb.2014.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahn JS, Osman F, Whitby MC. Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast. The EMBO J 2005; 24:2011-23; PMID:15889146; http://dx.doi.org/ 10.1038/sj.emboj.7600670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev 2005; 19:1905-19; PMID:16103218; http://dx.doi.org/ 10.1101/gad.337205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lambert S, Watson A, Sheedy DM, Martin B, Carr AM. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 2005; 121:689-702; PMID:15935756; http://dx.doi.org/ 10.1016/j.cell.2005.03.022 [DOI] [PubMed] [Google Scholar]
- 7. Hill TM, Marians KJ. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Nat Acad Sci USA 1990; 87:2481-5; PMID:2181438; http://dx.doi.org/ 10.1073/pnas.87.7.2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hill TM, Henson JM, Kuempel PL. The terminus region of the Escherichia coli chromosome contains two separate loci that exhibit polar inhibition of replication. Proc Nat Acad Sci USA 1987; 84:1754-8; PMID:3550796; http://dx.doi.org/ 10.1073/pnas.84.7.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Larsen NB, Sass E, Suski C, Mankouri HW, Hickson ID. The Escherichia coli Tus-Ter replication fork barrier causes site-specific DNA replication perturbation in yeast. Nat Commun 2014; 5:3574; PMID:24705096; http://dx.doi.org/ 10.1038/ncomms4574 [DOI] [PubMed] [Google Scholar]
- 10. Willis NA, Chandramouly G, Huang B, Kwok A, Follonier C, Deng C, Scully R. BRCA1 controls homologous recombination at Tus/Ter-stalled mammalian replication forks. Nature 2014; 510:556-9; PMID:24776801; http://dx.doi.org/ 10.1038/nature13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol cell 2003; 12:1525-36; PMID:14690605; http://dx.doi.org/ 10.1016/S1097-2765(03)00456-8 [DOI] [PubMed] [Google Scholar]
- 12. Easton DF, Deffenbaugh AM, Pruss D, Frye C, Wenstrup RJ, Allen-Brady K, Tavtigian SV, Monteiro AN, Iversen ES, Couch FJ, et al. . A systematic genetic assessment of 1433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet 2007; 81:873-83; PMID:17924331; http://dx.doi.org/ 10.1086/521032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liberi G, Cotta-Ramusino C, Lopes M, Sogo J, Conti C, Bensimon A, Foiani M. Methods to study replication fork collapse in budding yeast. Method Enzymol 2006; 409:442-62; PMID:16793417; http://dx.doi.org/ 10.1016/S0076-6879(05)09026-9 [DOI] [PubMed] [Google Scholar]
- 14. Mulugu S, Potnis A, Shamsuzzaman, Taylor J, Alexander K, Bastia D. Mechanism of termination of DNA replication of Escherichia coli involves helicase-contrahelicase interaction. Proc Nat Acad Sci USA 2001; 98:9569-74; PMID:11493686; http://dx.doi.org/ 10.1073/pnas.171065898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bastia D, Zzaman S, Krings G, Saxena M, Peng X, Greenberg MM. Replication termination mechanism as revealed by Tus-mediated polar arrest of a sliding helicase. Proc Nat Acad Sci U S A 2008; 105:12831-6; PMID:18708526; http://dx.doi.org/ 10.1073/pnas.0805898105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bizard AH, Hickson ID. The dissolution of double holliday junctions. Cold Spring Harb Perspect Biol 2014; 6:a016477; PMID:24984776; http://dx.doi.org/ 10.1101/cshperspect.a016477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev 2005; 19:339-50; PMID:15687257; http://dx.doi.org/ 10.1101/gad.322605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mankouri HW, Ashton TM, Hickson ID. Holliday junction-containing DNA structures persist in cells lacking Sgs1 or Top3 following exposure to DNA damage. Proc Nat Acad Sci U S A 2011; 108:4944-9; PMID:21383164; http://dx.doi.org/ 10.1073/pnas.1014240108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lambert S, Mizuno K, Blaisonneau J, Martineau S, Chanet R, Freon K, Murray JM, Carr AM, Baldacci G. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol Cell 2010; 39:346-59; PMID:20705238; http://dx.doi.org/ 10.1016/j.molcel.2010.07.015 [DOI] [PubMed] [Google Scholar]
- 20. Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 2006; 21:15-27; PMID:16387650; http://dx.doi.org/ 10.1016/j.molcel.2005.11.015 [DOI] [PubMed] [Google Scholar]
- 21. Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 2010; 17:1305-11; PMID:20935632; http://dx.doi.org/ 10.1038/nsmb.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mulcair MD, Schaeffer PM, Oakley AJ, Cross HF, Neylon C, Hill TM, Dixon NE. A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell 2006; 125:1309-19; PMID:16814717; http://dx.doi.org/ 10.1016/j.cell.2006.04.040 [DOI] [PubMed] [Google Scholar]
- 23. Leipe DD, Aravind L, Grishin NV, Koonin EV. The bacterial replicative helicase DnaB evolved from a RecA duplication. Genome Res 2000; 10:5-16; PMID:10645945 [PubMed] [Google Scholar]
- 24. Kaplan DL, Davey MJ, O'Donnell M. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J Biol Chem 2003; 278:49171-82; PMID:13679365; http://dx.doi.org/ 10.1074/jbc.M308074200 [DOI] [PubMed] [Google Scholar]
- 25. Henderson TA, Nilles AF, Valjavec-Gratian M, Hill TM. Site-directed mutagenesis and phylogenetic comparisons of the Escherichia coli Tus protein: DNA-protein interactions alone can not account for Tus activity. Mol Genet Genomics: MGG 2001; 265:941-53; PMID:11523786; http://dx.doi.org/ 10.1007/s004380100501 [DOI] [PubMed] [Google Scholar]
- 26. Daee DL, Myung K. Fanconi-like crosslink repair in yeast. Genome Integrity 2012; 3:7; PMID:23062727; http://dx.doi.org/ 10.1186/2041-9414-3-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McHugh PJ, Ward TA, Chovanec M. A prototypical Fanconi anemia pathway in lower eukaryotes? Cell Cycle (Georgetown, Tex) 2012; 11:3739-44; PMID:22895051; http://dx.doi.org/ 10.4161/cc.21727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Valjavec-Gratian M, Henderson TA, Hill TM. Tus-mediated arrest of DNA replication in Escherichia coli is modulated by DNA supercoiling. Mol Microbiol 2005; 58:758-73; PMID:16238625; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04860.x [DOI] [PubMed] [Google Scholar]
- 29. Ivessa AS, Zhou JQ, Schulz VP, Monson EK, Zakian VA. Saccharomyces Rrm3p, a 5' to 3' DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev 2002; 16:1383-96; PMID:12050116; http://dx.doi.org/ 10.1101/gad.982902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nickoloff JA, Chen EY, Heffron F. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc Nat Acad Sci U S A 1986; 83:7831-5; PMID:3020559; http://dx.doi.org/ 10.1073/pnas.83.20.7831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Plessis A, Perrin A, Haber JE, Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics 1992; 130:451-60; PMID:1551570 [DOI] [PMC free article] [PubMed] [Google Scholar]