Abstract
Early growth response gene 1 (Egr1), a zinc finger transcriptional factor, plays an important role in regulating cell proliferation, differentiation and angiogenesis. Current data have shown that Egr1 is involved in follicular development, ovulation, luteinization and placental angiogenesis. However, the expression, regulation and function of Egr1 in mouse uterus during embryo implantation and decidualization are poorly understood. Here we showed that Egr1 was strongly expressed in the subluminal stroma surrounding the implanting blastocyst on day 5 of pregnancy. Injection of Egr1 siRNA into the mouse uterine horn could obviously reduce the number of implanted embryos and affect the uterine vascular permeability. Further study found that Egr1 played a role through influencing the expression of cyclooxygenase-2 (Cox-2), microsomal prostaglandin E synthase 1 (mPGES-1), vascular endothelial growth factor (Vegf), transformation related protein 53 (Trp53) and matrix metallopeptidase 9 (Mmp9) genes in the process of mouse embryo implantation. Growth hormone (GH) and insulin-like growth factor 1 (IGF-1) might direct the expression of Egr1 in the uterine stromal cells. Under in vivo and in vitro artificial decidualization, Egr1 expression was significantly decreased. Overexpression of Egr1 downregulated the expression of decidual marker decidual/trophoblast PRL-related protein (Dtprp) in the uterine stromal cells, while inhibition of Egr1 upregulated the expression of Dtprp under in vitro decidualization. Estrogen and progesterone could regulate the expression of Egr1 in the ovariectomized mouse uterus and uterine stromal cells. These results suggest that Egr1 may be essential for embryo implantation and decidualization.
Keywords: Egr1, implantation, decidualization, uterus, mouse
Introduction
Implantation is a crucial step in the establishment of pregnancy and requires a functional embryo at the blastocyst developmental stage and a synchronized transformation of uteri into a receptive stage.1-5 Inadequate or inappropriate implantation can result in an increase of the risk of early pregnancy loss.5 Thus implantation failure is considered a major cause of infertility in healthy women.3,6 For the couples undergoing in vitro fertilization and embryo transfer cycles, implantation rates remain disappointingly low, probably owing to non-synchronization of blastocyst activation with uterine receptivity.2-4 Although molecular and genetic evidences have provided valuable insights into the mechanisms of implantation with respect to ovarian hormones, cytokines, growth factors, adhesion molecules and transcription factors, there is limited information regarding the nature of implantation.1-4
Early growth response 1 (Egr1), also referred to as NGFI-A, Zif268, TIS8, or krox-24, was a zinc finger transcription factor and originally identified in quiescent fibroblasts as an immediate-early gene that was induced by serum.7-9 This factor was subsequently shown to be rapidly activated by various extracellular stimuli, such as growth factors, cytokines, hormones and environmental stresses.8-10 Simultaneously, activated Egr1 could recognize and bind the GC-rich consensus sequence GCG(T/G)GGGCG in the promoter region of various target genes, and regulate cell proliferation, differentiation and apoptosis in the nervous system, immune system, cardiovascular system, reprodu-ctive system, etc.8,9,11-13 Accumulating data had shown that Egr1 was expressed in the granulosa and thecal cells of ovarian follicles, corpus luteum, and the expression was elevated in the granulosa cells of follicles after hCG or FSH treatment, in the corpus luteum during PGF2α-induced luteolysis in vivo and in PGF2α-treated luteal cells in vitro.14-18 Egr1-deficient female mice were infertile due to lack of mature follicles, ovulation and luteinization.12,18 These evidences indicated that Egr1 might play a central role in the control of ovarian function. However, the functions of Egr1 on embryo implantation and decidualization are still unknown. Only several studies found that Egr1 was expressed in the uterus of human, mouse and rat.19-21 According to our (unpublished) microarray data, Egr1 expression was increased fold 12 at implantation sites compared to inter-implantation sites. Therefore, we hypothesized that Egr1 might be important for mouse implantation and decidualization. The purpose of this study is to investigate the expression, regulation and function of Egr1 gene in mouse uterus during the peri-implantation period.
Results
Egr1 mRNA expression during early pregnancy
In situ hybridization was used to examine the spatial distribution of Egr1 mRNA in mouse uterus. A low level of Egr1 mRNA signal was detected in the uteri from days 1 to 4 of pregnancy (Fig. 1B). On day 5 of pregnancy when embryo implanted, Egr1 mRNA signal was strongly observed in the subluminal stroma immediately surrounding the implanting blastocyst (Fig. 1C). Once the DIG-labeled Egr1 antisense probe was replaced with DIG-labeled Egr1 sense probe, there was no corresponding signal in the uterus on day 5 of pregnancy (Fig. 1A). On days 6-8 of pregnancy, Egr1 mRNA was expressed in the decidua, and its expression scope was expanded along with the development of decidua (Fig. 1D–F). In addition, Egr1 mRNA was found in the embryos from days 6 to 8 of pregnancy by in situ hybridization (Fig. 1D–F).
Figure 1.
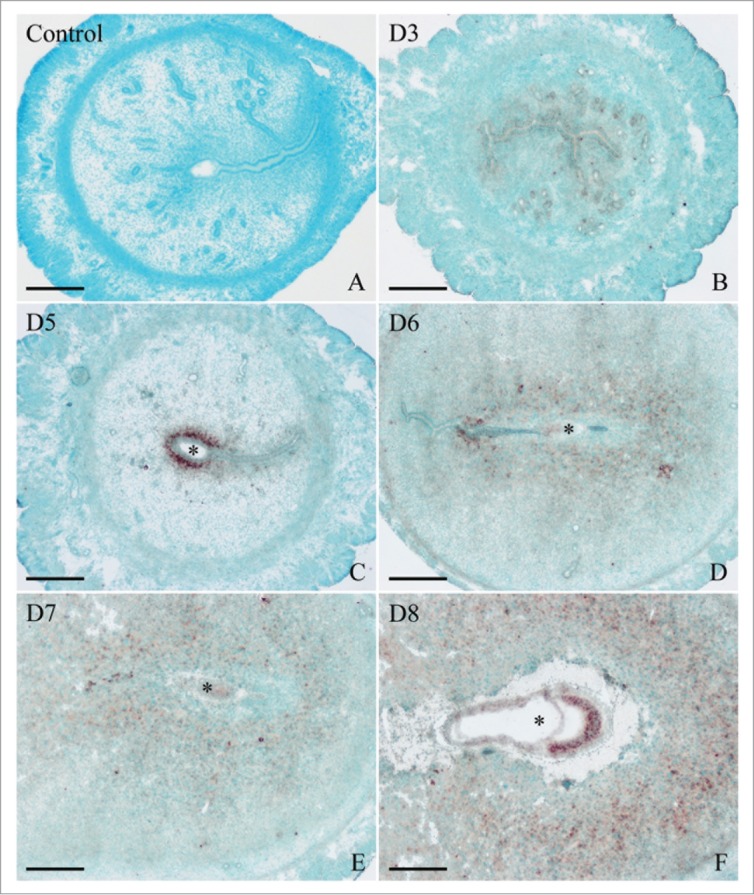
In situ hybridization of Egr1 expression in mouse uteri during early pregnancy on days 3 (B), 5 (C), 6 (D), 7 (E), and 8 (F). No hybridization signals were seen in mouse uterus on day 7 of pregnancy when DIG-labeled Egr1 sense probe was used to replace the antisense probe as a negative control (A). D stands for day of pregnancy. Asterisks indicate embryo. Bar = 60 μm.
To quantify Egr1 mRNA expression, real-time PCR was performed. A significantly higher level of Egr1 expression was detected on day 5 of pregnancy, although Egr1 expression was seen through days 1-8 (Fig. 2).
Figure 2.

Real-time PCR analysis of Egr1 expression in mouse uterus on days 1-8 during pregnancy. Data are shown mean ± SEM. Different letters on two bars show a significant difference between these two groups.
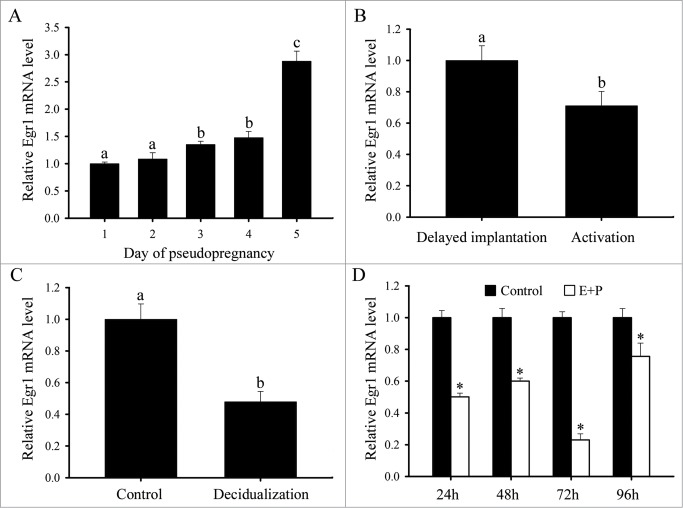
Egr1 mRNA expression during pseudopregnancy
To determine whether Egr1 expression was dependent on embryos, in situ hybridization was performed to examine Egr1 expression during pseudopregnancy. Egr1 mRNA was weakly expressed in the uteri from days 1 to 5 of pseudopregnancy (Fig. 3A and B). Real-time PCR results showed that Egr1 mRNA expression was gradually enhanced in the uteri from days 1 to 5 of pseudopregnancy and reached a peak on day 5 of pseudopregnancy (Fig. 4A).
Figure 3.

In situ hybridization of Egr1 expression in mouse uteri. (A) Day 3 of pseudopregnancy (PD3). (B) Day 5 of pseudopregnancy (PD5). (C) Delayed implantation (Delay). (D) Activation of delayed implantation by estrogen (Activation). (E) Uninjected uterine horn of control (Control). (F) Uterine horn under artificial decidualization (Decidualization). Asterisks indicate embryo. Bar = 60 μm.
Figure 4.
Real-time PCR analysis of Egr1 expression in mouse uteri. (A) Real-time PCR analysis of Egr1 expression in mouse uterus on days 1-5 of pseudopregnancy. Different letters on 2 bars show a significant difference between these 2 groups. (B) Real-time PCR analysis of Egr1 expression in mouse uterus during delayed implantation and activation. (C) Real-time PCR analysis of Egr1 expression in mouse uterus under artificial decidualization. (D) Real-time PCR analysis of Egr1 expression in in vitro decidualization of uterine stromal cells. Asterisks denote significance (P < 0.05) from the control group. E stands foRestrogen; P stands for progesterone.
Egr1 mRNA expression during delayed implantation and activation
In order to see whether Egr1 expression was dependent on activation status of blastocyst, a delayed implantation model was used. Under delayed implantation, Egr1 mRNA was barely detectable in mouse uterus (Fig. 3C). When delayed implantation was terminated by estrogen treatment and embryo implanted, Egr1 mRNA was strongly expressed in the subluminal stromal cells surrounding the implanting blastocyst (Fig. 3D). However, a relatively high level of Egr1 mRNA expression was found in the delayed uterus compared with the activated implantation uterus by real-time PCR (Fig. 4B).
Egr1 mRNA expression under artificial decidualization
To address whether Egr1 expression in decidua was dependent on the presence of embryos, we examined its expression under artificially induced decidualization in vivo. After the pseudopregnant uterine horn was induced to be artificially decidualized by injecting sesame oil into the uterine lumen, Egr1 mRNA signal was detectable in the decidualized cells and uninjected control uterus (Fig. 3E and F). By real-time PCR analysis, Egr1 expression was decreased in decidualized uterus compared with the control uterus (Fig. 4C).
Egr1 mRNA expression under in vitro decidualization
Primary stromal cells isolated from mouse uteri on day 4 of pregnancy were treated with the combination of estrogen and progesterone to induce in vitro decidualization. It has been shown that decidual/trophoblast PRL-related protein (Dtprp) is a reliable marker for decidualization in mice. In our study, the expression of Dtprp was significantly elevated from 24 to 96 h of culture,22 indicating that our system for in vitro decidualization was successful. On the contrary, Egr1 expression was significantly declined from 24 to 96 h after in vitro decidualization (Fig. 4D).
Regulation of steroid hormone on Egr1 mRNA expression
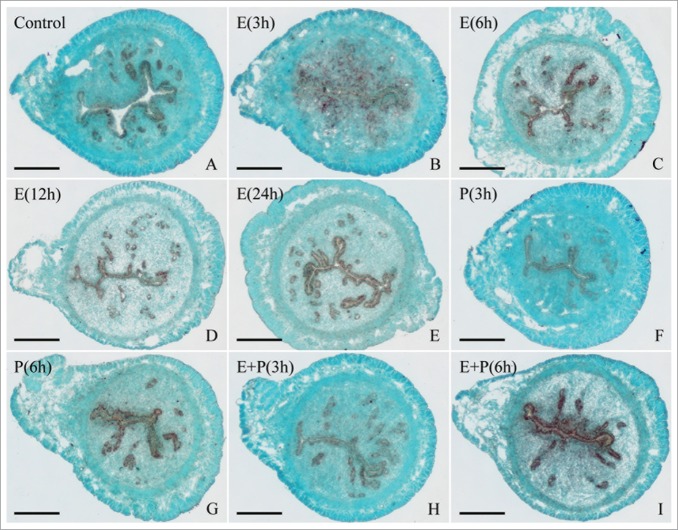
As estrogen and progesterone were essential for embryo implantation and decidualization, ovariectomized mice were used to examine whether Egr1 expression was regulated by ovarian steroid hormones. The in situ hybridization results showed that Egr1 mRNA signal was detected in the uterine glandular epithelium of ovariectomized mice (Fig. 5A). After the ovariectomized mice were treated with estrogen, Egr1 mRNA was mainly localized in the luminal and glandular epithelium and its expression reached the highest level at 6 h (Fig. 5B–E). The similar expression pattern was also observed in the mice treated with progesterone or a combination of estrogen and progesterone (Fig. 5F–I). Additionally, estrogen could induce the expression of Egr1 mRNA in the uterine stromal cells at 3 h (Fig. 5B). By real-time analysis, Egr1 mRNA expression was increased in ovariectomized mice uterus after injection of estrogen, reached a peak at 3 h, then declined and reached the lowest level at 24 h (Fig. 6A). Injection of progesterone resulted in a decline in uterine Egr1 mRNA level which reached a nadir at 3 h (Fig. 6B). After a combined injection of estrogen and progesterone, the Egr1 expression pattern in uterus was similar to those of estrogen treatment alone (Fig. 6C).
Figure 5.
In situ hybridization of Egr1 expression in ovariectomized mouse uteri after injection of estrogen, progesterone or a combination of estrogen and progesterone for 0 (control), 3, 6, 12 and 24 h. Bar = 60 μm.
Figure 6.
Hormonal regulation of Egr1 expression. (A) Real-time PCR analysis of Egr1 expression in ovariectomized mouse uterus afteRestrogen treatments for 0, 1, 3, 6, 12 and 24 h. Different letters on 2 bars show a significant difference between these 2 groups. (B) Egr1 expression in ovariectomized mouse uterus after injection of progesterone. (C) Egr1 expression in ovariectomized mouse uterus after injection of estrogen plus progesterone. (D) Egr1 expression in uterine stromal cells afteRestrogen treatments for 0, 3, 6, 12 and 24 h. (E) Egr1 expression in uterine stromal cells after progesterone treatments for 0, 3, 6, 12 and 24 h. (F) Egr1 expression after stromal cells were treated with estrogen or both estrogen and ICI 182,780, progesterone or both progesterone and RU486 for 24 h. Asterisks denote significance (P < 0.05) from the control group.
In the in vitro cultured stromal cells, Egr1 mRNA expression was gradually decreased after treatment of progesterone and reached the minimum at 24 h (Fig. 6E). After pretreatment with progesterone receptor antagonist RU486, a significantly higher level of Egr1 expression was detected at 24 h compared with that treated by progesterone only (Fig. 6F). Estrogen treatment resulted in an increase of Egr1 mRNA level in uterine stromal cells at 3 h and followed by a decline reaching a nadir at 24 h (Fig. 6D). Moreover, the inhibitory effect of estrogen on Egr1 was rescued by the addition of estrogen receptor (ER) antagonist ICI 182,780 (Fig. 6F).
Egr1 mRNA expression in the oocytes and preimplantation embryos
The RT-PCR was performed to examine the expression of Egr1 and Gapdh in the mouse oocytes and embryos at the stages of zygote, 2-cell, 4-cell, 8-cell, morula and blastocyst, respectively. Gapdh was used to normalize cDNA preparations. The predicted 452 bp fragment was seen with Gapdh primers. However, no Egr1 mRNA was detected in the oocytes and preimplantation mouse embryos (data not shown).
Effects of Egr1 on embryo implantation
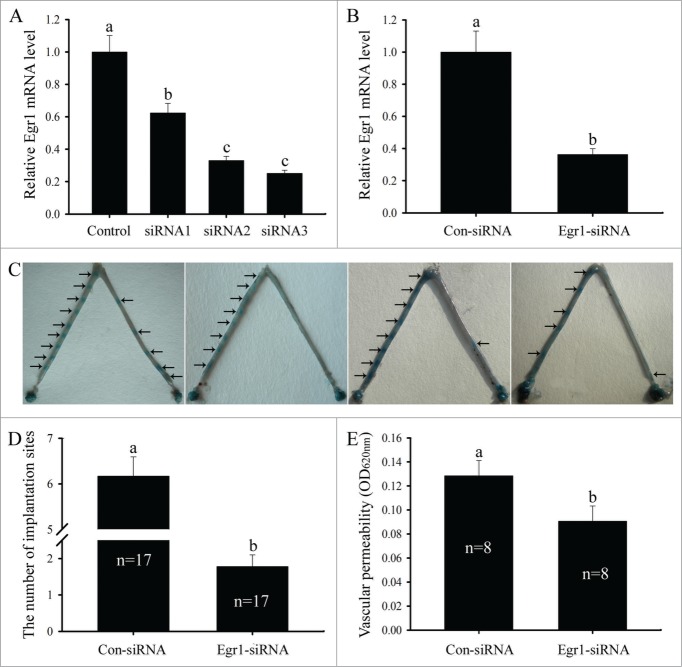
To see whether Egr1 was required for mouse embryo implantation, intrauterine blocking treatments were performed by siRNA injection. First, we synthesized Egr1 siRNA duplexes and nonspecific scrambled siRNA, transfected the uterine stromal cells and examined the expression of Egr1 gene by real-time PCR. The results showed that Egr1 siRNA 1, 2 or 3 could suppress the expression of Egr1 mRNA in uterine stromal cells compared with scrambled siRNA, and Egr1 siRNA 3 showed obvious inhibition effect (Fig. 7A). Therefore, Egr1 siRNA 3 was selected to knock down Egr1 gene in the following study. Moreover, the efficacy of Egr1 siRNA 3 in inhibiting the expression of Egr1 mRNA in vivo was also confirmed. Egr1 mRNA level in the uteri injected with Egr1 siRNA were remarkably lower than that in the contralateral horn treated with scrambled siRNA (Fig. 7B). Likewise, the number of implantation sites on day 5 of pregnancy was significantly reduced after the intrauterine injection of Egr1-specific siRNA on day 3 of pregnancy compared to that of scrambled siRNA group (Fig. 7C and D).
Figure 7.
Effects of Egr1 on embryo implantation and vascular permeability. (A) Effects of Egr1 siRNAs on Egr1 mRNA expression in the uterine stromal cells. After transfection with control siRNA, Egr1 siRNA 1, siRNA 2 or siRNA 3, Egr1 mRNA expression was determined by real-time PCR. (B) Effects of Egr1 siRNA on Egr1 mRNA expression at the implantation sites. After intrauterine injection of control siRNA (Con-siRNA) or Egr1 siRNA, Egr1 mRNA expression was determined by real-time PCR. (C, D) Effects of Egr1 on embryo implantation. After the intrauterine injection of control siRNA (left-side uterine horn) or Egr1 siRNA (right-side uterine horn), the number of implantation sites was evaluated. Arrows show the implantation sites. n stands for the total number of intrauterine-injected mice. (E) Effects of Egr1 on vascular permeability of implantation sites. Different letters on two bars show a significant difference between these 2 groups.
Effects of Egr1 on vascular permeability
Because endometrial vascular permeability has been considered as a marker for embryo implantation,23 intrauterine blocking treatments were used to examine whether Egr1 could regulate the vascular permeability of mouse implantation sites. After the intrauterine injection of Egr1-specific siRNA, vascular permeability of the implantation sites on day 5 of pregnancy was significantly declined compared to that of scrambled siRNA group (Fig. 7E).
Regulation of GH and IGF-1 on Egr1 mRNA expression
It has been shown that growth hormone (GH) and insulin-like growth factor 1 (IGF-1) are involved in the process of embryo implantation.1,4,25 Therefore, cultured uterine stromal cells were treated with GH (100 ng/ml) or IGF-1 (100 ng/ml) to observe the effects of GH and IGF-1 on Egr1 expression. The results showed that GH and IGF-1 could stimulate the expression of Egr1 mRNA in the uterine stromal cells (Fig. 8A).
Figure 8.

Effects of GH, IGF-1 and 8-Br-cAMP on Egr1 expression in the uterine stromal cells. (A) Egr1 expression in the uterine stromal cells treated with GH or IGF-1. Asterisks denote significance (P<0.05) from the control group. (B) Egr1 expression in the uterine stromal cells after 8-Br-cAMP treatment.
Effects of Egr1 on decidualization
To investigate the effects of Egr1 on decidualization, we observed the effects of Egr1 on the expression of decidual/trophoblast PRL-related protein (Dtprp) which was an established molecular marker for decidualization. After pc-Egr1 transfection, Egr1 mRNA expression was significantly increased in the uterine stromal cells compared with control (Fig. 9A), indicating that Egr1 overexpression plasmid was available. However, overexpression of Egr1 downregulated the expression of Dtprp gene in the uterine stromal cells (Fig. 9B). After the intrauterine injection of Egr1 siRNA, Dtprp expression was upregulated at implantation sites compared with control (Fig. 9C). Interestingly, Dtprp mRNA level did not show any obvious change in the uterine stromal cells after transfection with Egr1 siRNA (Fig. 9D). However, Dtprp mRNA level was significantly elevated when the uterine stromal cells were firstly induced for in vitro decidualization for 24 h, followed by Egr1 siRNA transfection (Fig. 9F). The similar results were also observed in the uterine stromal cells firstly transfected with Egr1 siRNA, followed by treatment with a combination of estrogen and progesterone for 24 h (Fig. 9H). In addition, inhibition of Egr1 with siRNA could decrease its expression under in vitro decidualization (Fig. 9E and G).
Figure 9.
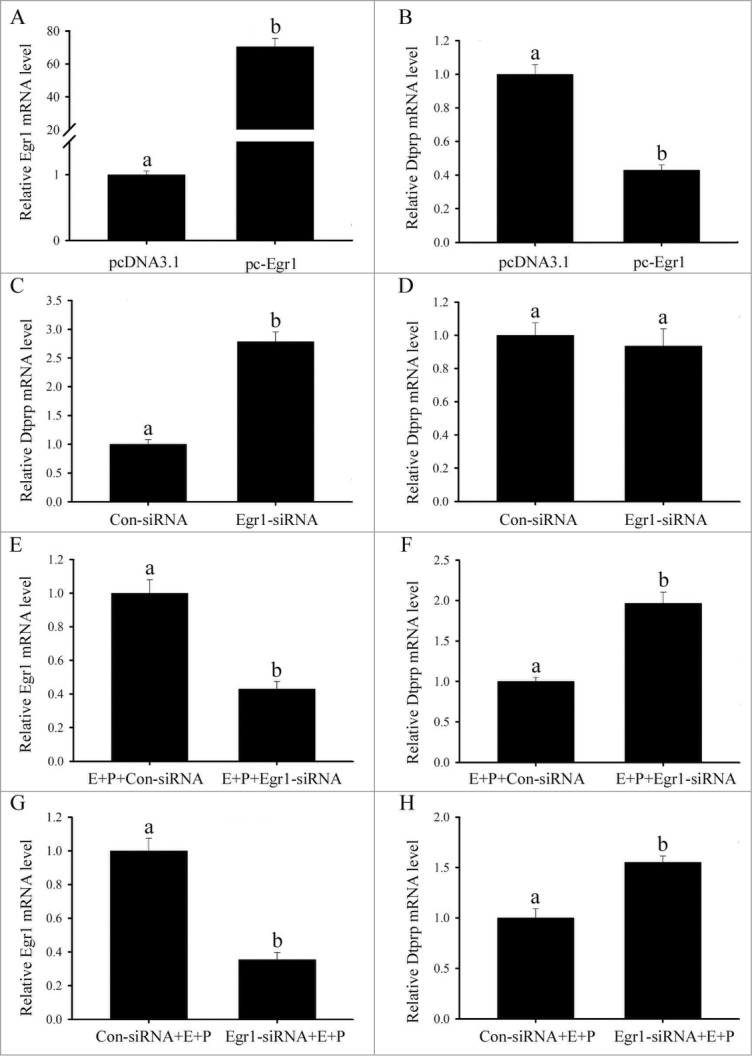
Effects of Egr1 on Dtprp expression. (A) Egr1 mRNA expression following Egr1 overexpression. (B) Effects of Egr1 overexpression on Dtprp mRNA expression. (C) Effects of Egr1 siRNA on Dtprp mRNA expression. After intrauterine injection of Egr1 siRNA, Dtprp mRNA expression was determined by real-time PCR. (D) Effects of Egr1 siRNA on Dtprp mRNA expression in the uterine stromal cells. After transfection with Egr1 siRNA, Dtprp mRNA expression was determined by real-time PCR. (E, F) Effects of Egr1 siRNA on Egr1 and Dtprp mRNA expression. After Egr1 siRNA was transfected in uterine stromal cells which were induced for in vitro decidualization for 24 h, Egr1 and Dtprp mRNA expression was determined by real-time PCR. (G, H) Effects of Egr1 siRNA on Egr1 and Dtprp mRNA expression. After transfection with control siRNA (Con-siRNA) or Egr1 siRNA for 6 h, the stromal cells were induced for in vitro decidualization for 24 h. Different letters on 2 bars show a significant difference between these 2 groups.
Regulation of cAMP and H89 on Egr1 mRNA expression
Because the elevated intracellular cAMP levels and sustained activation of the protein kinase A (PKA) pathway are essential for decidualization,26 we treated the uterine stromal cells with cAMP analog 8-bromoadenosine-cAMP (8-Br-cAMP, 500 μM) and PKA inhibitor H-89 (10 μM) to examine the effects of cAMP and H-89 on Egr1 expression. The results showed that Dtprp mRNA expression was gradually increased after 8-Br-cAMP treatment and reached the peak level at 24 h.24 However, Egr1 mRNA level was obviously unchanged in the uterine stromal cells treated by 8-Br-cAMP (Fig. 8B) and similar result was also observed after H89 treatment (data not shown).
Regulation of Egr1 on Cox-2, mPGES-1, Vegf, Trp53 and Mmp9 mRNA expression
In the in vitro cultured stromal cells, overexpression of Egr1 could raise the expression of cyclooxygenase-2 (Cox-2), microsomal prostaglandin E synthase 1 (mPGES-1), vascular endothelial growth factor (Vegf), transformation related protein 53 (Trp53) and matrix metallopeptidase 9 (Mmp9) genes (Fig. 10A). On the contrary, inhibition of Egr1 with the intrauterine injection of Egr1-specific siRNA could result in a decline in uterine Cox-2, mPGES-1, Vegf, Trp53 and Mmp9 mRNA levels at implantation sites (Fig. 10B). The similar results were also found in the cultured uterine stromal cells after transfection with Egr1 siRNA (Fig. 10C).
Figure 10.
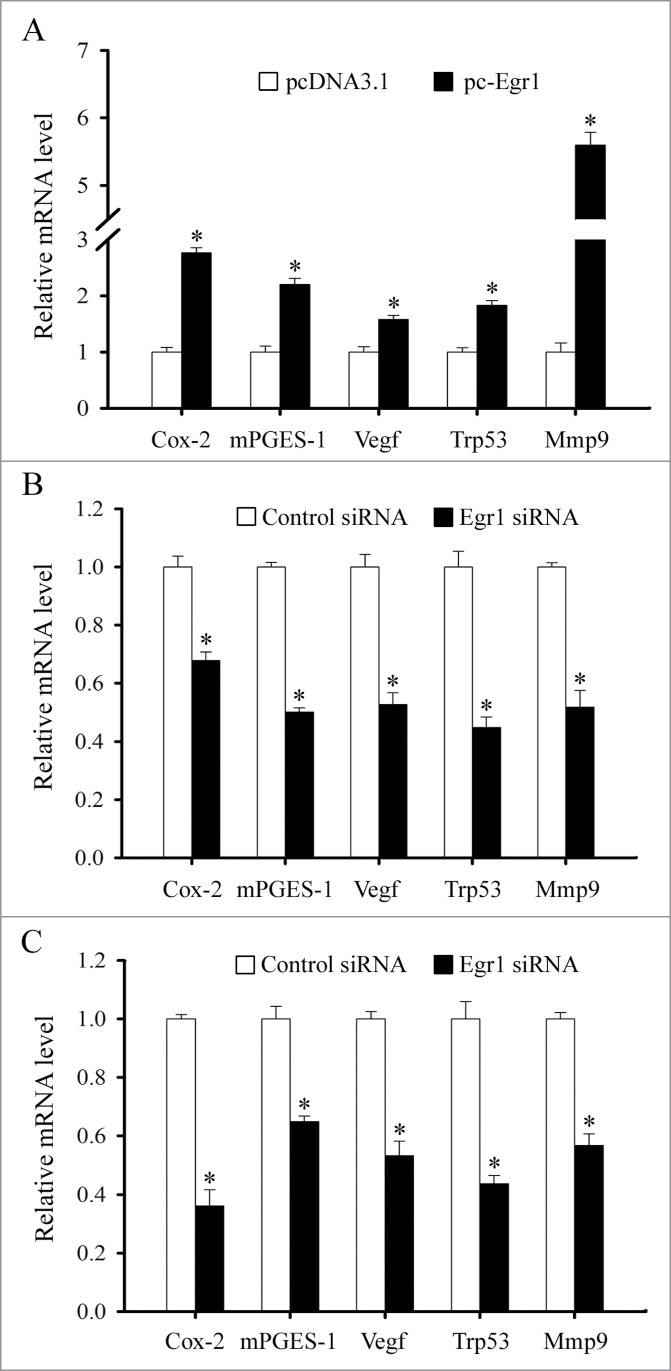
Regulation of Egr1 on Cox-2, mPGES-1, Vegf, Trp53 and Mmp9 expression. (A) Effects of Egr1 overexpression on Cox-2, mPGES-1, Vegf, Trp53 and Mmp9 expression. After transfected with control plasmid (empty pcDNA3.1 vector) or Egr1 overexpression plasmid (pc-Egr1) in the uterine stromal cells, the expression levels of Cox-2, mPGES-1, Vegf, Trp53 and Mmp9 were determined by real-time PCR. (B) Effects of Egr1 siRNA on Cox-2, mPGES-1, Vegf, Trp53 and Mmp9 expression at implantation sites. After intrauterine injection of Egr1 siRNA, the expression of Cox-2, mPGES-1, Vegf, Trp53 and Mmp9 was determined by real-time PCR. (C) Effects of Egr1 siRNA on Cox-2, mPGES-1, Vegf, Trp53 and Mmp9 expression in the uterine stromal cells. After transfected with Egr1 siRNA, the expression of Cox-2, mPGES-1, Vegf, Trp53 and Mmp9 was determined by real-time PCR. Asterisks denote significance (P < 0.05) from the control group.
Discussion
Egr1 is necessary for normal reproductive development and function.12,27 However, it remains unknown whether Egr1 is crucial for embryo implantation and decidualization. The present study was undertaken to investigate the expression and regulation of Egr1 in mouse uterus during the peri-implantation period in order to provide insight into the physiological function of Egr1 during embryo implantation and decidualization. Our results showed that Egr1 mRNA was strongly detected in the subluminal stroma surrounding the implanting blastocyst on day 5 of pregnancy, but not in the uterus on day 5 of pseudopregnancy. The similar expression pattern was also observed in the estrogen-activated implantation sites rather than in the progesterone-primed delayed implantation uterus. These results suggest that Egr1 might be important for mouse embryo implantation and its expression at the implantation site was dependent upon the presence of active blastocysts. However, Egr1 mRNA was not expressed in the mouse preimplantation embryos. After intrauterine injection of Egr1 siRNA, the number of implantation sites was markedly reduced, which further confirmed the effect of Egr1 on mouse embryo implantation. Previous study found that Egr1 acted as a transcription factor through the binding to GC-rich elements in the promoter region of target genes.8,9 In human melanoma A375-C6 cells, Egr1 directly bound to the promoter of Trp53 gene and enhanced the expression of Trp53 which was critical for blastocyst implantation because the number of implantation sites was significantly reduced in Trp53-deficient female mice.28,29 In this study, Trp53 expression was upregulated by Egr1 overexpression in the uterine stromal cells and downregulated by Egr1 siRNA at implantation sites and in the uterine stromal cells, indicating that Egr1 might regulate the process of embryo implantation through influencing the expression of Trp53. Simultaneously, Egr1 transactivated the promoter of the Mmp9 gene in HeLa cells and induced the transcription of Mmp9 which was also expressed in the subluminal stromal cells surrounding the implanting blastocyst on day 5 of pregnancy and thought to be a key mediator for matrix degradation during implantation.1,30,31 The present results found that Egr1 might also modulate the expression of Mmp9 gene during mouse embryo implantation.
Increased vascular permeability and angiogenesis are essential for successful implantation.1,2,32 After the intrauterine injection of Egr1 siRNA, vascular permeability of implantation sites on day 5 of pregnancy was significantly declined, demonstrating that Egr1 might affect the vascular permeability during mouse embryo implantation. Numerous data declared that endometrial vascular permeability and angiogenesis were profoundly influenced by prostaglandins (PGs) which were the product of a cascade of enzymes such as Cox and PG synthases.23,33 Cox-2 was an inducible rate-limiting enzyme in the biosynthesis of PGs and could convert arachidonic acid into PGH2, a common precursor of all PGs.1,33 The targeted disruption of Cox-2 in mice led to the failure of implantation.34 In RAW 264.7 cells, Egr1 overexpression was able to increase the luciferase activity driven by Cox-2 promoter.35 Likewise, overexpression of Egr1 could also elevate the expression of Cox-2 in uterine stromal cells while inhibition of Egr1 with siRNA could decrease its expression. Intrauterine injection of Egr1 siRNA could also result in a decline in Cox-2 mRNA level at implantation sites. mPGES-1 was a terminal prostanoid synthase and could catalyze the isomerization of PGH2 to PGE2 which was essential for mouse implantation.1,33,36 Egr1 bound to the proximal GC box in the mPGES-1 promoter region and facilitated its expression.35,37 During embryo implantation, Egr1 might also stimulate the expression of mPGES-1 gene. Further studies found that Cox-2 and mPGES-1 could promote angiogenesis via the Vegf gene which was a key factor involved in endometrial vascular permeability and angiogenesis.32,36,38,39 Inhibition of Vegf could significantly reduce the number of implantation sites.38 The evidence presented in this study demonstrated that Egr1 might also regulate the expression of Vegf gene in the uterine stromal cells and at implantation sites. Taken together, these results suggest that Egr1 might direct vascular permeability and angiogenesis by affecting the expression of Cox-2, mPGES-1 and Vegf genes during mouse embryo implantation.
Uterine stromal cell decidualization is a prerequisite for successful embryo implantation.1-4 On days 6-8 of pregnancy, Egr1 mRNA signal was observed in the decidua. The similar result was also found in decidualized cells under artificial decidualization. However, real-time PCR results showed that Egr1 mRNA was lowly expressed under in vivo and in vitro decidualization. These results imply that Egr1 can inhibit the process of mouse decidualization. Indeed, inhibition of Egr1 could stimulate the expression of decidual marker Dtprp under in vitro decidualization while overexpression of Egr1 could reduce the expression of Dtprp, which further verify the effect of Egr1 on decidualization. Additionally, cAMP might regulate the decidualization of endometrial stromal cells in a time-dependent manner.26,40 Increased intracellular cAMP by forskolin treatment is sufficient to mediate induction of Egr1 in granulosa cells.15 Mutation of the proximal putative cAMP response element of Egr1 promoter resulted in a significant decrease in the Egr1 transcription while activation of the cAMP system could increase Egr1 promoter activity.41,42 However, Egr1 mRNA expression level was not obviously changed in the uterine stromal cells treated by cAMP analog 8-Br-cAMP. The reason was likely that Egr1 might be modified by cAMP at stages later than 24 h after 8-Br-cAMP treatment.
It has been showed that Egr1 could be immediately stimulated by ovarian estrogen and progesterone which were essential for embryo implantation and decidualization.3,4,19,43-45 Estrogen could simulate the expression of Egr1 in the rodent uterus, breast and endometrial cancer cells.19,21,43,44 In the Egr1 promoter region, there were two estrogen response element half-sites, and estrogen might transactivate Egr1 promoter.44,46,47 In this study, Egr1 mRNA was highly expressed in the uterine luminal epithelium of ovariectomized mice by in situ hybridization afteRestrogen injection. However, real-time PCR result showed that Egr1 mRNA expression was gradually decreased from 6 to 24 h afteRestrogen injection. The discrepancies suggested that Egr1 was also expressed in the other cell constituents of uterus where estrogen might inhibit the expression of Egr1 gene. Indeed, estrogen treatment resulted in a decline of Egr1 mRNA level in the uterine stromal cells from 6 to 24 h. Moreover, the inhibitory effect of estrogen on Egr1 was rescued by the addition of ER antagonist ICI 182,780. ERα and ERβ could induce transcriptional activation of the Egr1 promoter in co-transfection experiments.46 These results suggest that estrogen regulation on Egr1 might be mediated by ER. Likewise, progesterone could upregulate the expression of Egr1 mRNA in the uterine luminal epithelium of ovariectomized mice and downregulate its expression in the uterine stromal cells. Simultaneously, Egr1 was also induced by GH which was a major regulator in the physiology of female reproduction, including regulation of oocyte maturation and fertilization, early embryo development, implantation, fetal/placental growth and development, and litter size.25,48-51 In poor-prognosis patients undertaking IVF, GH supplementation could improve oocyte quality, implantation and pregnancy productivity rates.52 This result showed that GH could modulate the expression of Egr1 in the uterine stromal cells. Moreover, the effects of GH on female reproduction were mediated by IGF-1.25,51 Indeed, IGF-1 could also enhance the expression of Egr1 in the uterine stromal cells. This data was consistent with those of previous studies in the cardiac fibroblasts, SH-SY5Y cells and vascular smooth muscle cells.53-55
In conclusion, Egr1 may play an important role during mouse embryo implantation and decidualization.
Materials and Methods
Animals
Matured mice (Kunming White strain) were caged in a controlled environment with a cycle of 14L:10D. All animal procedures were approved by the Institutional Animal Care and Use Committee of Jilin University. To confirm reproducibility of results, at least three mice per group were used in each stage or treatment in this study.
Pregnancy and pseudopregnancy
Adult female mice were mated with fertile or vasectomized males of the same strain to induce pregnancy or pseudopregnancy by co-caging, respectively (day 1 = day of vaginal plug). On days 14, pregnancy was confirmed by recovering embryos from the oviducts or uterus. The implantation sites on day 5 were identified by intravenous injection of 0.1 ml of 1% Chicago blue (Sigma, St. Louis, MO) in 0.85% sodium chloride.
Artificial induced decidualization
Artificial decidualization was induced by intraluminally infusing 25 μl of sesame oil into one uterine horn on day 4 of pseudopregnancy, while the contralateral uninjected horn served as a control. The uteri were collected on day 8 of pseudopregnancy. Decidualization was confirmed by weighing the uterine horn and by histological examination of uterine sections.
Steroid hormonal treatments
Mature female mice were ovariectomized and, after 2 weeks, given a single sc injection of estrogen (100 ng/mouse) or progesterone (2 mg/mouse). Uteri then were collected at 1, 3, 6, 12 and 24 h after steroid treatment. All steroids were dissolved in sesame oil and injected subcutaneously. Controls received the vehicle only (0.1 ml/mouse).
Collection of mouse oocytes and preimplantation embryos
Mature female mice were superovulated with an intraperitoneal injection of 5 IU equine chorionic gonadotrophin followed by administration of 5 IU hCG at 48 h later. Mature oocytes were collected from the oviduct by puncturing the ampulla at 13 h after hCG administration. In order to collect preimplantation embryos, the female mice were caged with fertile male mice after hCG injection. The embryos at the stages of zygote, 2-cell, 4-cell, 8-cell, morula and blastocyst were recovered by flushing oviducts or uterus at 23, 42, 54, 68, 80, and 92 h post-hCG injection. Oocytes and zygotes were digested with 300 IU hyaluronidase in Dulbecco's phosphate-buffered saline (D-PBS) to remove cumulus cells. All the oocytes and embryos were washed 5 times by D-PBS, and then treated with TPIΠYPE reagent with an addition of 100 μg yeast tRNA (Boehringer Mannheim) into each group as a carrier RNA. A total of 100 oocytes or embryos at each stage were used for RNA extraction in each group.
In situ hybridization
Total RNAs from the mouse uteri were reverse-transcribed and amplified with Egr1 primers. Egr1 forward primer 5′- CCCAGTGGCTACCTCCTACC and reverse primer 5′- CCTGGGAGAAAAGGTCGCTG were designed according to Mus musculus early growth response 1 gene (GenBank accession number NM_007913). The amplified fragment (259 bp) of Egr1 was cloned into pGEM-T plasmid (pGEM-T Vector System 1, Promega, Madison, WI) and verified by sequencing. Egr1-containing plasmid was amplified with the primers for T7 and SP6 to prepare templates for labeling. Digoxigenin (DIG)-labeled antisense and sense cRNA probes were transcribed in vitro using a DIG RNA labeling kit (Roche Diagnostics GmbH, Mannheim, Germany).
Frozen sections (10 μm) were mounted on 3-aminopropyltriethoxy-silane (Sigma)-coated slides and fixed in 4% paraformaldehyde solution in PBS. Hybridization was performed as described previously.24 Sections were counterstained with 1% methyl green in 0.12 M glacial acetic acid. The sense probe was also hybridized and served as a negative control. There was no detectable signal from sense probes.
Real-time PCR
Total RNAs from mouse uteri or cultured cells were isolated using TRIPURE reagent according to the manufacturer's instructions (Roche) and reverse-transcribed into cDNA using M-MLV reverse-transcriptase (Promega). Reverse transcriptase was performed at 42°C for 60 min with 2 μg total RNA in 25 μl volume. For real-time PCR, cDNA was amplified using FS Universal SYBR Green Real Master (Roche) on BIO-RAD CFX96TM Real Time Detection System. The conditions used for real-time PCR were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. All reactions were run in triplicate. The result was analyzed using CFX Manager Software. After analysis using the 2−ΔΔCt method, data were normalized to Gapdh expression. Primer sequences for real-time PCR were listed in Table 1.
Table 1.
Primers for real-time PCR
| Gene | Primer sequence | Accession number | Size (bp) |
|---|---|---|---|
| Early growth response 1 (Egr1) | TTGTGGCCTGAACCCCTTTT | NM_007913 | 166bp |
| AGATGGGACTGCTGTCGTTG | |||
| Cyclooxygenase-2 (Cox-2) | CATCCCCTTCCTGCGAAGTT | NM_011198 | 178bp |
| CATGGGAGTTGGGCAGTCAT | |||
| Microsomal prostaglandin E synthase 1 (mPGES-1) | TCCTCGGCTTCGTGTACTCA | NM_022415 | 157bp |
| GAGAACTGGGCCAGGACATAG | |||
| Vascular endothelial growth factor (Vegf) | ACGTCAGAGAGCAACATCACC | NM_001025257 | 90bp |
| CTGTGCTGTAGGAAGCTCATCTC | |||
| Transformation related protein 53 (Trp53) | CCATGGCCCCTGTCATCTTT | NM_011640 | 124bp |
| TGAGGGGAGGAGAGTACGTG | |||
| Matrix metallopeptidase 9 (Mmp9) | GCACCTCCCACTATGTGTCC | NM_013599 | 209bp |
| CAAGGATTGTCTGCCGGACT | |||
| Decidual/trophoblast PRL-related protein (Dtprp) | AGCCAGAAATCACTGCCACT | NM_010088 | 119bp |
| TGATCCATGCACCCATAAAA | |||
| Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) | GCCTTCCGTGTTCCTACCC | NM_008084 | 102bp |
| TGCCTGCTTCACCACCTTC |
RT-PCR
Total RNAs were extracted from preimplantation embryos with TRIPURE reagent, digested with RQ1 DNase I and reverse-transcribed into cDNA with M-MLV Reverse Transcriptase (Promega). The amplified PCR fragments for Egr1 were separated in 1.5% agarose gel electrophoresis and stained with ethidium bromide, and the bands were quantitated by optical density using the UVP laboratory imaging and analysis system (UVP, Inc., Upland, CA). The band densities for Egr1 were normalized to Gapdh expression. The PCR primers for mouse Gapdh were as follows: forward primer 5′-ACCACAGTCCATGCCATCAC and reverse primer 5′-TCCACCACCCTGTTGCTGTA.
Isolation of uterine stromal cells and in vitro decidualization
Uterine stromal cells from day 4 of pregnancy were isolated and cultured as previously described.22 Uterine stromal cells were induced for in vitro decidualization with fresh medium supplemented with progesterone (1 μM) and estrogen (10 nM) in DMEM-F12 with 2% charcoal-treated FBS (Biological Industries Ltd., Kibbutz Beit Hemeek, Israel).
Steroid hormonal treatments in vitro
Cultured stromal cells were treated with 100 nM of progesterone, or 0.1 nM of estrogen, respectively. For further studies, cells were pretreated with RU486 (1 μM), or ICI 182,780 (100 nM) for 2 h before the addition of progesterone oRestrogen, respectively. Then cells were collected at 3, 6, 12, and 24 h for further quantitative analysis by real-time PCR. All steroids and antagonists were dissolved in ethanol. Controls received the vehicle only.
Plasmid construction and transfection
Full-length Egr1 cDNA fragment was amplified by PCR from the uterus of pregnancy mouse using the following primers with Hind III/xhoI restriction sites: 5′-AAGCTT (Hind III) ATGGCAGCGGCCAAGGCCGAGA; 5′-CTCGAG (xhoI) TTAGCAAATTTCAATTGTCCTGG. The amplified product was purified and cloned into pGEM-T vector. Both pGEM-T-Egr1 and pcDNA3.1 vector were cut by Hind III/xhoI (TaKaRa, Dalian, China) at 37°C for 1h, and then the fragment was ligated into pcDNA3.1 with T4 ligase (Promega) at 4°C overnight to construct pcDNA-Egr1 (pc-Egr1). An empty pcDNA3.1 expression vector was served as control.
Transfection of uterine stromal cells was performed according to the manufacturer's protocol for lipofectamine 2000 (Invitrogen). After transfection with control plasmid (empty pcDNA3.1 vector) or pc-Egr1 plasmid, stromal cells from day 4 pregnancy mice were collected at 24 h.
RNA interference
The small-interfering RNA (siRNA) duplexes for targeting Egr1 as well as a scrambled sequence (control siRNA duplex, negative control) were designed and synthesized by GenePharma. The sequences were shown as follows: Egr1 siRNA 1 sense: 5′- GGACAAGAAAGCAGACAAATT, antisense: 5′-UUUGUCUGCUUUCUUGUCCTT; Egr1 siRNA 2 sense: 5′-CUGACAUCGCUCUGAAUAATT, antisense: 5′-UUAUUCAGAGCGAUGUCAGTT; Egr1 siRNA 3 sense: 5′-UCCAGCUGCUUCAUCGUCUTT, antisense: 5′- AGACGAUGAAGCAGCUGGATT; nonspecific scrambled siRNA (negative control) sense: 5′-UUCUCCGAACGUGUCACGUTT, antisense: 5′-ACGUGACACGUUCGGAGAATT. Transfections for siRNA were performed according to Lipofectamine 2000 protocol. After transfection with Egr1 siRNA, uterine stromal cells from day 4 of pregnancy mice were collected or induced for in vitro decidualization for 24 h. Meanwhile, Egr1 siRNA was also transfected in the uterine stromal cells which were treated with a combination of estrogen and progesterone for 24 h.
For treatment of early pregnant mice with siRNA, mice were anesthetized on day 3 of pregnancy and given an intrauterine injection with the Egr1 siRNA suspension contained 10 μM Egr1 siRNA, 10% Lipofectamine 2000 and 50% physiological saline, while the contralateral uterine horn received equal volume of scrambled siRNA suspension. On day 5 of pregnancy, implantation sites were visualized after tail vein injection of Chicago blue dye solution before the mice were killed and uteri were excised. Numbers of implantation sites were recorded.
Measurement of vascular permeability of mouse implantation sites
Vascular permeability was assayed as described previously.23 Briefly, the implantation sites on day 5 were identified by intravenous injection of 1 ml of 1% (w/v) Chicago blue solution in 0.85% (w/v) NaCl. The same weight of implantation sites were collected in 200 ml formamide. The optical density at 620 nm was measured after 120 h extraction.
Statistics
Values were reported as mean ± SEM. The significance of difference was analyzed by one-way ANOVA or paired t-test using the SPSS software program (SPSS Inc., Chicago). The differences were considered significance at P < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was financially supported by National Natural Science Foundation of China (31101778 and 31372390), Special Funds for Scientific Research on Public Causes (201303119) and National Key Basic Research Program of China (2011CB100805).
References
- 1. Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, Wang H. Molecular cues to implantation. EndocrRev 2004; 25:341–73; PMID: 15180948; http://dx.doi.org/10.1210/er.2003-0020 [DOI] [PubMed] [Google Scholar]
- 2. Wang H, Dey SK. Roadmap to embryo implantation: clues from mouse models. Nat RevGenet 2006; 7:185–99; PMID: 16485018; http://dx.doi.org/10.1038/nrg1808 [DOI] [PubMed] [Google Scholar]
- 3. Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. NatMed 2012; 18:1754–67; PMID: 23223073; http://dx.doi.org/10.1038/nm.3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang S, Lin H, Kong S, Wang S, Wang H, Wang H, Armant DR. Physiological and molecular determinants of embryo implantation. Mol AspectsMed 2013; 34:939–80; PMID: 23290997; http://dx.doi.org/10.1016/j.mam.2012.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cuman C, Menkhorst E, Winship A, Van Sinderen M, Osianlis T, Rombauts LJ, Dimitriadis E. Fetal-maternal communication: the role of Notch signaling in embryo implantation. Reproduction 2014; 147:75–86; http://dx.doi.org/10.1530/REP-13-0474 [DOI] [PubMed] [Google Scholar]
- 6. Melford SE, Taylor AH, Konje JC. Of mice and (wo)men: factors influencing successful implantation including endocannabinoids. Hum ReprodUpdate 2014; 20:415–28; PMID: 24306146; http://dx.doi.org/10.1093/humupd/dmt060 [DOI] [PubMed] [Google Scholar]
- 7. Sukhatme VP, Cao XM, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB, Curran T, et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellulardepolarization. Cell 1988; 53:37–43; PMID: 3127059; http://dx.doi.org/10.1016/0092-8674(88)90485-0 [DOI] [PubMed] [Google Scholar]
- 8. Gitenay D, Baron VT. Is EGR1 a potential target for prostate cancer therapy? FutureOncol 2009; 5:993–1003; PMID: 19792968; http://dx.doi.org/10.2217/fon.09.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng JC, Chang HM, Leung PC. Egr-1 mediates epidermal growth factor-induced downregulation of E-cadherin expression via Slug in humanovarian cancer cells. Oncogene 2013; 32:1041–9; PMID: 22508482; http://dx.doi.org/10.1038/onc.2012.127 [DOI] [PubMed] [Google Scholar]
- 10. Gashler A, Sukhatme VP. Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res MolBiol 1995; 50:191–224; PMID: 7754034; http://dx.doi.org/10.1016/S0079-6603(08)60815-6 [DOI] [PubMed] [Google Scholar]
- 11. Cao X, Mahendran R, Guy GR, Tan YH. Detection and characterization of cellular EGR-1 binding to its recognition site. J BiolChem 1993; 268:16949–57; PMID: 8349585. [PubMed] [Google Scholar]
- 12. Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt MA, Rao CV, Charnay P. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. MolEndocrinol 1998; 12:107–22; PMID: 9440815; http://dx.doi.org/10.1210/mend.12.1.0049 [DOI] [PubMed] [Google Scholar]
- 13. Sevetson BR, Svaren J, Milbrandt J. A novel activation function for NAB proteins in EGR-dependent transcription of the luteinizing hormone beta gene. J BiolChem 2000; 275:9749–57; PMID: 10734128; http://dx.doi.org/10.1074/jbc.275.13.9749 [DOI] [PubMed] [Google Scholar]
- 14. Espey LL, Ujioka T, Russell DL, Skelsey M, Vladu B, Robker RL, Okamura H, Richards JS. Induction of early growth response protein-1 gene expression in the rat ovary in response to an ovulatory dose of human chorionic gonadotropin. Endocrinology 2000; 141:2385–91; PMID: 10875238. [DOI] [PubMed] [Google Scholar]
- 15. Russell DL, Doyle KM, Gonzales-Robayna I, Pipaon C, Richards JS. Egr-1 induction in rat granulose cells by follicle-stimulating hormone and luteinizing hormone: combinatorial regulation by transcription factors cyclic adenosine 3’,5’-monophosphate regulatory element binding protein, serum response factor, sp1, and early growth response factor-1. MolEndocrinol 2003; 17:520–33; PMID: 12554779; http://dx.doi.org/10.1210/me.2002-0066 [DOI] [PubMed] [Google Scholar]
- 16. Sayasith K, Brown KA, Lussier JG, Doré M, Sirois J. Characterization of bovine early growth response factor-1 and its gonadotropin-dependent regulation in ovarian follicles prior to ovulation. J MolEndocrinol 2006; 37:239–50; PMID: 17032742; http://dx.doi.org/10.1677/jme.1.02078 [DOI] [PubMed] [Google Scholar]
- 17. Hou X, Arvisais EW, Jiang C, Chen DB, Roy SK, Pate JL, Hansen TR, Rueda BR, Davis JS. Prostaglandin F2alpha stimulates the expression and secretion of transforming growth factor B1 via induction of the early growth response 1 gene (EGR1) in the bovine corpus luteum. MolEndocrinol 2008; 22:403–14; PMID: 17916653; http://dx.doi.org/10.1210/me.2007-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carletti MZ, Christenson LK. Rapid effects of LH on gene expression in the mural granulosa cells of mouse periovulatory follicles. Reproduction 2009; 137:843–55; PMID: 19225042; http://dx.doi.org/10.1530/REP-08-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Suva LJ, Harm SC, Gardner RM, Thiede MA. In vivo regulation of Zif268 messenger RNA expression by 17 beta-estradiol in the rat uterus. MolEndocrinol 1991; 5:829–35; PMID: 1717835; http://dx.doi.org/10.1210/mend-5-6-829 [DOI] [PubMed] [Google Scholar]
- 20. Shozu M, Murakami K, Segawa T, Kasai T, Ishikawa H, Shinohara K, Okada M, Inoue M. Decreased expression of early growth response-1 and its role in uterine leiomyoma growth. CancerRes 2004; 64:4677–84; PMID: 15231681; http://dx.doi.org/10.1158/0008-5472.CAN-03-0560 [DOI] [PubMed] [Google Scholar]
- 21. Ivanga M, Labrie Y, Calvo E, Belleau P, Martel C, Luu-The V, Morissette J, Labrie F, Durocher F. Temporal analysis of E2 transcriptional induction of PTP and MKP and downregulation of IGF-I pathway keycomponents in the mouse uterus. PhysiolGenomics 2007; 29:13–23; PMID: 17361005; http://dx.doi.org/10.1152/physiolgenomics.00291.2005 [DOI] [PubMed] [Google Scholar]
- 22. Li DD, Gao YJ, Tian XC, Yang ZQ, Cao H, Zhang QL, Guo B, Yue ZP. Differential expression and regulation of Tdo2 during mouse decidualization. JEndocrinol 2013; 220:73–83; PMID: 24190896; http://dx.doi.org/10.1530/JOE-13-0429 [DOI] [PubMed] [Google Scholar]
- 23. Diao HL, Zhu H, Ma H, Tan HN, Cong J, Su RW, Yang ZM. Rat ovulation, implantation and decidualization are severely compromised by COX-2 inhibitors. FrontBiosci 2007; 12:3333–42; PMID: 17485303; http://dx.doi.org/10.2741/2316 [DOI] [PubMed] [Google Scholar]
- 24. Tian XC, Wang QY, Li DD, Wang ST, Yang ZQ, Guo B, Yue ZP. Differential expression and regulation of Cryab in mouse uterus during preimplantation period. Reproduction 2013; 145:577–85; PMID: 23579188; http://dx.doi.org/10.1530/REP-13-0042 [DOI] [PubMed] [Google Scholar]
- 25. Zaczek D, Hammond J, Suen L, Wandji S, Service D, Bartke A, Chandrashekar V, Coschigano K, Kopchick J. Impact of growth hormone resistance on female reproductive function: new insights from growth hormone receptor knockout mice. BiolReprod 2002; 67:1115–24; PMID: 12297526; http://dx.doi.org/10.1095/biolreprod67.4.1115 [DOI] [PubMed] [Google Scholar]
- 26. Gellersen B, Brosens J. Cyclic AMP and progesterone receptor cross-talk in human endometrium: a decidualizing affair. JEndocrinol 2003; 178:357–72; PMID: 12967329; http://dx.doi.org/10.1677/joe.0.1780357 [DOI] [PubMed] [Google Scholar]
- 27. Lee SL, Sadovsky Y, Swirnoff AH, Polish JA, Goda P, Gavrilina G, Milbrandt J. Luteinizing hormone deficiency and female infertility in mice lacking the transcription factor NGFI-A (Egr-1). Science 1996; 273:1219–21; PMID: 8703054; http://dx.doi.org/10.1126/science.273.5279.1219 [DOI] [PubMed] [Google Scholar]
- 28. Nair P, Muthukkumar S, Sells SF, Han SS, Sukhatme VP, Rangnekar VM. Early growth response-1-dependent apoptosis is mediated by p53. J BiolChem 1997; 272:20131–8; PMID: 9242687; http://dx.doi.org/10.1074/jbc.272.32.20131 [DOI] [PubMed] [Google Scholar]
- 29. Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature 2007; 450:721–4; PMID: 18046411; http://dx.doi.org/10.1038/nature05993 [DOI] [PubMed] [Google Scholar]
- 30. Hu SJ, Ren G, Liu JL, Zhao ZA, Yu YS, Su RW, Ma XH, Ni H, Lei W, Yang ZM. MicroRNA expression and regulation in mouse uterus during embryo implantation. J BiolChem 2008; 283:23473–84; PMID: 18556655; http://dx.doi.org/10.1074/jbc.M800406200 [DOI] [PubMed] [Google Scholar]
- 31. Shin SY, Kim JH, Baker A, Lim Y, Lee YH. Transcription factor Egr-1 is essential for maximal matrix metalloproteinase-9 transcription by tumor necrosis factor alpha. Mol CancerRes 2010; 8:507–19; PMID: 20332214; http://dx.doi.org/10.1158/1541-7786.MCR-09-0454 [DOI] [PubMed] [Google Scholar]
- 32. Matsumoto H, Ma WG, Daikoku T, Zhao X, Paria BC, Das SK, Trzaskos JM, Dey SK. Cyclooxygenase-2 differentially directs uterine angiogenesis during implantation in mice. J BiolChem 2002; 277:29260–7; PMID: 12034746; http://dx.doi.org/10.1074/jbc.M203996200 [DOI] [PubMed] [Google Scholar]
- 33. Kennedy TG, Gillio-Meina C, Phang SH. Prostaglandins and the initiation of blastocyst implantation and decidualization. Reproduction 2007; 134:635–43; PMID: 17965253; http://dx.doi.org/10.1530/REP-07-0328 [DOI] [PubMed] [Google Scholar]
- 34. Lim H, Paria BC, Das SK, Dinchuk JE, Langenbach R, Trzaskos JM, Dey SK. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell 1997; 91:197–208; PMID: 9346237; http://dx.doi.org/10.1016/S0092-8674(00)80402-X [DOI] [PubMed] [Google Scholar]
- 35. Díaz-Muñoz MD, Osma-García IC, Cacheiro-Llaguno C, Fresno M, Iñiguez MA. Coordinated up-regulation of cyclooxygenase-2 and microsomal prostaglandin E synthase 1 transcription by nuclear factor kappa B and early growth response-1 in macrophages. CellSignal 2010; 22:1427–36; PMID: 20546888; http://dx.doi.org/10.1016/j.cellsig.2010.05.011 [DOI] [PubMed] [Google Scholar]
- 36. Howe LR, Subbaramaiah K, Kent CV, Zhou XK, Chang SH, Hla T, Jakobsson PJ, Hudis CA, Dannenberg AJ. Genetic deletion of microsomal prostaglandin E synthase-1 suppresses mouse mammary tumor growth and angiogenesis. Prostaglandins Other LipidMediat 2013; 106:99–105; PMID: 23624019; http://dx.doi.org/10.1016/j.prostaglandins.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Naraba H, Yokoyama C, Tago N, Murakami M, Kudo I, Fueki M, Oh-Ishi S, Tanabe T. Transcriptional regulation of the membrane-associated prostaglandin E2 synthase gene. Essential role of thetranscription factor Egr-1. J BiolChem 2002; 277:28601–8; PMID: 12034740; http://dx.doi.org/10.1074/jbc.M203618200 [DOI] [PubMed] [Google Scholar]
- 38. Rockwell LC, Pillai S, Olson CE, Koos RD. Inhibition of vascular endothelial growth factor/vascular permeability factor action blocks estrogen-induced uterine edema and implantation in rodents. BiolReprod 2002; 67:1804–10; PMID: 12444056; http://dx.doi.org/10.1095/biolreprod.102.006700 [DOI] [PubMed] [Google Scholar]
- 39. Numao A, Hosono K, Suzuki T, Hayashi I, Uematsu S, Akira S, Ogino Y, Kawauchi H, Unno N, Majima M. The inducible prostaglandin E synthase mPGES-1 regulates growth of endometrial tissues and angiogenesisin a mouse implantation model. Biomed Pharmacother 2011; 65:77–84; PMID: 21247731; http://dx.doi.org/10.1016/j.biopha.2010.12.008 [DOI] [PubMed] [Google Scholar]
- 40. Logan PC, Ponnampalam AP, Steiner M, Mitchell MD. Effect of cyclic AMP and estrogen/progesterone on the transcription of DNA methyltransferases during the decidualization of human endometrial stromal cells. Mol HumReprod 2013; 19:302–12; PMID: 23233487; http://dx.doi.org/10.1093/molehr/gas062 [DOI] [PubMed] [Google Scholar]
- 41. Horton CD, Halvorson LM. The cAMP signaling system regulates LHbeta gene expression: roles of early growth response protein-1, SP1 and steroidogenic factor-1. J MolEndocrinol 2004; 32:291–306; PMID: 14766009; http://dx.doi.org/10.1677/jme.0.0320291 [DOI] [PubMed] [Google Scholar]
- 42. Kang JH, Kim MJ, Jang HI, Koh KH, Yum KS, Rhie DJ, Yoon SH, Hahn SJ, Kim MS, Jo YH. Proximal cyclic AMP response element is essential for exendin-4 induction of rat EGR-1 gene. Am J Physiol EndocrinolMetab 2007; 292:E215–22; PMID: 16926376; http://dx.doi.org/10.1152/ajpendo.00181.2006 [DOI] [PubMed] [Google Scholar]
- 43. Chen CC, Lee WR, Safe S. Egr-1 is activated by 17beta-estradiol in MCF-7 cells by mitogen-activated protein kinase-dependent phosphorylation of ELK-1. J CellBioche 2004; 93:1063–74; http://dx.doi.org/10.1002/jcb.20257 [DOI] [PubMed] [Google Scholar]
- 44. Vivacqua A, Romeo E, De Marco P, De Francesco EM, Abonante S, Maggiolini M. GPER mediates the Egr-1 expression induced by 17β-estradiol and 4-hydroxitamoxifen in breast and endometrial cancer cells. Breast Cancer ResTreat 2012; 133:1025–35; PMID: 22147081; http://dx.doi.org/10.1007/s10549-011-1901-8 [DOI] [PubMed] [Google Scholar]
- 45. Gonzalez CR, Vallcaneras SS, Calandra RS, Gonzalez Calvar SI. Involvement of KLF14 and egr-1 in the TGF-beta1 action on Leydig cell proliferation. Cytokine 2013; 61:670–5; PMID: 23317878; http://dx.doi.org/10.1016/j.cyto.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 46. de Jager T, Pelzer T, Müller-Botz S, Imam A, Muck J, Neyses L. Mechanisms of estrogen receptor action in the myocardium. Rapid gene activation via the ERK1/2 pathway and serum response elements. J BiolChem 2001; 276:27873–80; PMID: 11335712; http://dx.doi.org/10.1074/jbc.M010984200 [DOI] [PubMed] [Google Scholar]
- 47. Kim JH, Jeong IY, Lim Y, Lee YH, Shin SY. Estrogen receptor beta stimulates Egr-1 transcription via MEK1/Erk/Elk-1 cascade in C6 glioma cells. BMBRep 2011; 44:452–7; PMID: 21777515; http://dx.doi.org/10.5483/BMBRep.2011.44.7.452 [DOI] [PubMed] [Google Scholar]
- 48. Gong TW, Meyer DJ, Liao J, Hodge CL, Campbell GS, Wang X, Billestrup N, Carter-Su C, Schwartz J. Regulation of glucose transport and c-fos and egr-1 expression in cells with mutated or endogenous growth hormone receptors. Endocrinology 1998; 139:1863–71; PMID: 9528972. [DOI] [PubMed] [Google Scholar]
- 49. Hodge C, Liao J, Stofega M, Guan K, Carter-Su C, Schwartz J. Growth hormone stimulates phosphorylation and activation of elk-1 and expression of c-fos, egr-1, and junB through activation of extracellular signal-regulated kinases 1 and 2. J BiolChem 1998; 273:31327–36; PMID: 9813041; http://dx.doi.org/10.1074/jbc.273.47.31327 [DOI] [PubMed] [Google Scholar]
- 50. Clarkson RW, Shang CA, Levitt LK, Howard T, Waters MJ. Ternary complex factors Elk-1 and Sap-1a mediate growth hormone-induced transcription of egr-1 (early growth response factor-1) in 3T3-F442A preadipocytes. MolEndocrinol 1999; 13:619–31; PMID: 10194767; http://dx.doi.org/10.1210/mend.13.4.0266 [DOI] [PubMed] [Google Scholar]
- 51. Hull KL, Harvey S. Growth hormone: roles in female reproduction. JEndocrinol 2001; 168:1–23; PMID: 11139766; http://dx.doi.org/10.1677/joe.0.1680001 [DOI] [PubMed] [Google Scholar]
- 52. Yovich JL, Stanger JD. Growth hormone supplementation improves implantation and pregnancy productivity rates for poor-prognosispatients undertaking IVF. Reprod BiomedOnline 2010; 21:37–49; PMID: 20457541; http://dx.doi.org/10.1016/j.rbmo.2010.03.013 [DOI] [PubMed] [Google Scholar]
- 53. van Eickels M, Vetter H, Grohé C. Angiotensin-converting enzyme (ACE) inhibition attenuates insulin-like growth factor-I (IGF-I) induced cardiac fibroblast proliferation. Br J Pharmacol 2000; 131:1592–6; PMID: 11139436; http://dx.doi.org/10.1038/sj.bjp.0703740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li H, Costantini C, Scrable H, Weindruch R, Puglielli L. Egr-1 and Hipk2 are required for the TrkA to p75(NTR) switch that occurs downstream of IGF1-R. NeurobiolAging 2009; 30:2010–20; PMID: 18378044; http://dx.doi.org/10.1016/j.neurobiolaging.2008.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Youreva V, Kapakos G, Srivastava AK. Insulin-like growth-factor-1-induced PKB signaling and Egr-1 expression is inhibited by curcumin in A-10 vascular smooth muscle cells. Can J PhysiolPharmacol 2013; 91:241–7; PMID: 23537438; http://dx.doi.org/10.1139/cjpp-2012-0267 [DOI] [PubMed] [Google Scholar]