Abstract
Molecular estimates of evolutionary timescales have an important role in a range of biological studies. Such estimates can be made using methods based on molecular clocks, including models that are able to account for rate variation across lineages. All clock models share a dependence on calibrations, which enable estimates to be given in absolute time units. There are many available methods for incorporating fossil calibrations, but geological and climatic data can also provide useful calibrations for molecular clocks. However, a number of strong assumptions need to be made when using these biogeographic calibrations, leading to wide variation in their reliability and precision. In this review, we describe the nature of biogeographic calibrations and the assumptions that they involve. We present an overview of the different geological and climatic events that can provide informative calibrations, and explain how such temporal information can be incorporated into dating analyses.
Keywords: molecular clock, divergence dating, calibration, biogeography, Bayesian phylogenetics
1. Introduction
Estimating evolutionary timescales is an important component of many biological investigations, including analyses of populations as well as those of much broader taxonomic scope [1]. Timescales can be estimated from genetic data using phylogenetic methods based on the molecular clock, a hypothesis that proposes a constancy of evolutionary rates among lineages [2]. There is now a wide range of clock models that are able to relax the assumption of rate constancy, allowing timescales to be estimated even when there is significant heterogeneity in rates across lineages [3].
Genetic data alone can only provide an indication of relative divergence times. In order to produce estimates of absolute evolutionary timescales, molecular clocks need to be calibrated [4]. This can be done using various forms of temporal information, such as the ages of fossils, geological events or ancient samples. In recent years, there have been substantial advances in methods for extracting calibrations from the fossil record. These include developments in modelling fossil-occurrence data, allowing palaeontological evidence to be used more effectively [5]. The fossil record is useful for calibrating nodes across a range of geological time depths, making it the most commonly used source of calibrating information [6]. However, fossils are often uninformative for shallow timeframes because of insufficient diagnostic morphological variation, especially when conspecific individuals or populations are being studied [7].
A less direct method of calibration can be used when evolutionary divergence events are able to be associated with geological or climatic events of known age. These biogeographic calibrations can be informative across a broad range of timeframes. They can be based on a wide variety of geological events, from very recent drivers of population subdivision to ancient tectonic events that occurred over a hundred million years ago [8]. The usage of biogeographic calibrations varies across analyses of different taxonomic groups, with their employment being most common among studies of invertebrates [6].
There are several strong assumptions that need to be made when employing calibrations based on geological or climatic events. Primarily for this reason, biogeographic calibrations have been viewed unfavourably by many commentators [6,8,9]. In addition, employing such calibrations would be inappropriate if the purpose of the analysis is to test the impacts of the geological or climatic events on which the calibrations are based [8]. This potential circularity renders the use of biogeographic calibrations invalid in some molecular ecological studies. If implemented and interpreted carefully however, biogeographic calibrations have the potential to make a valuable and effective contribution to other studies that involve molecular dating.
In this review, we examine biogeographic calibrations in theory and in practice. We present a broad outline of the various geological and climatic events that can be used to calibrate molecular clock models. We also explain the assumptions that are made when formulating these calibrations in dating analyses. Finally, we discuss how biogeographic calibrations can be implemented in molecular dating studies.
2. The nature of biogeographic calibrations
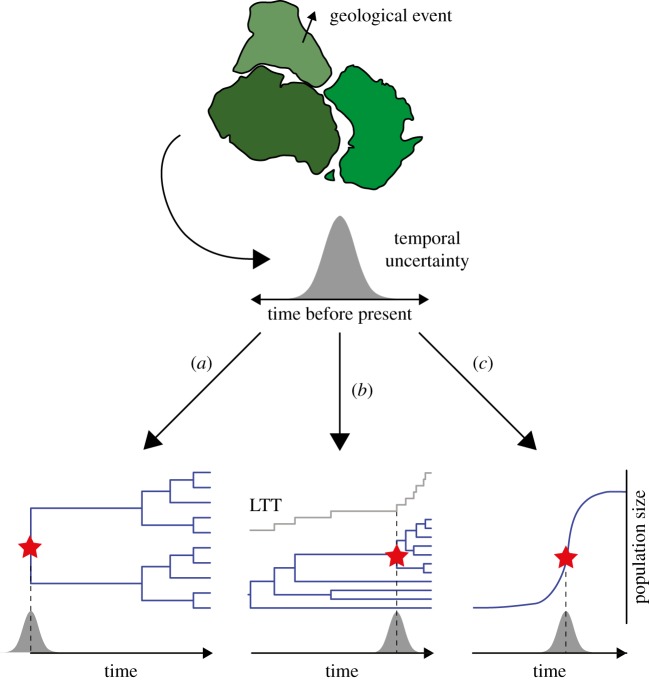
Significant geological and climatic events can drive the divergence of populations and species, producing evolutionary impacts that can be detected using genetic methods. Such events can be used to provide biogeographic calibrations for molecular clocks (figure 1), but they all share a number of key assumptions. Specifically, it is assumed that the biogeographic event has had measurable evolutionary and genetic impacts, and that it can be dated independently. The degree to which these assumptions are justified has substantial bearing on the reliability of the calibrations, with consequences for the accuracy of estimates of evolutionary rates and timescales [10].
Figure 1.
Biogeographic calibrations can be used in a number of ways. If a geological or climatic event has had an evolutionary or demographic impact, it can be used to calibrate molecular clocks. The age of the biogeographic event must be estimated by independent means, with the associated uncertainty being taken into account. Based on this information, an age constraint or prior distribution can be applied to (a) a divergence event in a phylogenetic tree or genealogy; (b) an estimated shift in diversification rate, as shown here in a lineages-through-time plot; or (c) an inferred change in the population size of a species. (Online version in colour.)
(a). Assumption 1: measurable evolutionary impact
Biogeographic calibrations are based on the assumption that a geological or climatic event has had an impact on populations or species. This impact needs to be measurable, such that it can be detected using phylogenetic or population-genetic methods. In a phylogenetic context, the biogeographic event is assumed to have affected the branching process, either by causing a lineage to split into two or by altering the rate of diversification. The divergence of two lineages can correspond to a speciation event or merely the separation of two subpopulations. Changes in diversification rates can be detected using phylogenetic methods if there is sufficient taxon sampling [11]. This can be done without calibrations, because relative node times are sufficient for detecting shifts in diversification rates.
When used for calibrating molecular clocks within populations, biogeographic events are sometimes assumed to have had a demographic rather than cladogenetic effect. This can involve population growth or spatial expansion, both of which can be detected using a range of statistics [12]. In addition, skyline-plot methods allow detailed inference of the population size through time [13]. A biogeographic calibration can be used if any changes in population size can be tied to a specific geological or climatic event [14].
(b). Assumption 2: age of biogeographic event
The second requirement of biogeographic calibrations is that the age of the geological or climatic event is known. In the case of geological events, this is usually informed by the application of radiometric dating methods to rocks and other materials. Although these techniques can sometimes yield precise date estimates of the material being analysed, the inferred dates need to be interpreted correctly in the geological context [15]. The dates of some geological events can be extremely difficult to establish with an acceptable degree of precision. Calibrations should also account for the prolonged nature of some geological events, whereby the development of a sustained barrier to gene flow would not have been immediate [16]. This has been a consistent source of uncertainty in studies of taxa in Australia and Zealandia, which comprises New Zealand, New Caledonia and various Australian external territories [17]. Historically, the most common explanation for closely related taxa in these regions is that rifting of Zealandia from Australia and Antarctica 82–85 million years ago (Ma) led to evolutionary divergence. However, recent geological data indicate that the rifting event occurred over a much longer time period than previously thought, beginning approximately 85 Ma and finishing about 52–55 Ma [18]. Uncertainty in the duration of geological events, particularly tectonic movements, is perhaps much greater than is generally acknowledged. The prolonged nature of these geological events needs to be taken into account in any corresponding biogeographic calibrations [19].
Even if the age of a geological or climatic event can be established with confidence, there can be a considerable lag in its biogeographic impact. Furthermore, determining the age of a biogeographic calibration can be particularly difficult if the associated geological or climatic event is not unique. For example, multiple glacial cycles occurred throughout the Quaternary [20], leading to repeated contraction and expansion of habitats. These also caused oscillations in sea level, causing land masses to be alternately connected and disconnected. Common practice has been to associate biogeographic events with the timing of the most recent occurrence of the geological or climatic event. For example, major expansion events in inferred population histories of Eurasian taxa are often assumed to have occurred after the Last Glacial Maximum 18 thousand years ago (ka), because signals from past demography are typically lost during severe population bottlenecks [21]. However, there is not always a compelling reason to assume that the most recent event was responsible for the divergence of populations; knowledge of multiple events must add to the uncertainty in the timing of the divergence.
Another source of uncertainty in the ages of biogeographic calibrations comes from the assumption of correspondence between population and genetic events. Most biogeographic calibrations are used to constrain the ages of nodes in phylogenetic trees. However, the evolutionary impact of the biogeographic event might not be closely associated with divergences at the genetic level. For example, genetic divergence occurs in the ancestral population prior to the separation of descendent populations [22]. This can lead to overestimation of evolutionary rates, because the age of the population split underestimates the age of the genetic divergence [23]. By contrast, ongoing gene flow can make it difficult to identify the exact timing of genetic divergence.
3. Sources of biogeographic calibrations
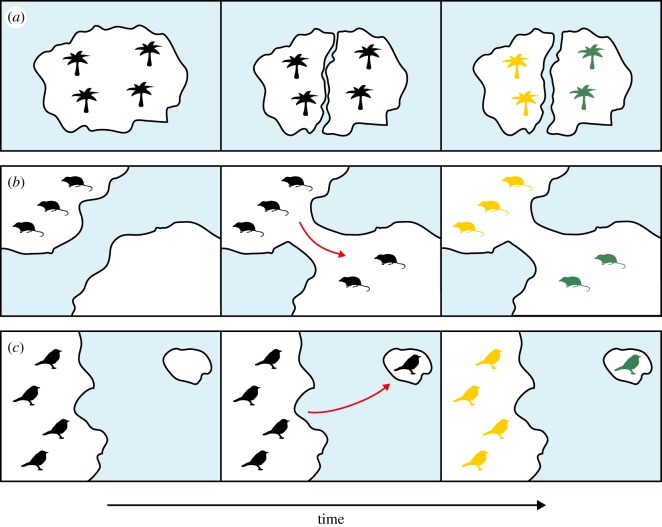
Biogeographic calibrations can be based on any event that causes a measurable change in gene flow or diversification rate. Here, we consider three broad types of biogeographic processes that promote allopatric speciation (figure 2): vicariance, geodispersal and biological dispersal. Some geological and climatic events can be associated with more than one of these biogeographic processes, depending on the taxa being considered.
Figure 2.
Three biogeographic processes that can be used to provide calibrations for molecular clocks. (a) Vicariance involves the formation of a barrier that causes a reduction in gene flow. Here, the fragmentation of a landmass causes a population of palms to split into two. (b) Geodispersal involves the removal of a barrier to gene flow, allowing movement into a new area. Here, the emergence of an isthmus allows a shrew to colonize a new land mass. (c) Biological dispersal involves the movement of an organism across a barrier. In this example, a passerine on the mainland disperses to an island and establishes a new population. (Online version in colour.)
(a). Vicariance
The process of vicariance involves the formation of a barrier that causes a reduction in gene flow (figure 2a). The barrier can be topographical, aquatic, oceanic, tectonic, environmental, climatic or biotic. In addition, the responses of different species to barriers to gene flow will depend on individual ecological preferences and climatic tolerances. In any case, these barriers to gene flow typically affect multiple taxa, leading to some degree of congruence between phylogenetic trees and area relationships [24]. The hypothesis of vicariance can be tested by comparing area relationships across multiple taxa, but this process can be misled by incomplete taxon sampling of extinct and extant lineages, errors in phylogenetic inference, and the effects of dispersal events [16,25].
Vicariance can be effected by the appearance of various terrestrial barriers, such as mountain ranges and plateaux. Climatic and anthropogenic factors can also hinder gene flow by changing the distribution of suitable habitat. Events creating aquatic and oceanic barriers include the formation of straits, ocean currents and deep trenches. Some of these can be caused by continental movements or by changes in sea level. One such event is the formation of the trench separating the eastern and western islands of the Aegean archipelago in the Mediterranean. This trench appeared about 9–12 Ma and has been used to calibrate evolutionary divergences in an exemplary analysis of six beetle genera [26].
(b). Geodispersal
Geodispersal refers to the removal of a barrier to gene flow, allowing range expansion (figure 2b). This can lead to rapid population growth, cladogenesis, or even an increase in the rate of speciation. As with vicariance, geodispersal is associated with events that can act on multiple taxa, potentially affecting the evolution of entire biotic assemblages [27].
Range expansion can be enabled by an increase in access to suitable habitat. This can occur as a result of changes in climatic conditions [28], or it can be due to geological events that alter physical connectivity. For example, the formation of land bridges enables terrestrial organisms to expand their ranges, potentially leading to cladogenesis. The formation of the Isthmus of Panama is a prominent example of using a dated land bridge to calibrate molecular clocks [29]. The isthmus created a connection between North and South America, allowing a large-scale exchange of fauna that is referred to as the Great American Biotic Interchange. This was thought to have occurred abruptly after the isthmus was raised and dry, some time between the Late Pliocene and Pleistocene [29]. However, emerging palaeontological and geological evidence points to a much larger window of uncertainty over when the land bridge formed [30], including the existence of an earlier land bridge in the middle Miocene [31]. The timescales of other tectonic instances of geodispersal, such as the early Miocene contact between the Sahul and Sunda shelves in Southeast Asia [32], also carry considerable uncertainty.
Among aquatic organisms, changes in riverine or lacustrine connections can be geologically dated and incorporated as time calibrations in a phylogenetic analysis [33,34]. River drainage patterns change when stream sections move the direction of their flow from one catchment to another, a phenomenon known as river capture. These events tend to occur much more rapidly than other geological transformations, meaning that there is considerably more precision in calibrations based on changes in riverine connections than on other geodispersal events [35].
Biogeographic calibrations can also be derived from climatic events that lead to substantial demographic changes or cladogenesis (figure 1). A prominent example is the dramatic climate cycles of the recent Quaternary glaciation (2.6 Ma to present), which are increasingly well documented through evidence from ice cores [36], providing a potentially rich source of calibration dates for shallow timescales. Postglacial population expansion in benthic or shallow-water marine organisms could have been caused by local inundation of the continental shelf by rising sea levels, which would have provided new habitat for colonization. High-resolution observational records of ancient sea levels are available over the last glacial cycle [37], allowing major inundation events to be dated accurately. Events of this kind that have been used for calibration include the postglacial flooding of the Sunda Shelf (14.58 ka [14]) and the flooding of the neighbouring East China Sea shelf during the last interglacial period (70–140 ka [38]).
(c). Biological dispersal
Biological dispersal involves the range expansion of a population or species across a barrier, allowing colonization of a new habitat (figure 2c). This might occur, for example, by transoceanic dispersal to a newly formed island. The capacity for dispersal varies across species, but dispersal events should be rare enough that the source and colonizing populations become genetically differentiated [39].
The formation of islands has often been used as a source of calibrating information based on biological dispersal. One of the most well-studied examples is the Hawaiian Islands, which have several features that make them especially suited to provide calibrations for molecular dating. The Hawaiian islands are widely accepted to have been formed along a ‘conveyor belt’, such that they are arranged linearly and sequentially by age, with the oldest islands located farthest from the hot spot that gave rise to them [40]. Radiometric (K-Ar) dating has provided precise estimates of the age of formation of each island, ranging from approximately 0.5 Ma for the island of Hawaii to 29.8 Ma for the Kure Atoll [41].
Divergences of extant island taxa are often assumed to have occurred at, or close to, the time of island formation. This requires that dispersal to islands occurred soon after their formation. Support for this assumption can be claimed when taxa follow the ‘rule of progression’ [42], which refers to congruence between the relative ages of islands and the branching structure in the phylogeny of the inhabitants. When the rule of progression is met, the ages of island formation can be used as calibrations for molecular dating analyses. Some taxa on the Hawaiian islands, such as Hypocosma moths [43], provide compelling examples of the progression rule. Nevertheless, colonization histories can sometimes be much more complex [44], potentially undermining the reliability of calibrations associated with island formation.
Biogeographic calibrations based on biological dispersal are subject to a number of potential confounding factors. One of the greatest risks is that evolutionary divergences can antedate the appearance of a new habitat. For example, lineages might have survived on ephemeral volcanic islands before achieving their distributions on extant islands [15,45]. In addition, dispersal can sometimes occur across barriers that were previously thought to be to be impassable [46].
4. Incorporating calibrations into a dating analysis
A wide range of phylogenetic methods are able to implement molecular clock models. Choosing a method for molecular dating depends on a number of factors, including the number of loci, number of taxa, degree of phylogenetic uncertainty and availability of computing resources [3]. The researcher must also decide on a method of incorporating the temporal information from biogeographic calibrations. In the vast majority of cases, calibrating information is used to constrain the age of a node in the phylogenetic tree. Traditionally, this was done by fixing the nodal age to an errorless point value. The use of such point calibrations is important in some methods, such as penalized likelihood [47], because it allows a unique solution to be identified in the dating analysis. However, omitting the uncertainty in calibrations is likely to lead to molecular date estimates that have an artificially high degree of precision. Instead, uncertainty in the calibrating information needs to be reflected in the age constraints [48]. In likelihood-based dating methods, calibrations can be used to set minimum and maximum bounds on node ages [47]. Soft bounds at the limits of the uniform distribution can be used to account for the possibility of errors in the chosen age constraints [49].
A more flexible approach can be taken in Bayesian methods, whereby the calibrating information can be used to specify the prior distributions of node ages [48]. This provides a means of incorporating uncertainty in the timing of geological events and their correspondence to biogeographic signals. In many cases, temporal uncertainty is most appropriately summarized in the form of a uniform prior distribution. For example, such a calibration prior might be used to account for the extended timeframe of some geological events, such as tectonic separations [19,34].
A more informative, non-uniform calibration prior can be used when there is evidence to suggest that the age of a node is more likely to take some values than others. A researcher might specify a normal prior to make an allowance for error around a most probable value, or when there is bidirectional uncertainty in the age of the node being calibrated [50]. Some well-considered examples of normal priors being used for calibrations have been those based on island colonization [43] and those used to reflect the uncertainty in the timing of geological events [26]. By contrast, exponential priors might be appropriate when the age of a node is likely to be very close to the timing of a particular geological or climatic event [48]. Such an assumption might be justified, for example, when there is evidence that the event in question has had a strong impact on gene flow. More flexible prior distributions, such as the lognormal and gamma, include parameters that allow them to take a range of shapes to reflect particular assumptions [48]. However, the timescale and impact of biogeographic events are rarely understood in sufficient detail to allow the parameters of these flexible prior distributions to be chosen with confidence.
In general, choosing the form of the prior distributions, and the values of their parameters, can be very difficult for biogeographic calibrations because of the diverse nature of the underlying processes. In this respect, they are at a distinct disadvantage compared with fossil calibrations, for which there have been various efforts to model preservation probabilities and to incorporate occurrence data [51]. One possible solution is to use a hierarchical Bayesian approach, whereby the user selects a hyperprior for the parameters of the calibration priors [52]. In any case, the representation of calibration uncertainty can have a substantial impact on the resulting estimates of divergence times [53].
A potential disadvantage of relying on biogeographic calibrations is that they are rarely available for multiple nodes in a phylogenetic tree of interest. By contrast, some analyses are able to incorporate large numbers of fossil calibrations or ancient samples. Increasing the number of calibrations in an analysis can have beneficial effects on the resulting estimates of divergence times. This is because the inclusion of multiple calibrations can reduce the influence of erroneous calibrations, reduce the average distance of nodes from calibrations and improve the robustness of estimates to clock-model misspecification [54].
A different approach needs to be taken when a biogeographic calibration is applied to a demographic event rather than to a specific node in a phylogenetic tree. Significant events include population expansions, contractions and subdivision. Changes in population size can be identified by examining inflection points in population skyline plots [55], plotting mismatch distributions [56] or comparing different demographic scenarios using approximate Bayesian computing [57]. In all of these cases, the uncertainty in the timing of the population event must be taken into account [14]. Biogeographic calibrations based on demographic events are used infrequently, but further developments in this area will be useful.
5. Concluding remarks
Biogeographic events are able to provide a rich source of calibrating information for molecular clocks, and can be particularly valuable when other calibrations are unavailable. However, the use of biogeographic calibrations should be accompanied by careful consideration of the assumptions made in their implementation, along with due acknowledgement of their uncertainty. A growing understanding of the potential and the limitations of biogeographic calibrations will help to improve the reliability of molecular estimates of evolutionary rates and timescales.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by the Australian Research Council (grant no. DP110100383 to S.Y.W.H.).
References
- 1.Arbogast BS, Edwards SV, Wakeley J, Beerli P, Slowinski JB. 2002. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu. Rev. Ecol. Syst. 33, 707–740. ( 10.1146/annurev.ecolsys.33.010802.150500) [DOI] [Google Scholar]
- 2.Zuckerkandl E, Pauling L. 1962. Molecular disease, evolution, and genic heterogeneity. In Horizons in biochemistry (eds Kasha M, Pullman B), pp. 189–225. New York, NY: Academic Press. [Google Scholar]
- 3.Ho SYW, Duchêne S. 2014. Molecular-clock methods for estimating evolutionary rates and timescales. Mol. Ecol. 23, 5947–5965. ( 10.1111/mec.12953) [DOI] [PubMed] [Google Scholar]
- 4.Donoghue PC, Benton MJ. 2007. Rocks and clocks: calibrating the tree of life using fossils and molecules. Trends Ecol. Evol. 22, 424–431. ( 10.1016/j.tree.2007.05.005) [DOI] [PubMed] [Google Scholar]
- 5.Heath TA, Huelsenbeck JP, Stadler T. 2014. The fossilized birth-death process for coherent calibration of divergence-time estimates. Proc. Natl Acad. Sci. USA 111, E2957–E2966. ( 10.1073/pnas.1319091111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hipsley CA, Muller J. 2014. Beyond fossil calibrations: realities of molecular clock practices in evolutionary biology. Front. Genet. 5, 138 ( 10.3389/fgene.2014.00138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shapiro B, Ho SYW. 2014. Ancient hyaenas highlight the old problem of estimating evolutionary rates. Mol. Ecol. 23, 499–501. ( 10.1111/mec.12621) [DOI] [PubMed] [Google Scholar]
- 8.Kodandaramaiah U. 2011. Tectonic calibrations in molecular dating. Curr. Zool. 57, 116–124. [Google Scholar]
- 9.Forest F. 2009. Calibrating the tree of life: fossils, molecules and evolutionary timescales. Ann. Bot. 104, 789–794. ( 10.1093/aob/mcp192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marko PB. 2002. Fossil calibration of molecular clocks and the divergence times of geminate species pairs separated by the Isthmus of Panama. Mol. Biol. Evol. 19, 2005–2021. ( 10.1093/oxfordjournals.molbev.a004024) [DOI] [PubMed] [Google Scholar]
- 11.Rabosky DL. 2014. Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS ONE 9, e89543 ( 10.1371/journal.pone.0089543) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Excoffier L, Foll M, Petit RJ. 2009. Genetic consequences of range expansions. Annu. Rev. Ecol. Evol. Syst. 40, 481–501. ( 10.1146/annurev.ecolsys.39.110707.173414) [DOI] [Google Scholar]
- 13.Ho SYW, Shapiro B. 2011. Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol. Ecol. Res. 11, 423–434. ( 10.1111/j.1755-0998.2011.02988.x) [DOI] [PubMed] [Google Scholar]
- 14.Crandall ED, Sbrocco EJ, DeBoer TS, Barber PH, Carpenter KE. 2012. Expansion dating: calibrating molecular clocks in marine species from expansions onto the Sunda Shelf following the Last Glacial Maximum. Mol. Biol. Evol. 29, 707–719. ( 10.1093/molbev/msr227) [DOI] [PubMed] [Google Scholar]
- 15.Heads M. 2005. Dating nodes on molecular phylogenies: a critique of molecular biogeography. Cladistics 21, 62–78. ( 10.1111/j.1096-0031.2005.00052.x) [DOI] [PubMed] [Google Scholar]
- 16.Upchurch P. 2008. Gondwanan break-up: legacies of a lost world? Trends Ecol. Evol. 23, 229–236. ( 10.1016/j.tree.2007.11.006) [DOI] [PubMed] [Google Scholar]
- 17.Grobys JWG, Gohl K, Eagles G. 2008. Quantitative tectonic reconstructions of Zealandia based on crustal thickness estimates. Geochem. Geophys. Geosyst. 9, 1 ( 10.1029/2007GC001691) [DOI] [Google Scholar]
- 18.Schellart WP, Lister GS, Toy VG. 2006. A Late Cretaceous and Cenozoic reconstruction of the Southwest Pacific region: tectonics controlled by subduction and slab rollback processes. Earth-Sci. Rev. 76, 191–233. ( 10.1016/j.earscirev.2006.01.002) [DOI] [Google Scholar]
- 19.Ericson PG, Klopfstein S, Irestedt M, Nguyen JMT, Nylander JA. 2014. Dating the diversification of the major lineages of Passeriformes (Aves). BMC Evol. Biol. 14, 8 ( 10.1186/1471-2148-14-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt G. 2000. The genetic legacy of the Quaternary ice ages. Nature 405, 907–913. ( 10.1038/35016000) [DOI] [PubMed] [Google Scholar]
- 21.Karl SA, Toonen RJ, Grant WS, Bowen BW. 2012. Common misconceptions in molecular ecology: echoes of the modern synthesis. Mol. Ecol. 21, 4171–4189. ( 10.1111/j.1365-294X.2012.05576.x) [DOI] [PubMed] [Google Scholar]
- 22.Edwards SV, Beerli P. 2000. Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution 54, 1839–1854. ( 10.1111/j.0014-3820.2000.tb01231.x) [DOI] [PubMed] [Google Scholar]
- 23.Hickerson MJ, Gilchrist MA, Takebayashi N. 2003. Calibrating a molecular clock from phylogeographic data: moments and likelihood estimators. Evolution 57, 2216–2225. ( 10.1111/j.0014-3820.2003.tb00234.x) [DOI] [PubMed] [Google Scholar]
- 24.Wiley EO. 1988. Vicariance biogeography. Annu. Rev. Ecol. Syst. 19, 513–542. ( 10.1146/annurev.es.19.110188.002501) [DOI] [Google Scholar]
- 25.Kodandaramaiah U. 2009. Use of dispersal-vicariance analysis in biogeography: a critique. J. Biogeogr. 37, 3–11. ( 10.1111/j.1365-2699.2009.02221.x) [DOI] [Google Scholar]
- 26.Papadopoulou A, Anastasiou I, Vogler AP. 2010. Revisiting the insect mitochondrial molecular clock: the mid-Aegean trench calibration. Mol. Biol. Evol. 27, 1659–1672. ( 10.1093/molbev/msq051) [DOI] [PubMed] [Google Scholar]
- 27.Lieberman BS. 1997. Early Cambrian paleogeography and tectonic history: a biogeographic approach. Geology 25, 1039–1042. () [DOI] [Google Scholar]
- 28.Baldwin BG, Sanderson MJ. 1998. Age and rate of diversification of the Hawaiian silversword alliance (Compositae). Proc. Natl Acad. Sci. USA 95, 9402–9406. ( 10.1073/pnas.95.16.9402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodburne MO. 2010. The Great American Biotic Interchange: dispersals, tectonics, climate, sea level and holding pens. J. Mammal Evol. 17, 245–264. ( 10.1007/s10914-010-9144-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leigh EG, O'Dea A, Vermeij GJ. 2014. Historical biogeography of the Isthmus of Panama. Biol. Rev. 89, 148–172. ( 10.1111/brv.12048) [DOI] [PubMed] [Google Scholar]
- 31.Montes C, et al. 2015. Middle Miocene closure of the Central American Seaway. Science 348, 226–229. ( 10.1126/science.aaa2815) [DOI] [PubMed] [Google Scholar]
- 32.Crayn DM, Costion C, Harrington MG. 2015. The Sahul-Sunda floristic exchange: dated molecular phylogenies document Cenozoic intercontinental dispersal dynamics. J. Biogeogr. 42, 11–24. ( 10.1111/jbi.12405) [DOI] [Google Scholar]
- 33.Peng Z, Ho SYW, Zhang Y, He S. 2006. Uplift of the Tibetan plateau: evidence from divergence times of glyptosternoid catfishes. Mol. Phylogenet. Evol. 39, 568–572. ( 10.1016/j.ympev.2005.10.016) [DOI] [PubMed] [Google Scholar]
- 34.Genner MJ, Seehausen O, Lunt DH, Joyce DA, Shaw PW, Carvalho GR, Turner GF. 2007. Age of cichlids: new dates for ancient lake fish radiations. Mol. Biol. Evol. 24, 1269–1282. ( 10.1093/molbev/msm050) [DOI] [PubMed] [Google Scholar]
- 35.Waters JM, Rowe DL, Apte S, King TM, Wallis GP, Anderson L, Norris RJ, Craw D, Burridge CP. 2007. Geological dates and molecular rates: rapid divergence of rivers and their biotas. Syst. Biol. 56, 271–282. ( 10.1080/10635150701313855) [DOI] [PubMed] [Google Scholar]
- 36.Stauffer B. 1999. Climate change: cornucopia of ice core results. Nature 399, 412–413. ( 10.1038/20807) [DOI] [Google Scholar]
- 37.Lambeck K, Esat TM, Potter EK. 2002. Links between climate and sea levels for the past three million years. Nature 419, 199–206. ( 10.1038/nature01089) [DOI] [PubMed] [Google Scholar]
- 38.He LJ, Zhang AB, Weese D, Li SF, Li JS, Zhang J. 2014. Demographic response of cutlassfish (Trichiurus japonicus and T. nanhaiensis) to fluctuating palaeo-climate and regional oceanographic conditions in the China seas. Sci. Rep. 4, 6380 ( 10.1038/srep06380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cook LG, Crisp MD. 2005. Directional asymmetry of long-distance dispersal and colonization could mislead reconstructions of biogeography. J. Biogeogr. 32, 741–754. ( 10.1111/j.1365-2699.2005.01261.x) [DOI] [Google Scholar]
- 40.Fleischer RC, McIntosh CE, Tarr CL. 1998. Evolution on a volcanic conveyor belt: using phylogeographic reconstructions and K-Ar-based ages of the Hawaiian Islands to estimate molecular evolutionary rates. Mol. Ecol. 7, 533–545. ( 10.1046/j.1365-294x.1998.00364.x) [DOI] [PubMed] [Google Scholar]
- 41.Price JP, Clague DA. 2002. How old is the Hawaiian biota? Geology and phylogeny suggest recent divergence. Proc. R. Soc. Lond. B 269, 2429–2435. ( 10.1098/rspb.2002.2175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hennig W. 1966. Phylogenetic systematics. Urbana, IL: University of Illinois Press. [Google Scholar]
- 43.Haines WP, Schmitz P, Rubinoff D. 2014. Ancient diversification of Hyposmocoma moths in Hawaii. Nat. Commun. 5, 3502 ( 10.1038/ncomms4502) [DOI] [PubMed] [Google Scholar]
- 44.Mairal M, Pokorny L, Aldasoro JJ, Alarcón M, Sanmartín I. 2015. Ancient vicariance and climate-driven extinction continental-wide disjunctions in Africa: the case of the Rand Flora genus Canarina (Campanulaceae). Mol. Ecol. 24, 1335–1354. ( 10.1111/mec.13114) [DOI] [PubMed] [Google Scholar]
- 45.Heads M. 2011. Old taxa on young islands: a critique of the use of island age to date island-endemic clades and calibrate phylogenies. Syst. Biol. 60, 204–218. ( 10.1093/sysbio/syq075) [DOI] [PubMed] [Google Scholar]
- 46.Vences M, Vieites DR, Glaw F, Brinkmann H, Kosuch J, Veith M, Meyer A. 2003. Multiple overseas dispersal in amphibians. Proc. R. Soc. Lond. B 270, 2435–2442. ( 10.1098/rspb.2003.2516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanderson MJ. 2002. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19, 101–109. ( 10.1093/oxfordjournals.molbev.a003974) [DOI] [PubMed] [Google Scholar]
- 48.Ho SYW, Phillips MJ. 2009. Accounting for calibration uncertainty in phylogenetic estimation of evolutionary divergence times. Syst. Biol. 58, 367–380. ( 10.1093/sysbio/syp035) [DOI] [PubMed] [Google Scholar]
- 49.Yang Z, Rannala B. 2006. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 23, 212–226. ( 10.1093/molbev/msj024) [DOI] [PubMed] [Google Scholar]
- 50.Ho SYW. 2007. Calibrating molecular estimates of substitution rates and divergence times in birds. J. Avian Biol. 38, 409–414. ( 10.1111/j.0908-8857.2007.04168.x) [DOI] [Google Scholar]
- 51.Nowak MD, Smith AB, Simpson C, Zwickl DJ. 2013. A simple method for estimating informative node age priors for the fossil calibration of molecular divergence time analyses. PLoS ONE 8, e66245 ( 10.1371/journal.pone.0066245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heath TA. 2012. A hierarchical Bayesian model for calibrating estimates of species divergence times. Syst. Biol. 61, 793–809. ( 10.1093/sysbio/sys032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inoue J, Donoghue PC, Yang Z. 2010. The impact of the representation of fossil calibrations on Bayesian estimation of species divergence times. Syst. Biol. 59, 74–89. ( 10.1093/sysbio/syp078) [DOI] [PubMed] [Google Scholar]
- 54.Duchêne S, Lanfear R, Ho SYW. 2014. The impact of calibration and clock-model choice on molecular estimates of divergence times. Mol. Phylogenet. Evol. 78, 277–289. ( 10.1016/j.ympev.2014.05.032) [DOI] [PubMed] [Google Scholar]
- 55.Hope AG, Ho SYW, Malaney JL, Cook JA, Talbot SL. 2014. Accounting for rate variation among lineages in comparative demographic analyses. Evolution 68, 2689–2700. ( 10.1111/evo.12469) [DOI] [PubMed] [Google Scholar]
- 56.Rogers AR, Harpending H. 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569. [DOI] [PubMed] [Google Scholar]
- 57.Bertorelle G, Benazzo A, Mona S. 2010. ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol. Ecol. 19, 2609–2625. ( 10.1111/j.1365-294X.2010.04690.x) [DOI] [PubMed] [Google Scholar]