Comment on" González-Prieto R, et al. c-Myc is targeted to the proteasome for degradation in a SUMOylation- dependent manner, regulated by PIAS1, SENP7 and RNF4. Cell Cycle 2015; 14(12):1859-72; PMID:25895136; http://dx.doi.org/10.1080/15384101.2015.1040965
The cellular Myc oncoprotein (c-Myc) is part of an intricate regulatory network controlling cell growth, cell proliferation, and apoptosis. Its proliferation-promoting activity is a reason why mutations leading to increased activity of MYC are frequently found in cancer cells and of critical importance for tumor maintenance and progression.1,2 Myc is a basic helix-loop-helix type transcription factor that, together with Max, binds to specific elements (E-boxes: CACGTG) in the promoters of many target genes usually acting as an activator of their transcription.2 Attempts to target this transcription factor therapeutically have been unsuccessful.1
Myc itself is regulated in a complex way at the levels of transcription and translation as well as by various post-translational modifications.2 Many studies have investigated the role of phosphorylation and ubiquitylation in controlling Myc activity and stability and various ubiquitin ligases and deubiquitylating enzymes have been implicated.2 The SCF Fbw7 ubiquitin-ligase, for example, recognizes Myc phosphorylated in a distinct way leading to its ubiquitin-dependent degradation by the proteasome.2
A recent study revealed that conjugation with small ubiquitin-related modifier (SUMO) is of critical importance for Myc-induced tumorigenesis, establishing a functional link between SUMO protein modification (sumoylation) and Myc activity.1 The identity of the underlying SUMO-modified proteins remained unknown. In an interesting new study published in a recent issue of Cell cycle, González-Prieto et al. show that Myc is directly regulated by sumoylation, adding another level of complexity to the post-translational control of Myc.3
PIAS1 is identified as the relevant Myc SUMO ligase, the activity of which is counteracted by the SENP7 SUMO deconjugating enzyme.3 A heterogeneous array of SUMO2-modified forms of Myc accumulated after proteasome inhibitor treatment3 suggesting that these forms are normally rapidly degraded by the proteasome in a process depending on SUMO-targeted ubiquitin ligases.4,5 Although His-SUMO2 pulldown experiments revealed high molecular weight modified forms of Myc, their appearance was only mildly affected by expressing a mutant SUMO2, in which all lysine residues were mutated (K0) to block chain formation (poly-sumolyation).3 Although a contribution of endogenous wild-type SUMO to chain formation in this experimental setup could not be entirely excluded, the heterogeneous nature of these conjugates suggested that Myc is sumoylated at multiple sites (multi-sumoylation). Consistent with this notion, 10 lysine residues of Myc were mapped as SUMO acceptor sites by mass spectrometry. Mutating all of them to arginine residues, surprisingly however, had very little effect on the efficiency of Myc sumoylation in vivo indicating that acceptor site selection in Myc by PIAS1 is quite promiscuous.
RNF4 was identified as the relevant ubiquitin ligase targeting SUMO-modified Myc to the proteasome3 (Fig. 1). Sumoylated Myc accumulated in substantial amounts upon RNF4 knockdown when cells were treated with proteasome inhibitor, whereas no such conjugates were detected without inhibitor treatment.3 The latter observation suggests that either other ubiquitin ligases contribute to targeting of sumoylated Myc, or that proteasome inhibition creates a stress that enhances Myc sumoylation, similar to the induction of c-Myb sumoylation by various forms of stress.7 While previous studies have pointed to SUMO chains as the main degron recognized by RNF4 (Fig. 1A),6 the data obtained with Myc and the SUMO2-K0 variant provide a first hint that RNF4 might also target multi-sumoylated proteins3 (Fig. 1B).
Figure 1.
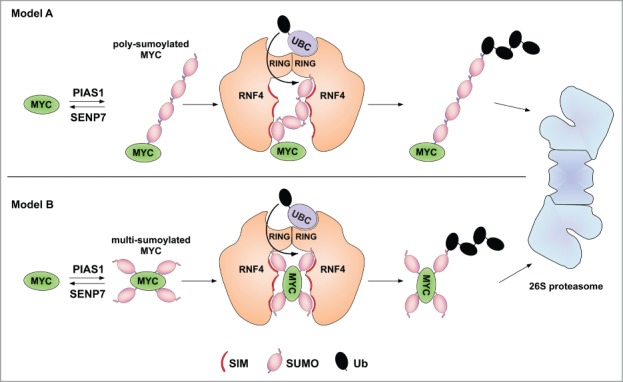
Proteolytic targeting of SUMO-modified cMyc. In (A), poly-sumoylation of Myc leads to recognition by RNF4 via multiple SUMO interaction motifs (SIMs). In (B), RNF4 recognizes multi-(mono)sumoylated Myc. Substrate binding may involve SIMs in both RNF4 subunits. Combinatorial binding via a SUMO chain and a mono-sumoylated lysine residues of the substrate are considerable as well (not depicted).
Whereas ubiquitylation of Myc in its transactivation domain not only acts to target it for degradation but also promotes activation of target gene expression,2 Myc sumoylation instead might inhibit target gene expression, as knockdown of PIAS1, which inhibits Myc sumoylation, increased Myc-dependent reporter gene activation.3 Kessler et al., by contrast, found that a subgroup of Myc target genes, instead of being induced, as in cells only overexpressing Myc, is repressed when sumoylation is inhibited simultaneously.1 These findings reflect the complexity of target regulation by Myc, to which sumoylation contributes. The observed promiscuous sumoylation site selection in Myc3 has prevented a selective inactivation of its sumoylation and therefore to differentiate direct effects of this modification from indirect ones stemming from sumoylation of other proteins. It thus remains a challenge for future studies to understand in more detail how and under which conditions the SUMO-modified forms of Myc and their proteolytic control contribute to the regulation of this system and to find possible ways to use this information for cancer therapy.
References
- 1.Kessler JD, et al.. Science 2012; 335:348-53; PMID:22157079; http://dx.doi.org/ 10.1126/science.1212728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farrell AS, Sears RC. Cold Spring Harb Perspect Med 2014; 4:a014365; PMID:24591536; http://dx.doi.org/ 10.1101/cshperspect.a014365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.González-Prieto R, et al.. Cell Cycle 2015; http://dx.doi.org/ 10.1080/15384101.2015.1040965 [DOI] [Google Scholar]
- 4.Uzunova K, et al.. J Biol Chem 2007; 282:34167-75; PMID:17728242; 14(12): 1859-72; PMID:25895136; http://dx.doi.org/ 10.1074/jbc.M706505200 [DOI] [PubMed] [Google Scholar]
- 5.Sriramachandran AM, Dohmen RJ. Biochim Biophys Acta 2014; 1843:75-85; PMID:24018209; http://dx.doi.org/ 10.1016/j.bbamcr.2013.08.022 [DOI] [PubMed] [Google Scholar]
- 6.Tatham MH, et al.. Nat Cell Biol 2008; 10:538-46; PMID:18408734; http://dx.doi.org/ 10.1038/ncb1716 [DOI] [PubMed] [Google Scholar]
- 7.Sramko M et al.. J Biol Chem 2006; 281:40065-75; PMID:17077080; http://dx.doi.org/ 10.1074/jbc.M609404200 [DOI] [PubMed] [Google Scholar]


