Abstract
Heparan sulfate (HS) polysaccharides are ubiquitous components of the cell surface and extracellular matrix of all multicellular animals, whereas heparin is present within mast cells and can be viewed as a more sulfated, tissue-specific, HS variant. HS and heparin regulate biological processes through interactions with a large repertoire of proteins. Owing to these interactions and diverse effects observed during in vitro, ex vivo and in vivo experiments, manifold biological/pharmacological activities have been attributed to them. The properties that have been thought to bestow protein binding and biological activity upon HS and heparin vary from high levels of sequence specificity to a dependence on charge. In contrast to these opposing opinions, we will argue that the evidence supports both a level of redundancy and a degree of selectivity in the structure–activity relationship. The relationship between this apparent redundancy, the multi-dentate nature of heparin and HS polysaccharide chains, their involvement in protein networks and the multiple binding sites on proteins, each possessing different properties, will also be considered. Finally, the role of cations in modulating HS/heparin activity will be reviewed and some of the implications for structure–activity relationships and regulation will be discussed.
Keywords: heparin, heparan sulfate, polysaccharide, protein binding, redundancy, cations
1. Introduction
1.1. Heparan sulfate and heparin similarities and differences
Heparan sulfate (HS) polysaccharides are a family of linear sulfated, heterogeneous polysaccharides found on the cell membrane and in the extracellular matrix as part of heparan sulfate proteoglycans (HSPGs). They are composed of repeating 1 → 4 linked disaccharide units, in which one monosaccharide is a α-d-glucosamine residue and the other an uronic acid (in a salt form—an uronate; figure 1). Heparin is a structurally similar polysaccharide found within mast cells as a component of serglycin proteoglycans and has been shown to differ in composition with several mammalian forms of HS [6]. Under the conventional definition, HS and heparin can be compared as follows: first, in heparin, the uronates are predominantly α-l-iduronate, whereas in HS, they are mainly its C-5 epimer, β-d-glucuronate. Second, in HS, the d-glucosamine residues are predominantly N-acetylated, whereas in heparin, they are N-sulfated. Finally, whereas at least 70–80% of heparin is composed of the disaccharide l-iduronate 2-O-sulfate α(1 → 4) d-glucosamine N,6-sulfate, in HS around 40–60% of the disaccharides consist of →4) d-glucuronate β (1 → 4) d-glucosamine (1→, that can be either N-acetylated or N-sulfated). Together, these structural characteristics make heparin more sulfated and, hence, more charged than HS [6–8]. Furthermore, HS also has a much higher maximum average molecular weight (ca 50 kDa) than heparin (ca 20 kDa) [9]. The differences in underlying composition, as well as between substitution pattern and domain structure, which could also lead to distinct glycosidic linkage geometry (torsion angles, phi and psi) and iduronate conformational equilibria (e.g. 1C4, 2S0, etc.) do, according to the conventional definitions, make HS fundamentally different to heparin. Furthermore, HS is considered typically to adopt longer, more flexible chains in solution than heparin [10,11]. It has become apparent, however, that the designations heparin or HS are less clear-cut than this description implies, and that polysaccharides isolated from some organisms appear to be hybrid [12,13]. Indeed, HS with features approaching heparin are produced in a variety of tissues [14–16] and cell types [17–19]. It is possible, therefore, to consider heparin a tissue-specific form of HS with the conventional definitions of heparin and HS residing towards two opposite extremes. More detailed studies across many species and tissues will provide a more complete picture of the extent of sulfation and structural diversity throughout the animal kingdom and a distinction from ‘heparin’, the finished pharmaceutical agent, must also be considered.
Figure 1.
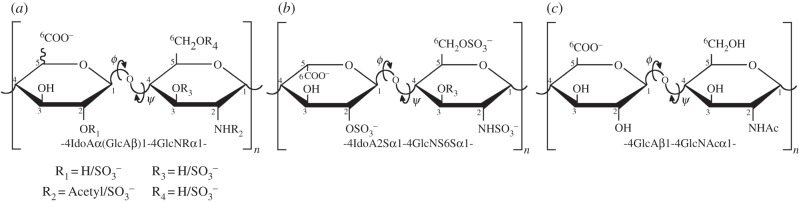
Structural features of HS and heparin. (a) Possible substitution patterns in heparin/HS. (b) The major disaccharide unit of heparin, which corresponds typically to 70–80% [1], although differences between sources, such as porcine mucosa and bovine lung are typical [2]. (c) The major repeating disaccharide unit of HS from, for example, porcine mucosa. Considerable variation in HS composition also occurs between species and tissues [3–5]. ϕ (phi) and ψ (psi) denote glycosidic linkage torsion angles.
1.2. Heparin sulfate/heparin structural diversity from biosynthesis
The biosynthesis of both heparin and HS, as the carbohydrate components of proteoglycans, starts in the endoplasmic reticulum and proceeds through the Golgi apparatus where several polymerases and modifying enzymes and their respective isoforms may act on different substrates, or sets of substrates. The biosynthetic process and its machinery have been studied extensively [20] and a sequential hierarchical order of events, in which selective enzymatic modifications are required to allow further editing of the polymer, are widely accepted. While this model does provide for some of the possible substitution patterns observed in both heparin and HS, several disaccharides and their respective substitution patterns cannot be explained within the scheme, for example IdoA-GlcNAc,6S, if the proposed order of events and their known specificities are respected rigidly [21]. Furthermore, unlike nucleic acids and proteins, HS and heparin biosynthesis is not template driven and the conventional experimental approaches of gene knockout and single enzyme inhibition do not provide conclusive answers, because compensatory mechanisms can be activated [22,23]. Further understanding of how the biosynthetic machinery functions, particularly at the sequence level, transcend the approaches cited above and the way in which such process are controlled is therefore still sought [20]. The process of biosynthesis results in the introduction of structural properties that can be viewed at a number of levels; the disaccharide composition, the order of disaccharides (sequence), the clustering of similar disaccharides within the chains (domains) and stereochemical variation arising from distinct monosaccharide composition, such as d-glucuronic acid and l-iduronic acid, all of which combine to bestow particular conformational, charge distribution and flexibility on a given sequence and, ultimately, define their biological function. It is worth emphasizing that structure, defined simply as saccharide sequence, has not been found to correspond to function in a simple manner.
1.3. Structural variation in heparan sulfate and heparin
Dynamic temporal and spatial variations in HS structure underpin the concept of the ‘heparanome’ [24]. Owing to the fact that HS occurs as an element of proteoglycans, the primary source of structural variation could arise from changes in proteoglycan expression. Nonetheless, changes in HS structural characteristics are thought to be more tissue specific rather than depending on the nature of its constituent proteoglycans [25]. One could assume that the overall HS assembly in similar tissues and/or cells are comparable but, such an assumption ignores this temporal variation within biological microenvironments. Substantial changes in both HS quantity and overall assembly in response to variation in environment have been shown in different cell cycle stages [26–29] and, in the case of heparin, shown to stimulate antithrombotic HS production by endothelial cells [17]. Heparin and HS can interact with a large number of proteins, which are being identified [30–32] The poly- and oligosaccharide chains can interact with a very large number of proteins, which are being identified [33,34], and are defined as heparin binding proteins (HBPs) usually employing heparin as an experimental proxy for HS, and are known as HBPs. Ultimately, changes in HS structure bestow the particular protein binding capabilities that are required for specific biological functions at different moments.
2. The aims and scope of the review
2.1. Studies of heparan sulfate and heparin interacting with proteins encompass two broad fields of research: the biochemical and the physico-chemical
There are two broad areas of research that relate to the interactions of HS/heparin with proteins. One area employs biochemical approaches, including protein assays, in vitro cell assays as well as in vivo and ex vivo perturbations which, only relatively rarely, incorporate studies of the effects of changes to the complex chemical environment in which the polysaccharides can be found, such as pH, ionic strength and types of associated cation. The other area covers physico-chemical techniques, including studies into the polyelectrolyte nature of the polysaccharides and the involvement of cation binding to HS/heparin chains, but only rarely investigates biological activity. As has been observed aptly by Kayitmazer et al. [30], there is often little communication between researchers in these two fields and those involved may employ very different experimental techniques. Debate regarding selectivity reflects not only this, but also the perspective, strategy and technical approaches employed. Thus, what may appear peripheral for one group may be considered ‘mainstream’ for the other.
Rather than attempt to review exhaustively the extensive biological literature in the heparin/HS field, which has been undertaken successfully elsewhere [3,31,35–42], the aim of this review is to focus on the findings from both biochemical approaches and physico-chemical studies, regarding HS/heparin polysaccharides and the relationship between sequence, structure and activity. It is not intended to restrict the review to recent articles, because the work in this area has been accumulating for decades and debate continues. Hence, relevant articles from across the breadth of the literature will be considered. A summary of the selected works, illustrated with examples from the most extensively studied systems, particularly relating to antithrombin (AT) and the fibroblast growth factor (FGF) protein family, will be made and a brief synthesis (§5), intended to be accessible to a wide audience and providing findings from both perspectives, will be attempted. It is hoped that the review will help to promote discussion regarding HS/heparin–protein interactions between researchers in both of these fields, and more widely, without particular favour towards either audience. Finally, aspects to which, it would seem, future efforts might be usefully directed will be suggested (§5.3).
2.2. The contrasting structure of oligo- and polysaccharides
Heparin and HS are expressed primarily as the polysaccharide component of HSPGs such as syndecans and glypicans in the case of HS, and serglycin in the case of heparin. They can also be subject to enzymatic cleavage to release free glycosaminoglycan (GAG) chains of varying lengths. The challenge has been to delineate the HS/heparin sequences that carry activity for a given protein and, eventually, a particular biological function. It has become apparent that activity in this system does not reside in unique HS/heparin sequences but, in a limited choice of potential binding sequences. Subtle relationships prevail between sulfation position and uronic acid identity on the one hand, and activity on the other hand. Because HS is relatively difficult to isolate from natural systems in substantial quantities, it is usually necessary to employ oligosaccharide fragments (§3.1) in isolated systems, so that binding properties to proteins can be interpreted yet, as intimated by the section above, there is evidence that oligosaccharides and polysaccharides can exhibit distinct properties [43–45]. Furthermore, the equivalence of the conformations that short sequences excised from the polysaccharide can adopt, compared with the intact parental polysaccharide [10,11] and the likely lack of domain structure and thermodynamic properties of the former, can be questioned. The manifold binding sites, which are characteristic of the polysaccharides, have usually been reduced substantially in oligosaccharides. These caveats are of less concern, however, if the aim is to obtain active structures for potential application as pharmaceutical agents, and it may be useful to distinguish this activity from attempts to understand the biological system per se, in which the polysaccharide component of HSPGs are usually involved. These two distinct aims can sometimes be conflated, one example being the frequently stated experimental aim of identifying the ‘minimal active sequence or fragment’. Having said this, the concentration of circulating oligosaccharide fragments derived from HS chains after heparanase degradation may be increased in certain circumstances, such as in the disease condition, mucopolysaccharidosis [46], may play a role in others [47] and disease outcome has been linked to heparanase in cancer [48,49].
3. The nature of specificity in heparan sulfate/protein interactions
An important contribution to the binding of heparin/HS polysaccharides to proteins involves binding of the negatively charged GAG to the amino acid residues lysine and arginine, and can also include protonated histidine residues at low pH values [50], as well as interactions with a range of other amino acids. A thorough analysis of the distribution and nature of these binding regions on the surfaces of the several hundred HS binding proteins so far identified will be interesting and warrants further investigation. Studies, which include the identification of interacting arginine side-chains considered to be stronger binders [51], will also be interesting to compare. Some of the data regarding binding specificity between heparin/HS and proteins were obtained using X-ray crystallography, in which small sugar fragments, usually heparin oligosaccharide fractions (obtained by size-exclusion chromatography and probably containing several different sequences) served as substitutes for the physiologically relevant polysaccharides. It must also be recalled that protein structural analysis by X-ray crystallography is subject inevitably to a number of limitations, despite its very high precision in measuring atomic position. These limitations include structural packing in which conformational artefacts can be created, incomplete solvation, which contrasts the normal physiological conditions, and the fact that crystallography cannot usually be employed to study protein–polysaccharide complexes nor, in many cases, analyse glycosylated and structurally disordered proteins. Moreover, heparin-binding processes are dynamic and protein structures obtained from crystallography can be viewed as ‘frozen’ in a particular conformation, potentially reporting conformations that may not represent the physiological situation.
3.1. Experimental tools: polysaccharides and oligosaccharides
The nature and selectivity of heparin and HS–protein interactions is the subject of widespread debate. Unfortunately, the experiments that could determine unequivocally the origins of specificity cannot be conducted at present for several reasons, which include the need to test vast numbers of structural variants in their pure forms [52]. It is the ability to separate these species in sufficient quantities when using natural HS as a source, despite some progress [53–56], rather than the problem of sequencing them per se, which hinders progress. Heparin is often employed experimentally as a proxy for HS on account of its structural similarity (§1) and is considerably more highly charged than HS. Its purification process, intended to achieve high anticoagulant potency also favours chains with certain characteristics, leading to a more homogeneous mixture than most HS, tending to activate in some systems more extensively than tissue-derived HS where the active entity may be a lower proportion of the more heterogeneous mixture. For this reason, and also because they offer a route by which the relationship between substitution pattern, (in terms of the prevailing statistical content of O-, N-sulfate and N-acetyl groups), with activity in polysaccharides, rather than oligosaccharides (§2.2) can be explored, chemically modified heparin derivatives have been used [57–59]. Using these in their polysaccharide forms, or as a means of generating oligosaccharides following enzyme digestion, there have been attempts to examine the structure–activity relationship of heparin/HS [59–62]. Oligosaccharides can be selected, by affinity chromatography or filter trapping with salt elution, thereby tending to select high charge interactions [63–66]. Searches are often made among sugar sequences representative of only a very limited subset of the total possible sequences because, usually, the available sugars derive either from HS of a particular tissue, or from heparin, both of which have limited structural diversity. Early work attempted to identify unique sequences responsible for protein binding, again employing affinity chromatography, and eluting with a salt gradient (e.g. in the case of FGF-2) [67] although it was realized that other sequences could exert an effect. Interpreting these results as providing evidence for preferred binding sequences [65,67] could lead to the potentially tautological argument that biological activity resides predominantly in the highly sulfated domains of HS. These results contrast with an earlier report identifying FGF-1 binding saccharides [68] and another, in which an HS structure described as the FGF-1 binding domain was identified [63] (§3.3).
3.2. Antithrombin-heparin pentasaccharide: an exception that does not prove the rule?
The early work on elucidating the relationship between HS/heparin structure with protein binding and activity concentrated on analysis of the heparin sequence capable of binding and activating the serine protease inhibitor AT. This approach employed the fractionation of heparin followed by affinity chromatography, revealing a pentasaccharide that bound a particular sequence in heparin with a high degree of selectivity over other sequences, while retaining its anticoagulant activity. The pentasaccharide was first suggested as that possessing highest affinity under the experimental conditions employed (elution in high salt), which seem likely to have been selective for highly charged species [69,70]. Despite these reservations, which were expressed in the original reports, over time, the interactions with AT and the pentasaccharide sequence within the heparin polysaccharide have tended to be viewed by some [71] as the unique binding structure redolent of the ‘lock and key’ hypothesis and, furthermore, one that was prototypical of all heparin and HS interactions with proteins in general. A more nuanced view is now prevailing (vide infra). Furthermore, such a sequence is yet to be found within HS—perhaps one of the natural ligands—and it is worth noting that, under normal physiological conditions, there may be very little, or no, circulating heparin, hence it is also important to distinguish biological anticoagulation from the pharmacological actions of anticoagulants. While the pentasaccharide sequence undoubtedly binds AT with high affinity and activates the protein, subsequent evidence has emerged suggesting that net charge plays a significant role in the affinity of heparin for AT (§4.1) and that the 3-O-sulfated group in the central glucosamine unit of the heparin binding pentasaccharide is not essential to activate AT [72,73]. In fact, other types of carbohydrate structures, distinct from GAGs, including those with low or no activity, have also been identified that can fulfil the structural requirements of AT binding [72] and a proposal has been made that the stabilization of AT is the key determinant of its activity [73]. It has also been shown that N-acetylation or N-sulfation is permitted in the substitution pattern of residues adjacent to and within, the first glucosamine residue, the pentasaccharide and that specific residues outside of the pentasaccharide region active for AT can influence AT activity [74–78]. This reinforces the idea that binding, even with low affinity, does not necessarily equate to activity. Furthermore, non-carbohydrate compounds can bind both the pentasaccharide binding site and the extended heparin binding sites of AT, leading to enhanced anticoagulant activity [79], suggesting that alternative classes of compounds may find therapeutic roles.
The structure of AT in the heparin-bound, intermediate state illustrates such issues. It was reported that it was difficult to conclude that the form of AT obtained represented the kinetic intermediate identified from solution based experiments [80]. A recent X-ray crystallographic study of the solvent accessible areas of AT and thrombin, suggested that, contrary to what had long been held to be the case, the nature of heparin binding to the two proteins is fundamentally different; the polysaccharide ligand being much less strongly held in the case of thrombin. This analysis was based on the differences in mobility of the two main amino acid side chains responsible for heparin binding (Arg and Lys) [81]. More recently, it has been shown that the reactive centre loop, exposed upon heparin-binding, and considered to act as ‘bait’ for proteases, can populate conformational states in which the Arg side chain is exposed to the solvent [82].
3.3. Heparan sulfate/heparin interactions with fibroblast growth factors: representative of typical protein–heparan sulfate/heparin interactions?
The other well-studied class of HBPs that interact primarily with HS, on the cell surface and in the extracellular space, are the FGFs and their cognate receptor tyrosine kinases (FGFRs). This family of signalling proteins forms complexes in which an FGF and an FGFR, or FGFRs (the nature of the signalling complex in vivo remains unclear) are brought together to form functioning signalling complexes in the presence of HS or heparin polysaccharides or oligosaccharides, acting as obligatory co-receptors. From the perspective of FGFR specificity, it was found that FGFR1 and FGFR2 had distinct kinetics and affinities for heparin, despite the apparent involvement of the same sulfate groups in heparin [83]. It is generally held that the naturally occurring co-receptor is HS but, heparin and its derivatives or fragments, and even analogous non-GAG structures are able to elicit both signalling and inhibitory activity [84].
In the literature relating to the HS/heparin sequences required for binding to FGFs, there are several claims to have uncovered ‘minimal binding sequences’ or similar, and it is important to assess these carefully. In most cases, the oligosaccharides identified originate from parent polysaccharides of rather restricted sequence diversity and this should temper the claims made. Furthermore, scrutiny of the methods employed to select bound saccharides reveals that they may rely on some form of salt elution, which could be biased towards those interactions involving charges and may be more likely to return highly charged species preferentially.
The employment of such a series of modified heparin derivatives, in which sequence diversity has been reduced by systematic desulfation, has also shown that no simple relationship exists between sulfation and charge distribution and their ability to support signalling with FGF-1 and -2 [84,43]. Furthermore, distinct carbohydrate structures can sometimes fulfil the role of HS/heparin, as has been shown for CS/DS extracted from brittlestars, which activated FGF-2 [85]. However, as was the case for AT, FGF-1 signalling correlated strongly with protein stabilization induced by the polysaccharides, whereas FGF-2 signalling did not [79]. For the FGFs, this complex situation is underpinned by the detection of widely ranging heparin binding properties among FGF subfamily members, which possess varied numbers of HBPs, identified by Lys residues (the chemistry of Arg residues proving thus far more problematic), that ranged from 1 (FGF-9) to 3 (FGF-1) and their binding properties (Kd values from 38 to 620 nM and kass varied over 20-fold) [86]. Different sources of HSPGs are known to exert distinct activities [87], and HS is both spatially and temporally regulated, controlling FGFR signalling [88]. Furthermore, it has been reported that the structural requirements for FGFR activation may be more relaxed than those required for FGF binding [89] and, that the position of an interacting sequence within the HS chain can also be important [90].
An alternative route to structurally varied fragments was provided by oligosaccharides obtained from chemically modified heparin. In one case, random distributions of sulfate groups were generated in heparin by chemical means, and the polysaccharides partially depolymerized using enzymes to generate deliberately as many sequences as possible and chains of varying lengths. The sized pools were then screened for a biological activity (not merely binding) in increasing order of their sizes, the first pool capable of activating was then selected and further fractionated by strong anion exchange chromatography and screened again, this time starting from the first eluted fractions, corresponding to the least charged oligosaccharides. In this way, the smallest and least charged oligosaccharides capable of activating were identified and characterized, revealing that in principle, oligosaccharides with very low levels of sulfation were capable of activating FGF-1 and FGFR2c [91]. The implications of this in biological systems have yet to be explored in detail.
Compared with the FGFs, the interactions involving their receptors have been relatively little studied, but the structural basis of interactions between either FGF and HS, or FGFR and HS is distinct [92,93] and glycosylation of the FGFR is known to alter its activity [94]. The interactions between FGF-2 and heparin-derived oligosaccharides (in the case of a hexasaccharide) have been studied by crystallography, suggesting interactions between asparagine and lysine (Asp 28, Asp 102, Lys 27, Lys 126, Lys 136) and glutamine (Gln 135) residues with the oligosaccharide [95]. Several complexes of FGF and FGF receptor (FGFR) have also been reported, for example, incorporating heparin-derived oligosaccharides in which the oligosaccharide contacts both FGF-1 units of the dimeric FGF–FGFR complex but, only one of the two FGFRs [96]. In another model [97], the complex has 2 : 2 : 2 stoichiometry, the heparin contacting both FGF and FGFR and the 6-O-sulfate reportedly playing a key role.
It is established that binding of HS or heparin to FGF or FGFR alone does not correspond with the activity of the signalling complex [84]. The formation of the ternary complex bestows a set of structural constrains that are likely to be more selective than those imposed by the binding of the polysaccharide to either the FGF or FGFR alone. Indeed, selecting structures in terms of their activity, which involves interactions with both an FGF and its receptor [98], has allowed some of the structural requirements of activation of FGF and its receptors by HS to be addressed [43,99]. More recently, it has been shown that, for FGF-1, even the GAG polarity may be essential for HS–FGF binding and signalling [100].
Tissue-derived HS chromatographic fractions and more recently, several synthetic and defined oligosaccharides [101–103] have also been generated, with the purpose of exploring structure–function relationships [4,12,43,64,66,104–109]. However, it is conceivable that the use of such fractions may misrepresent activities performed by stretches of the full-length polysaccharide.
3.4. Interactions with protein networks
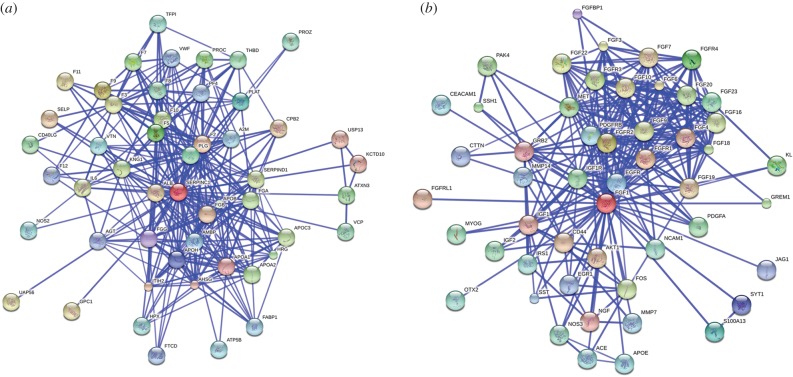
Heparin and HS are also endowed with a variety of functions regulated by their distribution in tissues [110] and facilitate a complex HS ‘interactome’ [33,34], the interactome of a molecule is the whole set of interactions a molecule is involved in. The HS interactome may have a wide influence on biological processes because, while HS interacts with a large variety of proteins [34,111], these HS-binding proteins themselves interact with a plethora of others, forming complex protein–protein interaction networks, in a way that their respective activities within a given network will change (figure 2). AT, for instance, when bound to heparin will have its interaction with factor II and X enhanced (figure 2a). Furthermore, once bound to HS, FGF-1 will bind to its cell-surface receptors, FGFR1 and others, eliciting a given cellular response (figure 2b). On the other hand, the function of biologically available heparin, in contrast to that supplied pharmaceutically, is not clear, and seems most likely to be linked to inflammatory responses when released by mast cells during inflammatory events [113]. Presumably, heparin will be available for protein binding to an interactome of comparable complexity, at least in the circulatory and lymphatic systems, as well as in tissues into which it can diffuse. Because HS is structurally diverse, its expression is regulated and linked to functional outcomes and the relationship between its structure and function is widely debated. Generally, there is little evidence of simple correlations between the sequence of particular disaccharide units and a given activity, and several perspectives have been presented in previous reviews [35–41,114]. The evidence suggests overwhelmingly that several saccharides with different disaccharide compositions, hence distinct sequences, can generate geometry and charge distributions appropriate to elicit similar biological effects. This implies that the requirement to bind proteins can be fairly relaxed but, it should be emphasized, by no means do all sequences behave similarly. Taking this into account and considering their potential sequence diversity, whose interactions are determined ultimately by the three-dimensional architecture arising from that sequence, the huge information content seemingly available in heparin and HS chains may be lower than previously thought [115]. Furthermore, individual HS/heparin structures are capable of interacting with more than one protein [116,117], which has led to the proposal that the relationship between heparin/HS structure and activity should be viewed at the level of their interactions with multiple proteins [116,118,119]. It remains an open question as to whether all heparin/HS sequences interact to some degree with proteins and whether synergy effects are relevant. If this is the case, the analysis of interactions throughout networks will be essential.
Figure 2.
Protein–protein interaction (PPI) network of heparin/HS binding proteins. (a) PPI network for AT. (b) PPI network for FGF-1. Networks were generated with STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) [112]. They are shown in the confidence view. Stronger associations are represented by thicker lines.
The nature of HBP-interaction networks in disease states has recently been addressed. Two studies concentrated on attempting to establish the network of proteins interacting with HS/heparin in angiogenesis. The first was based on an analysis of the affinity and kinetics [120], and the second on an analysis of the supposed HS sulfation requirements of proteins involved in angiogenesis and construction of a connectivity map [121]. In another approach, the mRNA expression of HBPs in various stages of pancreatic disease were used to identify relevant HBP networks, which exhibited distinct characteristics for the various disease states [32]. In other work, microarray analysis of proteins involved in breast cancer showed that the expression of 105 proteins from a total of 1357 genes were influenced by heparin and these related to tumourigenicity [122].
4. Physico-chemical approaches
4.1. Fundamentals of polysaccharide–protein interactions: the polysaccharide as polyanion
One approach to understanding interactions between polysaccharides and proteins is to explore the fundamental properties of the species involved. This is the basis of the approach adopted by Dubin, Seyrek and co-workers, who have applied concepts from polymer and polyelectrolyte chemistry to their analyses [123] as a means of examining the interaction between the highly negatively charged polysaccharides and their protein binding partners, considered as charges in a given space. For example, interaction between AT and heparin bound on an affinity column exhibited a continuous, broad maximum in the apparent binding, rather than showing any abrupt changes (indicative of ‘high’ and ‘low’ affinity fractions), as the ionic strength and pH required for elution from an affinity column was varied, suggesting relatively low specificity [124,125]. A degree of complementarity between the pattern of charges on the protein and those of the heparin was suggested as explaining selectivity [120]. In a study of the interaction between a polyanionic sulfated model compound and β-lactoglobulin, employing capillary electrophoresis and small angle neutron scattering, increasing the chain flexibility of the saccharide was found to increase binding when the overall negative charge of the polymer dominated [124]. A similar property was uncovered relating to interactions between heparin and BSA [126] and is analogous to the effect of ionic strength on proteins binding to DNA [127]. Treating the interacting species as polyanions demonstrated that the overall attractive and repulsive forces participating in an interaction were important to consider. These depended on polymer chain length and charge density but the ionic conditions in the buffer were also critical [126]. In the case of AT, a low affinity binding fraction from heparin was found to contain low charge species [125] and Kd values, which peaked at particular salt concentrations, were very similar for heparin derivatives of similar charge [128]. Furthermore, a correlation between the charge density of both low molecular weight heparin and dermatan sulfate with apparent affinity was noted, leading to the suggestion that there are multiple binding sites on the polysaccharide chain for AT [125] and arguing against exquisite specificity. In a development of this theme, longer range interactions, which had hitherto been thought to be of relatively minor importance, were then considered in conjunction with modelling studies, supporting the idea that the strongest binding occurs when the heparin/HS polyanion sequences are able to bind the protein in such a way as to optimize both attractive and repulsive electrostatic forces, hence, minimize global energy. Furthermore, simulations showed that there remained some conformational freedom in the bound carbohydrate chain, even when bound with high affinity, and there was evidence of multiple binding sites with different affinities [123]. It is important to attempt to emulate the ionic conditions that pertain physiologically in such investigations because it was observed that, employing ITC [129], only 30% of binding free energy was provided by charge–charge interactions.
4.2. Cation binding to heparan sulfate/heparin and its effects
The polyanionic nature of heparin and HS, arising from carboxylate groups, O- and N-sulfates, dictates that these polysaccharides must be associated with a large number of cations in solution. In evolutionary terms, it has been suggested that one of the first functions for such molecules may have been in metal ion sequestration and transfer [130]. Measuring the concentrations of cations in cells and tissues is not only extremely difficult, particularly given the likely variations over small distances, their temporal variation and regulation, as well as cell compartmentalization, but is compounded by the dearth of techniques able to make such measurements. Despite these difficulties, some information has emerged. It is known that physiological cation concentrations can affect heparin/HS activity significantly [131] and can vary, for example, for K+ from 3.0 mM extracellularly to 150 mM intracellularly which may be relevant if HS can gain entry into the cell, as has been suggested [132]. However, it is likely that, for example in the vicinity of ion channels, considerable variation in local concentration could prevail.
There are several examples of the effects of bound cations on the activity of GAGs. For example, the ratio of AT III/thrombin activation varied according to the identity of the cation, when present at 100 mM [133] and rarer cations, such as Cu2+ have also been found to increase during tumourigenesis and angiogenesis [134,135]. Studies of the structural effects of cation binding to heparin/HS saccharides using NMR include their influence on heparin [136], which indicated delocalized, long-range interactions with carboxylate groups for Na+, Mg2+ and Ca2+ at low pH values that, for Na+ and Mg2+, were also maintained at higher pH values. However, it has been noted [137] that not all heparin–cation interactions may be described in terms of relatively simple charge-density considerations, as described by Manning [138], because diffusion rates of cations were shown to be sensitive to the concentration of Na+ ions [139] and heparin exhibited an apparently lower charge density than predicted [140]. Obvious questions concerning the observation of altered properties and activities of heparin/HS with cations include whether such effects arise from the consequences of changing the charge distribution and/or the resulting altered conformation and flexibility. Detailed studies of small oligosaccharide model compounds have been conducted, in which a specific binding site was proposed between glucosamine and iduronate residues for Ca2+ ion binding, stiffening the molecule. However, for Na+ and Mg2+, no preferred binding site was apparent [141]. These findings agree with observations made of a heparin derivative (2-de-O-sulfated), in which the binding of divalent cations stiffened the heparin chain considerably, deduced from altered NMR T2 relaxation measurements. Binding of Ca2+ also changed substantially the conformation across glycosidic linkages, as well as the equilibrium of chair and skew-boat forms of the iduronate residues [142]. The effects of cation binding on conformation and dynamics have also been analysed with heparin-derived oligosaccharides possessing different substitution patterns, which exhibited considerable anisotropy and distinct internal motions. Furthermore, the recognition of HS by phage display antibodies can be altered by exposure to cations [143]. Altering the predominant cation from Na+ to Ca2+ induced conformational changes in the 2-O-sulfated iduronic acid residue, which was transferred to the conformation of the adjacent glucosamine moiety [144]. It is worth noting, however, that the validity of extrapolating results from small molecules to large polyanions has been questioned [125].
The consequences of altering the cation form of heparin/HS on biological activity can also be substantial, which has been known for some time in the FGF/FGFR signalling field [142,145]. Changes in signalling ability can be induced by cations bound to heparin/HS analogues. For example, for one chemically modified heparin analogue of HS, conversion of the predominant cation to Cu2+ employing cation exchange resin resulted in a complex active in FGF-2/R1c signalling [142], while changing the cation from Na+ to Cu2+ of another analogue, converted it from signalling to inhibitory [146], and involved a structural change at the level of FGF–HS interaction as measured in solution by circular dichroism spectroscopy. It has also been shown that Ca2+ is an essential activator of a heparin degrading enzyme, heparinase I, in which the heparin–Ca2+ complex is the true enzyme substrate, whereas Ca2+-free heparin is a competitive inhibitor [147].
Annexin V, an abundant anticoagulant and phospholipid binding protein binds HS in a calcium ion-dependent manner, binding via two binding sites on opposite faces of the protein [148,149]. The phospholipid binding and calcium-dependent annexin I has been reported as requiring N- and 2-O-sulfate groups for binding [150]. The influence of cations can also vary. For example, heparin stimulates the activation of human plasminogen (Pg) by tissue-type Pg activator and in this, HS differs from heparin, while the effect of the polysaccharide depends on the ionic strength, and can range from stimulatory to inhibitory [151]. A final example of the involvement of cations is the prion–protein interaction with low molecular weight heparin and HS, which occurs through the formation of oligomeric complexes stabilized by Cu(II) bridges. At low pH, this interaction involves protonated histidine residues but, at higher pH values the GAG binds the histidine-bound Cu(II) ion [50].
Understandably, owing to lack of sensitive and accessible instrumentation for the detection of cations in biological environments, the influence of cations is frequently ignored. Nevertheless, the examples highlighted indicate that cation–heparin/HS interactions need to be considered, if possible, when dissecting biological activities. A final consideration is the possible effect of cation binding on the protein. One protein, which has several pairs of acidic residues exposed on its surface, with distinct spacings, suitable for cation sequestration evident from the crystallographic model, is lysozyme [152]. Again, whether these examples can be extrapolated to physiological conditions is not known. Nonetheless, accumulating in vitro data suggest that it may be so, but conclusive answers may only become available after further technological development.
4.3. Other physico-chemical approaches
A combination of other physical chemical techniques including polarized light microscopy, reflection anisotropy and terahertz (1012 Hz) absorbance spectroscopies have been used to examine the properties of physiologically relevant cationic forms of heparin (Na+, Ca2+, Mg2+, Cu2+, Zn2+), showing that heparin chains in several cation forms (100 mg ml−1, in water) do not appear to exhibit substantial molecular ordering. Furthermore, heparin samples showed considerable absorption of terahertz radiation in contrast to neutral dextran samples, indicating that heparin can occupy new vibrational modes as the temperature increases while the uncharged dextrans for the most part, simply move more quickly [153], implying potential temperature sensitivity in HS–cation and HS–protein interactions that has been little studied. The interactions between heparin and Cu2+ ions in particular stand out for their high degree of regioselectivity within the heparin chain. A combination of NMR, FTIR and EPR spectroscopies showed that the ion binds preferentially between the iduronate residue and the adjacent 6-O-sulfated glucosamine, adopting tetragonal coordination involving the carboxylic acid, the 6-O-sulfate, the ring oxygen of the iduronate and the glycosidic oxygen [154].
5. Concluding remarks: a degree of redundancy and selectivity
5.1. Summary of findings
The findings regarding the relationship between structure and activity can be summarized and a synthesis of the findings from the biochemical evidence and the physico-chemical approaches may be attempted. There are observations showing that several diverse carbohydrate scaffolds can support activity with particular proteins and that protein binding does not equate to activity directly. Indeed, inactive [155] and inhibitory HS structures have been described for some activities [107,146] but, neither binding nor activity depends on sequences and charge alone [107,146], although charge undoubtedly plays an important role bringing the interacting species into proximity [156,157]. However, activity does not depend on the presence of particular ‘key sulfates’ [158] yet, paradoxically, there is a sizeable and presumably energetically costly biosynthesis apparatus for HS and heparin [159], the regulation of which is currently unclear.
5.2. What happens as polysaccharide and protein approach each other?
At long range, heparin/HS represents a highly charged target for protein binding, that attracts the positively charged binding sites of proteins via lysine and arginine (as well as potentially, under appropriate pH conditions, histidine). As such, different HS binding proteins can bind with relatively low affinity to common sequences and translocate across HS chains [116]. As the distance between the two interacting species decreases, however, a degree of charge and shape complementarity may come into play, strong binding resulting from optimization of attractive and repulsive forces through compatible charge distribution and shape complementarity, leading to more protein-specific, sustained binding [116,160]. Some conformational change is usually evident in the protein and, although not often studied, also seems likely in the polysaccharide. The role of solvent water and the thermodynamic consequences of releasing solvated cations may also be important and would warrant further study. An additional consideration that may be expected to affect protein binding is the existence of multiple polysaccharide chains on, or near, the cell surface, whether they originate from the same HSPG, or separate HSPGs. The importance of multivalency in cooperative glycan–protein interactions has also been demonstrated [161].
Understandably, the emphasis has usually been on studying interactions between heparin/HS and individual proteins in isolated systems. However, it has been shown that heparin/HS interacts with hundreds of proteins, many of them associated with the extracellular matrix and involved in intercellular signalling [34]. According to most of the observations reported to date, there is little evidence for a high degree of sequence specificity, rather, support for selectivity with some redundancy, yet biosynthetic machinery comprising around 20 enzymes exists for HS/heparin biosynthesis. Ultimately, structure drives function, yet, ‘structure’ cannot be simplified to primary composition, sequence and substitution patterns. Perhaps, as for proteins, several of the binding modes may be explained by three-dimensional architecture in which—for HS/heparin—several sequences can lead to similar topological chemical entities; hence, the considerable redundancy observed.
5.3. A link to cation concentration and transport?
A key question in HS/heparin–protein interactions that remains largely unanswered concerns the role of cations, which can alter the activity of the molecule. If cations are not linked to the regulation of HS biosynthesis in some way, but simply encounter HS/heparin and these encounters modify activity essentially at random, then the nature of heparin/HS interactions with proteins seems even more loosely regulated. To begin to resolve this, it may be necessary to consider the interplay between the network of proteins interacting with HS/heparin and their relation to both the biosynthetic machinery and mechanisms for the regulation of cations. An appreciation of the effects of structural modification to HS/heparin on this network may reveal that particular regions of the protein network are affected. What limited work has taken place in this area has established the significant differences in the effect of calcium, lowering the affinity of heparin for thrombin [162,133] and among the signalling network of the FGF–FGFR system with heparin as the polysaccharide cofactor [118,119].
These ideas do go some way to explaining how, when an agent such as heparin is added to the body, despite (or perhaps because of) the hundreds of potential interactions that are possible in the circulatory and lymph systems and elsewhere following diffusion into tissues, very few side-effects are observed. Notwithstanding, the relatively low number of cases of such problems as heparin-induced thrombocytopenia, that can occur with prolonged heparin administration, the potentially adverse effects of adding pharmaceutical heparin may be absorbed largely by the network of proteins with which it interacts. It will be interesting to see whether particular ‘zonal regulation’ of the signalling network takes place, when it is perturbed by addition of HS, or its modification, or whether a simple altering of the signalling intensity throughout the network is observed.
Acknowledgements
Support from the CAPES (E.A.Y., M.A.L. and H.B.N.) and FAPESP (M.A.L. and H.B.N.) is gratefully acknowledged. M.C.Z. is recipient of a PhD Fellowship from CNPq.
Authors' contributions
M.A.L., A.K.P., T.R.R. and E.A.Y. conceived the review, whereas all the authors contributed to research, writing and editing.
Competing interests
We declare have no competing interests.
Funding
We received no funding for this study.
References
- 1.Perlin AS, Mackie DM, Dietrich CP. 1971. Evidence for a (1→4)-linked 4-O-(-l-idopyranosyluronic acid 2-sulfate)-(2-deoxy-2-sulfoamino-d-glucopyranosyl 6-sulfate) sequence in heparin. Long-range H-H coupling in 4-deoxy-hex-4-enopyranosides. Carbohydr. Res. 18, 185–194. ( 10.1016/S0008-6215(00)80341-9) [DOI] [PubMed] [Google Scholar]
- 2.Casu B, et al. 1996. Characterization of sulfation patterns of beef and pig mucosal heparins by nuclear magnetic resonance spectroscopy. Arzneimittelforschung 46, 472–477. [PubMed] [Google Scholar]
- 3.Rabenstein DL. 2002. Heparin and heparan sulfate: structure and function. Nat. Prod. Rep. 19, 312–331. ( 10.1039/b100916h) [DOI] [PubMed] [Google Scholar]
- 4.Bianchini P, Osima B, Parma B, Nader HB, Dietrich CP. 1985. Lack of correlation between ‘in vitro’ and ‘in vivo’ antithrombotic activity of heparin fractions and related compounds. Heparan sulfate as an antithrombotic agent ‘in vivo’. Thromb. Res. 40, 597–607. ( 10.1016/0049-3848(85)90298-1) [DOI] [PubMed] [Google Scholar]
- 5.Dietrich CP, et al. 1998. Structure of heparan sulfate: identification of variable and constant oligosaccharide domains in eight heparan sulfates of different origins. Cell Mol. Biol. (Noisy-le-grand). 44, 417–429. [PubMed] [Google Scholar]
- 6.Gallagher JT, Walker A. 1985. Molecular distinctions between heparan sulphate and heparin. Analysis of sulphation patterns indicates that heparan sulphate and heparin are separate families of N-sulphated polysaccharides. Biochem. J. 230, 665–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casu B. 1989. Structure of heparin and heparin fragments. Ann. NY Acad. Sci. 556, 1–17. ( 10.1111/j.1749-6632.1989.tb22485.x) [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg RD, Lam L. 1979. Correlation between structure and function of heparin. Proc. Natl Acad. Sci. USA 76, 1218–1222. ( 10.1073/pnas.76.3.1218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulloy B, Gray E, Barrowcliffe TW. 2000. Characterization of unfractionated heparin: comparison of materials from the last 50 years. Thromb. Haemost. 84, 1052–1056. [PubMed] [Google Scholar]
- 10.Khan S, Fung KW, Rodriguez E, Patel R, Gor J, Mulloy B, Perkins SJ. 2013. The solution structure of heparan sulfate differs from that of heparin: implications for function. J. Biol. Chem. 288, 27 737–27 751. ( 10.1074/jbc.M113.492223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan S, Gor J, Mulloy B, Perkins SJ. 2010. Semi-rigid solution structures of heparin by constrained X-ray scattering modelling: new insight into heparin–protein complexes. J. Mol. Biol. 395, 504–521. ( 10.1016/j.jmb.2009.10.064) [DOI] [PubMed] [Google Scholar]
- 12.Brito AS, et al. 2014. A non-hemorrhagic hybrid heparin/heparan sulfate with anticoagulant potential. Carbohydr. Polym. 99, 372–378. ( 10.1016/j.carbpol.2013.08.063) [DOI] [PubMed] [Google Scholar]
- 13.Parra A, Veraldi N, Locatelli M, Fini M, Martini L, Torri G, Sangiorgi L, Bisio A. 2012. Heparin-like heparan sulfate from rabbit cartilage. Glycobiology 22, 248–257. ( 10.1093/glycob/cwr143) [DOI] [PubMed] [Google Scholar]
- 14.Dietrich CP, Nader HB, Perlin AS. 1975. The heterogeneity of heparan sulfate from beef-lung tissue: p.m.r.-spectral evidence. Carbohydr. Res. 41, 334–338. ( 10.1016/S0008-6215(00)87036-6) [DOI] [PubMed] [Google Scholar]
- 15.Lyon M, Deakin JA, Gallagher JT. 1994. Liver heparan sulfate structure. A novel molecular design. J. Biol. Chem. 269, 11 208–11 215. [PubMed] [Google Scholar]
- 16.Vongchan P, Warda M, Toyoda H, Toida T, Marks RM, Linhardt RJ. 2005. Structural characterization of human liver heparan sulfate. Biochim. Biophys. Acta 1721, 1–8. ( 10.1016/j.bbagen.2004.09.007) [DOI] [PubMed] [Google Scholar]
- 17.Nader HB, Dietrich CP, Buonassisi V, Colburn P. 1987. Heparin sequences in the heparan sulfate chains of an endothelial cell proteoglycan. Proc. Natl Acad. Sci. USA 84, 3565–3569. ( 10.1073/pnas.84.11.3565) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao C, Shi X, White M, Huang Y, Hartshorn K, Zaia J. 2013. Comparative glycomics of leukocyte glycosaminoglycans. FEBS J. 280, 2447–2461. ( 10.1111/febs.12231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stringer SE, Mayer-Proschel M, Kalyani A, Rao M, Gallagher JT. 1999. Heparin is a unique marker of progenitors in the glial cell lineage. J. Biol. Chem. 274, 25 455–25 460. ( 10.1074/jbc.274.36.25455) [DOI] [PubMed] [Google Scholar]
- 20.Kreuger J, Kjellen L. 2012. Heparan sulfate biosynthesis: regulation and variability. J. Histochem. Cytochem. 60, 898–907. ( 10.1369/0022155412464972) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudd TR, Yates EA. 2012. A highly efficient tree structure for the biosynthesis of heparan sulfate accounts for the commonly observed disaccharides and suggests a mechanism for domain synthesis. Mol. Biosyst. 8, 1499–1506. ( 10.1039/c2mb25019e) [DOI] [PubMed] [Google Scholar]
- 22.Bain LJ, Feldman RA. 2003. Altered expression of sulfotransferases, glucuronosyltransferases and mrp transporters in FVB/mrp1−/− mice. Xenobiotica 33, 1173–1183. ( 10.1080/00498250310001609138) [DOI] [PubMed] [Google Scholar]
- 23.Lamanna WC, Frese MA, Balleininger M, Dierks T. 2008. Sulf loss influences N-, 2-O-, and 6-O-sulfation of multiple heparan sulfate proteoglycans and modulates fibroblast growth factor signaling. J. Biol. Chem. 283, 27 724–27 735. ( 10.1074/jbc.M802130200) [DOI] [PubMed] [Google Scholar]
- 24.Turnbull J, Powell A, Guimond S. 2001. Heparan sulfate: decoding a dynamic multifunctional cell regulator. Trends Cell Biol. 11, 75–82. ( 10.1016/S0962-8924(00)01897-3) [DOI] [PubMed] [Google Scholar]
- 25.Kato M, Wang H, Bernfield M, Gallagher JT, Turnbull JE. 1994. Cell surface syndecan-1 on distinct cell types differs in fine structure and ligand binding of its heparan sulfate chains. J. Biol. Chem. 269, 18 881–18 890. [PubMed] [Google Scholar]
- 26.Kraemer PM, Tobey RA. 1972. Cell-cycle dependent desquamation of heparan sulfate from the cell surface. J. Cell Biol. 55, 713–717. ( 10.1083/jcb.55.3.713) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blair OC, Sartorelli AC. 1984. Incorporation of 35S-sulfate and 3H-glucosamine into heparan and chondroitin sulfates during the cell cycle of B16-F10 cells. Cytometry 5, 281–288. ( 10.1002/cyto.990050311) [DOI] [PubMed] [Google Scholar]
- 28.Porcionatto MA, Pinto CR, Dietrich CP, Nader HB. 1994. Heparan sulfate proteoglycan and control of cell proliferation: enhanced synthesis induced by phorbol ester (PMA) during G(1)-phase. Braz. J. Med. Biol. Res. 27, 2185–2190. [PubMed] [Google Scholar]
- 29.Porcionatto MA, Moreira CR, Lotfi CF, Armelin HA, Dietrich CP, Nader HB. 1998. Stimulation of heparan sulfate proteoglycan synthesis and secretion during G1 phase induced by growth factors and PMA. J. Cell. Biochem. 70, 563–572. () [DOI] [PubMed] [Google Scholar]
- 30.Kayitmazer AB, Seeman D, Minsky BB, Dubin PL, Xu Y. 2013. Protein–polyelectrolyte interactions. Soft Matter 9, 2553–2583. ( 10.1039/c2sm27002a) [DOI] [Google Scholar]
- 31.Mulloy B, Forster MJ. 2000. Conformation and dynamics of heparin and heparan sulfate. Glycobiology 10, 1147–1156. ( 10.1093/glycob/10.11.1147) [DOI] [PubMed] [Google Scholar]
- 32.Nunes QM, Mournetas V, Lane B, Sutton R, Fernig DG, Vasieva O. 2013. The heparin-binding protein interactome in pancreatic diseases. Pancreatology 13, 598–604. ( 10.1016/j.pan.2013.08.004) [DOI] [PubMed] [Google Scholar]
- 33.Ori A, Wilkinson MC, Fernig DG. 2008. The heparanome and regulation of cell function: structures, functions and challenges. Front. Biosci. 13, 4309–4338. ( 10.2741/3007) [DOI] [PubMed] [Google Scholar]
- 34.Ori A, Wilkinson MC, Fernig DG. 2011. A systems biology approach for the investigation of the heparin/heparan sulfate interactome. J. Biol. Chem. 286, 19 892–19 904. ( 10.1074/jbc.M111.228114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell AK, Yates EA, Fernig DG, Turnbull JE. 2004. Interactions of heparin/heparan sulfate with proteins: appraisal of structural factors and experimental approaches. Glycobiology 14, R17–R30. ( 10.1093/glycob/cwh051) [DOI] [PubMed] [Google Scholar]
- 36.Xu D, Esko JD. 2014. Demystifying heparan sulfate–protein interactions. Annu. Rev. Biochem. 83, 129–157. ( 10.1146/annurev-biochem-060713-035314) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreuger J, Spillmann D, Li JP, Lindahl U. 2006. Interactions between heparan sulfate and proteins: the concept of specificity. J. Cell Biol. 174, 323–327. ( 10.1083/jcb.200604035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallagher JT. 2001. Heparan sulfate: growth control with a restricted sequence menu. J. Clin. Invest. 108, 357–361. ( 10.1172/JCI13713) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capila I, Linhardt RJ. 2002. Heparin–protein interactions. Angew. Chem. 41, 391–412. () [DOI] [PubMed] [Google Scholar]
- 40.Lindahl U, Li JP. 2009. Interactions between heparan sulfate and proteins–design and functional implications. Int. Rev. Cell. Mol. Biol. 276, 105–159. ( 10.1016/S1937-6448(09)76003-4) [DOI] [PubMed] [Google Scholar]
- 41.Nugent MA, Zaia J, Spencer JL. 2013. Heparan sulfate–protein binding specificity. Biochemistry (Mosc). 78, 726–735. ( 10.1134/S0006297913070055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez S, Mulloy B. 2005. Prospects for glycoinformatics. Curr. Opin. Struct. Biol. 15, 517–524. ( 10.1016/j.sbi.2005.08.005) [DOI] [PubMed] [Google Scholar]
- 43.Gambarini AG, Miyamoto CA, Lima GA, Nader HB, Dietrich CP. 1993. Mitogenic activity of acidic fibroblast growth factor is enhanced by highly sulfated oligosaccharides derived from heparin and heparan sulfate. Mol. Cell. Biochem. 124, 121–129. ( 10.1007/BF00929204) [DOI] [PubMed] [Google Scholar]
- 44.Kinsella L, Chen HL, Smith JA, Rudland PS, Fernig DG. 1998. Interactions of putative heparin-binding domains of basic fibroblast growth factor and its receptor, FGFR-1, with heparin using synthetic peptides. Glycoconj. J. 15, 419–422. ( 10.1023/A:1006986104865) [DOI] [PubMed] [Google Scholar]
- 45.Rahmoune H, Chen HL, Gallagher JT, Rudland PS, Fernig DG. 1998. Interaction of heparan sulfate from mammary cells with acidic fibroblast growth factor (FGF) and basic FGF. Regulation of the activity of basic FGF by high and low affinity binding sites in heparan sulfate. J. Biol. Chem. 273, 7303–7310. ( 10.1074/jbc.273.13.7303) [DOI] [PubMed] [Google Scholar]
- 46.Mason K, Meikle P, Hopwood J, Fuller M. 2014. Distribution of heparan sulfate oligosaccharides in murine mucopolysaccharidosis type IIIA. Metabolites 4, 1088–1100. ( 10.3390/metabo4041088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li JP, Galvis ML, Gong F, Zhang X, Zcharia E, Metzger S, Vlodavsky I, Kisilevsky R, Lindahl U. 2005. In vivo fragmentation of heparan sulfate by heparanase overexpression renders mice resistant to amyloid protein A amyloidosis. Proc. Natl Acad. Sci. USA 102, 6473–6477. ( 10.1073/pnas.0502287102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. 2004. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J. Natl Cancer Inst. 96, 1219–1230. ( 10.1093/jnci/djh230) [DOI] [PubMed] [Google Scholar]
- 49.Cohen I, Pappo O, Elkin M, San T, Bar-Shavit R, Hazan R, Peretz T, Vlodavsky I, Abramovitch R. 2006. Heparanase promotes growth, angiogenesis and survival of primary breast tumors. Int. J. Cancer 118, 1609–1617. ( 10.1002/ijc.21552) [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez-Iglesias R, Pajares MA, Ocal C, Espinosa JC, Oesch B, Gasset M. 2002. Prion protein interaction with glycosaminoglycan occurs with the formation of oligomeric complexes stabilized by Cu(II) bridges. J. Mol. Biol. 319, 527–540. ( 10.1016/S0022-2836(02)00341-8) [DOI] [PubMed] [Google Scholar]
- 51.Fromm JR, Hileman RE, Caldwell EE, Weiler JM, Linhardt RJ. 1995. Differences in the interaction of heparin with arginine and lysine and the importance of these basic amino acids in the binding of heparin to acidic fibroblast growth factor. Arch. Biochem. Biophys. 323, 279–287. ( 10.1006/abbi.1995.9963) [DOI] [PubMed] [Google Scholar]
- 52.Sterner E, et al. 2014. Fibroblast growth factor-based signaling through synthetic heparan sulfate blocks copolymers studied using high cell density three-dimensional cell printing. J. Biol. Chem. 289, 9754–9765. ( 10.1074/jbc.M113.546937) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holman J, Skidmore MA, Rudd TR, Yates EA. 2010. The latent ampholytic nature of glycosaminoglycan (GAG) oligosaccharides facilitates their separation by isoelectric focusing. Anal. Methods 2, 1550–1554. ( 10.1039/c0ay00340a) [DOI] [Google Scholar]
- 54.Huang R, Liu J, Sharp JS. 2013. An approach for separation and complete structural sequencing of heparin/heparan sulfate-like oligosaccharides. Anal. Chem. 85, 5787–5795. ( 10.1021/ac400439a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei W, Miller RL, Leary JA. 2013. Method development and analysis of free HS and HS in proteoglycans from pre- and postmenopausal women: evidence for biosynthetic pathway changes in sulfotransferase and sulfatase enzymes. Anal. Chem. 85, 5917–5923. ( 10.1021/ac400690g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ucakturk E, Cai C, Li L, Li G, Zhang F, Linhardt RJ. 2014. Capillary electrophoresis for total glycosaminoglycan analysis. Anal. Bioanal. Chem. 406, 4617–4626. ( 10.1007/s00216-014-7859-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Inoue Y, Nagasawa K. 1976. Selective N-desulfation of heparin with dimethyl sulfoxide containing water or methanol. Carbohydr. Res. 46, 87–95. ( 10.1016/S0008-6215(00)83533-8) [DOI] [PubMed] [Google Scholar]
- 58.Rej RN, Ludwig-Baxter KG, Perlin AS. 1991. Sulfation of some chemically-modified heparins. Formation of a 3-sulfate analog of heparin. Carbohydr. Res. 210, 299–310. ( 10.1016/0008-6215(91)80130-F) [DOI] [PubMed] [Google Scholar]
- 59.Yates EA, Santini F, Guerrini M, Naggi A, Torri G, Casu B. 1996. 1H and 13C NMR spectral assignments of the major sequences of twelve systematically modified heparin derivatives. Carbohydr. Res. 294, 15–27. ( 10.1016/S0008-6215(96)00213-3) [DOI] [PubMed] [Google Scholar]
- 60.Paredes-Gamero EJ, et al. 2012. Chemical reduction of carboxyl groups in heparin abolishes its vasodilatory activity. J. Cell. Biochem. 113, 1359–1367. ( 10.1002/jcb.24008) [DOI] [PubMed] [Google Scholar]
- 61.Powell AK, Ahmed YA, Yates EA, Turnbull JE. 2010. Generating heparan sulfate saccharide libraries for glycomics applications. Nat. Protoc. 5, 821–833. ( 10.1038/nprot.2010.17) [DOI] [PubMed] [Google Scholar]
- 62.Uniewicz KA, Ori A, Ahmed YA, Yates EA, Fernig DG. 2014. Characterisation of the interaction of neuropilin-1 with heparin and a heparan sulfate mimetic library of heparin-derived sugars. PeerJ. 2, e461 ( 10.7717/peerj.461) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kreuger J, Prydz K, Pettersson RF, Lindahl U, Salmivirta M. 1999. Characterization of fibroblast growth factor 1 binding heparan sulfate domain. Glycobiology 9, 723–729. ( 10.1093/glycob/9.7.723) [DOI] [PubMed] [Google Scholar]
- 64.Kreuger J, Salmivirta M, Sturiale L, Gimenez-Gallego G, Lindahl U. 2001. Sequence analysis of heparan sulfate epitopes with graded affinities for fibroblast growth factors 1 and 2. J. Biol. Chem. 276, 30 744–30 752. ( 10.1074/jbc.M102628200) [DOI] [PubMed] [Google Scholar]
- 65.Maccarana M, Casu B, Lindahl U. 1993. Minimal sequence in heparin/heparan sulfate required for binding of basic fibroblast growth factor. J. Biol. Chem. 268, 23 898–23 905. ( 10.1007/bf01209909) [DOI] [PubMed] [Google Scholar]
- 66.Turnbull JE, Fernig DG, Ke Y, Wilkinson MC, Gallagher JT. 1992. Identification of the basic fibroblast growth factor binding sequence in fibroblast heparan sulfate. J. Biol. Chem. 267, 10 337–10 341. [PubMed] [Google Scholar]
- 67.Turnbull JE, Gallagher JT. 1993. Heparan sulphate: functional role as a modulator of fibroblast growth factor activity. Biochem. Soc. Trans. 21, 477–482. [DOI] [PubMed] [Google Scholar]
- 68.Guerrini M, et al. 2002. Minimal heparin/heparan sulfate sequences for binding to fibroblast growth factor-1. Biochem. Biophys. Res. Commun. 292, 222–230. ( 10.1006/bbrc.2002.6634) [DOI] [PubMed] [Google Scholar]
- 69.Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. 1985. Contribution of monosaccharide residues in heparin binding to antithrombin III. Biochemistry 24, 6723–6729. ( 10.1021/bi00344a063) [DOI] [PubMed] [Google Scholar]
- 70.Atha DH, Lormeau JC, Petitou M, Rosenberg RD, Choay J. 1987. Contribution of 3-O- and 6-O-sulfated glucosamine residues in the heparin-induced conformational change in antithrombin III. Biochemistry 26, 6454–6461. ( 10.1021/bi00394a024) [DOI] [PubMed] [Google Scholar]
- 71.Esko JD, Lindahl U. 2001. Molecular diversity of heparan sulfate. J. Clin. Invest. 108, 169–173. ( 10.1172/JCI200113530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chavante SF, Brito AS, Lima M, Yates E, Nader H, Guerrini M, Torri G, Bisio A. 2014. A heparin-like glycosaminoglycan from shrimp containing high levels of 3-O-sulfated d-glucosamine groups in an unusual trisaccharide sequence. Carbohydr. Res. 390, 59–66. ( 10.1016/j.carres.2014.03.002) [DOI] [PubMed] [Google Scholar]
- 73.Lima MA, et al. 2013. Antithrombin stabilisation by sulfated carbohydrates correlates with anticoagulant activity. Med. Chem. Commun. 4, 870–873. ( 10.1039/c3md00048f) [DOI] [Google Scholar]
- 74.Guerrini M, et al. 2013. An unusual antithrombin-binding heparin octasaccharide with an additional 3-O-sulfated glucosamine in the active pentasaccharide sequence. Biochem. J. 449, 343–351. ( 10.1042/BJ20121309) [DOI] [PubMed] [Google Scholar]
- 75.Guerrini M, Guglieri S, Beccati D, Torri G, Viskov C, Mourier P. 2006. Conformational transitions induced in heparin octasaccharides by binding with antithrombin III. Biochem. J. 399, 191–198. ( 10.1042/BJ20060656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guerrini M, et al. 2008. Antithrombin-binding octasaccharides and role of extensions of the active pentasaccharide sequence in the specificity and strength of interaction. Evidence for very high affinity induced by an unusual glucuronic acid residue. J. Biol. Chem. 283, 26 662–26 675. ( 10.1074/jbc.M801102200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guerrini M, Mourier PA, Torri G, Viskov C. 2014. Antithrombin-binding oligosaccharides: structural diversities in a unique function? Glycoconj. J. 31, 409–416. ( 10.1007/s10719-014-9543-9) [DOI] [PubMed] [Google Scholar]
- 78.Viskov C, et al. 2013. Heparin dodecasaccharide containing two antithrombin-binding pentasaccharides: structural features and biological properties. J. Biol. Chem. 288, 25 895–25 907. ( 10.1074/jbc.M113.485268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Henry BL, Connell J, Liang A, Krishnasamy C, Desai UR. 2009. Interaction of antithrombin with sulfated, low molecular weight lignins: opportunities for potent, selective modulation of antithrombin function. J. Biol. Chem. 284, 20 897–20 908. ( 10.1074/jbc.M109.013359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson DJ, Huntington JA. 2003. Crystal structure of antithrombin in a heparin-bound intermediate state. Biochemistry 42, 8712–8719. ( 10.1021/bi034524y) [DOI] [PubMed] [Google Scholar]
- 81.Mosier PD, Krishnasamy C, Kellogg GE, Desai UR. 2012. On the specificity of heparin/heparan sulfate binding to proteins. Anion-binding sites on antithrombin and thrombin are fundamentally different. PLoS ONE 7, e48632 ( 10.1371/journal.pone.0048632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Toth L, Fekete A, Balogh G, Bereczky Z, Komaromi I. 2014. Dynamic properties of the native free antithrombin from molecular dynamics simulations: computational evidence for solvent-exposed Arg393 side chain. J. Biomol. Struct. Dyn. 8, 1–14. [DOI] [PubMed] [Google Scholar]
- 83.Powell AK, Fernig DG, Turnbull JE. 2002. Fibroblast growth factor receptors 1 and 2 interact differently with heparin/heparan sulfate. Implications for dynamic assembly of a ternary signaling complex. J. Biol. Chem. 277, 28 554–28 563. ( 10.1074/jbc.M111754200) [DOI] [PubMed] [Google Scholar]
- 84.Rudd TR, et al. 2010. Comparable stabilisation, structural changes and activities can be induced in FGF by a variety of HS and non-GAG analogues: implications for sequence–activity relationships. Org. Biomol. Chem. 8, 5390–5397. ( 10.1039/c0ob00246a) [DOI] [PubMed] [Google Scholar]
- 85.Ramachandra R, et al. 2014. Brittlestars contain highly sulfated chondroitin sulfates/dermatan sulfates that promote fibroblast growth factor 2-induced cell signaling. Glycobiology 24, 195–207. ( 10.1093/glycob/cwt100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xu R, et al. 2012. Diversification of the structural determinants of fibroblast growth factor-heparin interactions: implications for binding specificity. J. Biol. Chem. 287, 40 061–40 073. ( 10.1074/jbc.M112.398826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang Z, Meyer K, Rapraeger AC, Friedl A. 2000. Differential ability of heparan sulfate proteoglycans to assemble the fibroblast growth factor receptor complex in situ. FASEB J. 14, 137–144. [DOI] [PubMed] [Google Scholar]
- 88.Allen BL, Rapraeger AC. 2003. Spatial and temporal expression of heparan sulfate in mouse development regulates FGF and FGF receptor assembly. J. Cell. Biol. 163, 637–648. ( 10.1083/jcb.200307053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Z, Verheyden JM, Hassell JA, Sun X. 2009. FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev. Cell 16, 607–613. ( 10.1016/j.devcel.2009.02.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jastrebova N, Vanwildemeersch M, Lindahl U, Spillmann D. 2010. Heparan sulfate domain organization and sulfation modulate FGF-induced cell signaling. J. Biol. Chem. 285, 26 842–26 851. ( 10.1074/jbc.M109.093542) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yates EA, Guimond SE, Turnbull JE. 2004. Highly diverse heparan sulfate analogue libraries: providing access to expanded areas of sequence space for bioactivity screening. J. Med. Chem. 47, 277–280. ( 10.1021/jm0309755) [DOI] [PubMed] [Google Scholar]
- 92.Ostrovsky O, Berman B, Gallagher J, Mulloy B, Fernig DG, Delehedde M, Ron D. 2002. Differential effects of heparin saccharides on the formation of specific fibroblast growth factor (FGF) and FGF receptor complexes. J. Biol. Chem. 277, 2444–2453. ( 10.1074/jbc.M108540200) [DOI] [PubMed] [Google Scholar]
- 93.Zhang H, Vutskits L, Pepper MS, Kiss JZ. 2003. VEGF is a chemoattractant for FGF-2-stimulated neural progenitors. J. Cell Biol. 163, 1375–1384. ( 10.1083/jcb.200308040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Duchesne L, Tissot B, Rudd TR, Dell A, Fernig DG. 2006. N-glycosylation of fibroblast growth factor receptor 1 regulates ligand and heparan sulfate co-receptor binding. J. Biol. Chem. 281, 27 178–27 189. ( 10.1074/jbc.M601248200) [DOI] [PubMed] [Google Scholar]
- 95.Faham S, Hileman RE, Fromm JR, Linhardt RJ, Rees DC. 1996. Heparin structure and interactions with basic fibroblast growth factor. Science 271, 1116–1120. ( 10.1126/science.271.5252.1116) [DOI] [PubMed] [Google Scholar]
- 96.Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. 2000. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407, 1029–1034. ( 10.1038/35039551) [DOI] [PubMed] [Google Scholar]
- 97.Schlessinger J, et al. 2000. Crystal structure of a ternary FGF–FGFR–heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750. ( 10.1016/S1097-2765(00)00073-3) [DOI] [PubMed] [Google Scholar]
- 98.Guimond S, Maccarana M, Olwin BB, Lindahl U, Rapraeger AC. 1993. Activating and inhibitory heparin sequences for FGF-2 (basic FGF). Distinct requirements for FGF-1, FGF-2, and FGF-4. J. Biol. Chem. 268, 23 906–23 914. [PubMed] [Google Scholar]
- 99.Naimy H, Buczek-Thomas JA, Nugent MA, Leymarie N, Zaia J. 2011. Highly sulfated nonreducing end-derived heparan sulfate domains bind fibroblast growth factor-2 with high affinity and are enriched in biologically active fractions. J. Biol. Chem. 286, 19 311–19 319. ( 10.1074/jbc.M110.204693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Munoz-Garcia JC, et al. 2014. Importance of the polarity of the glycosaminoglycan chain on the interaction with FGF-1. Glycobiology 24, 1004–1009. ( 10.1093/glycob/cwu071) [DOI] [PubMed] [Google Scholar]
- 101.Liu J, Linhardt RJ. 2014. Chemoenzymatic synthesis of heparan sulfate and heparin. Nat. Prod. Rep. 31, 1676–1685. ( 10.1039/C4NP00076E) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu Y, Pempe EH, Liu J. 2012. Chemoenzymatic synthesis of heparin oligosaccharides with both anti-factor Xa and anti-factor IIa activities. J. Biol. Chem. 287, 29 054–29 061. ( 10.1074/jbc.M112.358523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hu YP, et al. 2012. Divergent synthesis of 48 heparan sulfate-based disaccharides and probing the specific sugar-fibroblast growth factor-1 interaction. J. Am. Chem. Soc. 134, 20 722–20 727. ( 10.1021/ja3090065) [DOI] [PubMed] [Google Scholar]
- 104.Chavante SF, Santos EA, Oliveira FW, Guerrini M, Torri G, Casu B, Dietrich CP, Nader HB. 2000. A novel heparan sulphate with high degree of N-sulphation and high heparin cofactor-II activity from the brine shrimp Artemia franciscana. Int. J. Biol. Macromol. 27, 49–57. ( 10.1016/S0141-8130(99)00114-2) [DOI] [PubMed] [Google Scholar]
- 105.Cosmi B, Fredenburgh JC, Rischke J, Hirsh J, Young E, Weitz JI. 1997. Effect of nonspecific binding to plasma proteins on the antithrombin activities of unfractionated heparin, low-molecular-weight heparin, and dermatan sulfate. Circulation 95, 118–124. ( 10.1161/01.CIR.95.1.118) [DOI] [PubMed] [Google Scholar]
- 106.Dietrich CP, et al. 1999. Structural features and anticoagulant activities of a novel natural low molecular weight heparin from the shrimp Penaeus brasiliensis. Biochim. Biophys. Acta 1428, 273–283. ( 10.1016/S0304-4165(99)00087-2) [DOI] [PubMed] [Google Scholar]
- 107.Guimond SE, Turnbull JE. 1999. Fibroblast growth factor receptor signalling is dictated by specific heparan sulphate saccharides. Curr. Biol. 9, 1343–1346. ( 10.1016/S0960-9822(00)80060-3) [DOI] [PubMed] [Google Scholar]
- 108.Pye DA, Vives RR, Hyde P, Gallagher JT. 2000. Regulation of FGF-1 mitogenic activity by heparan sulfate oligosaccharides is dependent on specific structural features: differential requirements for the modulation of FGF-1 and FGF-2. Glycobiology 10, 1183–1192. ( 10.1093/glycob/10.11.1183) [DOI] [PubMed] [Google Scholar]
- 109.Walker A, Turnbull JE, Gallagher JT. 1994. Specific heparan sulfate saccharides mediate the activity of basic fibroblast growth factor. J. Biol. Chem. 269, 931–935. [PubMed] [Google Scholar]
- 110.Dreyfuss JL, Regatieri CV, Jarrouge TR, Cavalheiro RP, Sampaio LO, Nader HB. 2009. Heparan sulfate proteoglycans: structure, protein interactions and cell signaling. Anais da Academia Brasileira de Ciencias 81, 409–429. ( 10.1590/S0001-37652009000300007) [DOI] [PubMed] [Google Scholar]
- 111.Lamanna WC, Kalus I, Padva M, Baldwin RJ, Merry CL, Dierks T. 2007. The heparanome—the enigma of encoding and decoding heparan sulfate sulfation. J. Biotechnol. 129, 290–307. ( 10.1016/j.jbiotec.2007.01.022) [DOI] [PubMed] [Google Scholar]
- 112.Snel B, Lehmann G, Bork P, Huynen MA. 2000. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28, 3442–3444. ( 10.1093/nar/28.18.3442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Straus AH, Sant'anna OA, Nader HB, Dietrich CP. 1984. An inverse relationship between heparin content and antibody response in genetically selected mice. Sex effect and evidence of a polygenic control for skin heparin concentration. Biochem. J. 220, 625–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mulloy B. 2005. The specificity of interactions between proteins and sulfated polysaccharides. Anais da Academia Brasileira de Ciencias 77, 651–664. ( 10.1590/S0001-37652005000400007) [DOI] [PubMed] [Google Scholar]
- 115.Rudd TR, Yates EA. 2010. Conformational degeneracy restricts the effective information content of heparan sulfate. Mol. Biosyst. 6, 902–908. ( 10.1039/b923519a) [DOI] [PubMed] [Google Scholar]
- 116.Chu CL, Goerges AL, Nugent MA. 2005. Identification of common and specific growth factor binding sites in heparan sulfate proteoglycans. Biochemistry 44, 12 203–12 213. ( 10.1021/bi050241p) [DOI] [PubMed] [Google Scholar]
- 117.Forsten-Williams K, Chu CL, Fannon M, Buczek-Thomas JA, Nugent MA. 2008. Control of growth factor networks by heparan sulfate proteoglycans. Ann. Biomed. Eng. 36, 2134–2148. ( 10.1007/s10439-008-9575-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ornitz DM, et al. 1996. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 271, 15 292–15 297. ( 10.1074/jbc.271.25.15292) [DOI] [PubMed] [Google Scholar]
- 119.Xu R, Rudd TR, Hughes AJ, Siligardi G, Fernig DG, Yates EA. 2013. Analysis of the fibroblast growth factor receptor (FGFR) signalling network with heparin as coreceptor: evidence for the expansion of the core FGFR signalling network. FEBS J. 280, 2260–2270. ( 10.1111/febs.12201) [DOI] [PubMed] [Google Scholar]
- 120.Peysselon F, Ricard-Blum S. 2014. Heparin–protein interactions: from affinity and kinetics to biological roles. Application to an interaction network regulating angiogenesis. Matrix Biol. 35, 73–81. ( 10.1016/j.matbio.2013.11.001) [DOI] [PubMed] [Google Scholar]
- 121.Chiodelli P, Bugatti A, Urbinati C, Rusnati M. 2015. Heparin/heparan sulfate proteoglycans glycomic interactome in angiogenesis: biological implications and therapeutical use. Molecules 20, 6342–6388. ( 10.3390/molecules20046342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chen Y, Scully M, Dawson G, Goodwin C, Xia M, Lu X, Kakkar A. 2013. Perturbation of the heparin/heparin-sulfate interactome of human breast cancer cells modulates pro-tumourigenic effects associated with PI3 K/Akt and MAPK/ERK signalling. Thromb. Haemost. 109, 1148–1157. ( 10.1160/TH12-12-0935) [DOI] [PubMed] [Google Scholar]
- 123.Seyrek E, Dubin P. 2010. Glycosaminoglycans as polyelectrolytes. Adv. Colloid Interface Sci. 158, 119–129. ( 10.1016/j.cis.2010.03.001) [DOI] [PubMed] [Google Scholar]
- 124.Hattori T, Bat-Aldar S, Kato R, Bohidar HB, Dubin PL. 2005. Characterization of polyanion–protein complexes by frontal analysis continuous capillary electrophoresis and small angle neutron scattering: effect of polyanion flexibility. Anal. Biochem. 342, 229–236. ( 10.1016/j.ab.2005.03.043) [DOI] [PubMed] [Google Scholar]
- 125.Seyrek E, Dubin PL, Henriksen J. 2007. Nonspecific electrostatic binding characteristics of the heparin–antithrombin interaction. Biopolymers 86, 249–259. ( 10.1002/bip.20731) [DOI] [PubMed] [Google Scholar]
- 126.Seyrek E, Dubin PL, Tribet C, Gamble EA. 2003. Ionic strength dependence of protein–polyelectrolyte interactions. Biomacromolecules 4, 273–282. ( 10.1021/bm025664a) [DOI] [PubMed] [Google Scholar]
- 127.Hart DJ, Speight RE, Cooper MA, Sutherland JD, Blackburn JM. 1999. The salt dependence of DNA recognition by NF-κB p50: a detailed kinetic analysis of the effects on affinity and specificity. Nucleic Acids Res. 27, 1063–1069. ( 10.1093/nar/27.4.1063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Desai UR, Petitou M, Bjork I, Olson ST. 1998. Mechanism of heparin activation of antithrombin: evidence for an induced-fit model of allosteric activation involving two interaction subsites. Biochemistry 37, 13 033–13 041. ( 10.1021/bi981426h) [DOI] [PubMed] [Google Scholar]
- 129.Thompson LD, Pantoliano MW, Springer BA. 1994. Energetic characterization of the basic fibroblast growth factor–heparin interaction: identification of the heparin binding domain. Biochemistry 33, 3831–3840. ( 10.1021/bi00179a006) [DOI] [PubMed] [Google Scholar]
- 130.Heidarieh M, Maragheh MG, Shamami MA, Behgar M, Ziaei F, Akbari Z. 2013. Evaluate of heavy metal concentration in shrimp (Penaeus semisulcatus) and crab (Portunus pelagicus) with INAA method. SpringerPlus 2, 72 ( 10.1186/2193-1801-2-72) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Landt M, Hortin GL, Smith CH, McClellan A, Scott MG. 1994. Interference in ionized calcium measurements by heparin salts. Clin. Chem. 40, 565–570. [PubMed] [Google Scholar]
- 132.Sepulveda-Diaz JE, et al. 2015. HS3ST2 expression is critical for the abnormal phosphorylation of tau in Alzheimer's disease-related tau pathology. Brain 138, 1339–1354. ( 10.1093/brain/awv056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Griffith MJ, Beavers G, Kingdon HS, Lundblad RL. 1980. Effect of monovalent cations on the heparin-enhanced antithrombin III/thrombin reaction. Thromb. Res. 17, 29–39. ( 10.1016/0049-3848(80)90291-1) [DOI] [PubMed] [Google Scholar]
- 134.Alessandri G, Raju K, Gullino PM. 1984. Angiogenesis in vivo and selective mobilization of capillary endothelium in vitro by heparin–copper complex. Microcirc. Endothel. Lymphatics 1, 329–346. [PubMed] [Google Scholar]
- 135.Finney L, et al. 2007. X-ray fluorescence microscopy reveals large-scale relocalization and extracellular translocation of cellular copper during angiogenesis. Proc. Natl Acad. Sci. USA 104, 2247–2252. ( 10.1073/pnas.0607238104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rabenstein DL, Robert JM, Peng J. 1995. Multinuclear magnetic resonance studies of the interaction of inorganic cations with heparin. Carbohydr. Res. 278, 239–256. ( 10.1016/0008-6215(95)00263-4) [DOI] [PubMed] [Google Scholar]
- 137.Grant D, Long WF, Williamson FB. 1992. A potentiometric titration study of the interaction of heparin with metal cations. Biochem. J. 285, 477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Manning GS. 1978. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q. Rev. Biophys. 11, 179–246. ( 10.1017/S0033583500002031) [DOI] [PubMed] [Google Scholar]
- 139.Ander P, Lubas W. 1981. Sodium ion diffusion in aqueous salt-free heparin solutions. Macromolecules 14, 1058–1061. ( 10.1021/ma50005a032) [DOI] [Google Scholar]
- 140.Tivant P, Turq P, Drifford M, Magdelenat H, Menez R. 1983. Effect of ionic strength on the diffusion coefficient of chondroitin sulfate and heparin measured by quasielastic light scattering. Biopolymers 22, 643–662. ( 10.1002/bip.360220209) [DOI] [Google Scholar]
- 141.Chevalier F, Angulo J, Lucas R, Nieto Pedro M, Martín-Lomas M. 2002. The heparin−Ca2+ interaction: structure of the Ca2+ binding site. Eur. J. Org. Chem. 2002, 2367–2376. () [DOI] [Google Scholar]
- 142.Rudd TR, et al. 2007. Influence of substitution pattern and cation binding on conformation and activity in heparin derivatives. Glycobiology 17, 983–993. ( 10.1093/glycob/cwm062) [DOI] [PubMed] [Google Scholar]
- 143.Solari V, Rudd TR, Guimond SE, Powell AK, Turnbull JE, Yates EA. 2015. Heparan sulfate phage display antibodies recognise epitopes defined by a combination of sugar sequence and cation binding. Org. Biomol. Chem. 13, 6066–6072. ( 10.1039/C5OB00564G) [DOI] [PubMed] [Google Scholar]
- 144.Hricovini M. 2011. Effect of solvent and counterions upon structure and NMR spin–spin coupling constants in heparin disaccharide. J. Phys. Chem. B 115, 1503–1511. ( 10.1021/jp1098552) [DOI] [PubMed] [Google Scholar]
- 145.Kan M, Wang F, To B, Gabriel JL, McKeehan WL. 1996. Divalent cations and heparin/heparan sulfate cooperate to control assembly and activity of the fibroblast growth factor receptor complex. J. Biol. Chem. 271, 26 143–26 148. ( 10.1074/jbc.271.42.26143) [DOI] [PubMed] [Google Scholar]
- 146.Guimond SE, et al. 2009. Cations modulate polysaccharide structure to determine FGF–FGFR signaling: a comparison of signaling and inhibitory polysaccharide interactions with FGF-1 in solution. Biochemistry 48, 4772–4779. ( 10.1021/bi802318z) [DOI] [PubMed] [Google Scholar]
- 147.Cordula CR, et al. 2014. On the catalytic mechanism of polysaccharide lyases: evidence of His and Tyr involvement in heparin lysis by heparinase I and the role of Ca2+. Mol. Biosyst. 10, 54–64. ( 10.1039/C3MB70370C) [DOI] [PubMed] [Google Scholar]
- 148.Capila I, Hernaiz MJ, Mo YD, Mealy TR, Campos B, Dedman JR, Linhardt RJ, Seaton BA. 2001. Annexin V–heparin oligosaccharide complex suggests heparan sulfate-mediated assembly on cell surfaces. Structure 9, 57–64. ( 10.1016/S0969-2126(00)00549-9) [DOI] [PubMed] [Google Scholar]
- 149.Capila I, VanderNoot VA, Mealy TR, Seaton BA, Linhardt RJ. 1999. Interaction of heparin with annexin V. FEBS Lett. 446, 327–330. ( 10.1016/S0014-5793(99)00245-8) [DOI] [PubMed] [Google Scholar]
- 150.Horlacher T, Noti C, de Paz JL, Bindschadler P, Hecht ML, Smith DF, Fukuda MN, Seeberger PH. 2011. Characterization of annexin A1 glycan binding reveals binding to highly sulfated glycans with preference for highly sulfated heparan sulfate and heparin. Biochemistry 50, 2650–2659. ( 10.1021/bi101121a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Young TN, Edelberg JM, Stack S, Pizzo SV. 1992. Ionic modulation of the effects of heparin on plasminogen activation by tissue plasminogen activator: the effects of ionic strength, divalent cations, and chloride. Arch. Biochem. Biophys. 296, 530–538. ( 10.1016/0003-9861(92)90607-X) [DOI] [PubMed] [Google Scholar]
- 152.Blake CCF, Koenig DF, Mair GA, North ACT, Phillips DC, Sarma VR. 1965. Structure of hen egg-white lysozyme: a three-dimensional Fourier synthesis at 2Å resolution. Nature 206, 757–761. ( 10.1038/206757a0) [DOI] [PubMed] [Google Scholar]
- 153.Holder GM, et al. 2012. Fundamental differences in model cell-surface polysaccharides revealed by complementary optical and spectroscopic techniques. Soft Matter 8, 6521–6527. ( 10.1039/c2sm25239b) [DOI] [Google Scholar]
- 154.Rudd TR, et al. 2008. Site-specific interactions of copper(II) ions with heparin revealed with complementary (SRCD, NMR, FTIR and EPR) spectroscopic techniques. Carbohydr. Res. 343, 2184–2193. ( 10.1016/j.carres.2007.12.019) [DOI] [PubMed] [Google Scholar]
- 155.Pye DA, Vives RR, Turnbull JE, Hyde P, Gallagher JT. 1998. Heparan sulfate oligosaccharides require 6-O-sulfation for promotion of basic fibroblast growth factor mitogenic activity. J. Biol. Chem. 273, 22 936–22 942. ( 10.1074/jbc.273.36.22936) [DOI] [PubMed] [Google Scholar]
- 156.Jemth P, Kreuger J, Kusche-Gullberg M, Sturiale L, Gimenez-Gallego G, Lindahl U. 2002. Biosynthetic oligosaccharide libraries for identification of protein-binding heparan sulfate motifs. Exploring the structural diversity by screening for fibroblast growth factor (FGF)1 and FGF2 binding. J. Biol. Chem. 277, 30 567–30 573. [DOI] [PubMed] [Google Scholar]
- 157.Kreuger J, Jemth P, Sanders-Lindberg E, Eliahu L, Ron D, Basilico C, Salmivirta M, Lindahl U. 2005. Fibroblast growth factors share binding sites in heparan sulphate. Biochem. J. 389, 145–150. ( 10.1042/BJ20042129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Catlow K, Deakin JA, Delehedde M, Fernig DG, Gallagher JT, Pavao MS, Lyon M. 2003. Hepatocyte growth factor/scatter factor and its interaction with heparan sulphate and dermatan sulphate. Biochem. Soc. Trans. 31, 352–353. ( 10.1042/BST0310352) [DOI] [PubMed] [Google Scholar]
- 159.Hagner-McWhirter A, Li JP, Oscarson S, Lindahl U. 2004. Irreversible glucuronyl C5-epimerization in the biosynthesis of heparan sulfate. J. Biol. Chem. 279, 14 631–14 638. ( 10.1074/jbc.M313760200) [DOI] [PubMed] [Google Scholar]
- 160.Duchesne L, Octeau V, Bearon RN, Beckett A, Prior IA, Lounis B, Fernig DG. 2012. Transport of fibroblast growth factor 2 in the pericellular matrix is controlled by the spatial distribution of its binding sites in heparan sulfate. PLoS Biol. 10, e1001361 ( 10.1371/journal.pbio.1001361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.de Paz JL, Noti C, Bohm F, Werner S, Seeberger PH. 2007. Potentiation of fibroblast growth factor activity by synthetic heparin oligosaccharide glycodendrimers. Chem. Biol. 14, 879–887. ( 10.1016/j.chembiol.2007.07.007) [DOI] [PubMed] [Google Scholar]
- 162.Speight MO, Griffith MJ. 1983. Calcium inhibits the heparin-catalyzed antithrombin III/thrombin reaction by decreasing the apparent binding affinity of heparin for thrombin. Arch. Biochem. Biophys. 225, 958–963. ( 10.1016/0003-9861(83)90111-X) [DOI] [PubMed] [Google Scholar]