Abstract
PPARγ2 is expressed almost exclusively in adipose tissue and plays a central role in adipogenesis. Despite intensive studies over the last 2 decades, the mechanism regulating the expression of the Pparg2 gene, especially the role of cis-regulatory elements, is still not completely understood. Here, we report a comprehensive investigation of the enhancer elements within the murine Pparg2 gene. Utilizing the combined techniques of sequence conservation analysis and chromatin marker examination, we identified a potent enhancer element that augmented the expression of a reporter gene under the control of the Pparg2 promoter by 20-fold. This enhancer element was first identified as highly conserved non-coding sequence 10 (CNS10) and was later shown to be enriched with the enhancer marker H3 K27 acetylation. Further studies identified a binding site for p300 as the essential enhancer element in CNS10. Moreover, p300 physically binds to CNS10 and is required for the enhancer activity of CNS10. The depletion of p300 by siRNA resulted in significantly impaired activation of Pparg2 at the early stages of 3T3-L1 adipogenesis. In summary, our study identified a novel enhancer element on the murine Pparg2 gene and suggested a novel mechanism for the regulation of Pparg2 expression by p300 in 3T3-L1 adipogenesis.
Keywords: adipogenesis, enhancer, H3 K27 acetylation, PPARg2, p300
Abbreviations
- CNS
conserved non-coding sequence
- TZDs
thiazolidinediones
- KAT
lysine acetyl-transferase
- DEX
dexamethasone
- IBMX
3-isobutyl-1-methylxanthine
- Rosi
rosiglitazone
- ChIP
chromatin immunoprecipitation
- GR
glucocorticoid receptor
- PPRE
PPAR-Response Elements
- TSS
transcription start site
Introduction
Peroxisome proliferator-activated receptor γ (PPARγ), a member of the hormone nuclear receptor super-family, plays a central role in regulating adipogenesis and adipocyte function.1-3 Being the only identified factor that is both necessary and sufficient to promote fat cell differentiation, PPARγ is considered the master regulator of adipogenesis.1 Once activated, PPARγ drives the expression of an array of downstream target genes to complete the maturation process of adipocytes.4 In addition to the well-established role of PPARγ in differentiated adipocytes, a recent study revealed that PPARγ is also detectable in adipocyte progenitor cells residing in the vasculature of white adipose tissue.5 These PPARγ-positive adipocyte progenitor cells give rise to the vast majority of mature adipocytes in the fat pad, which suggests that PPARγ may also play a role in the lineage commitment of adipose cells. In mammals, PPARγ exists in 2 isoforms, PPARγ1 and PPARγ2, that are generated by alternative promoter usage and mRNA splicing of the same gene.6-8 The amino acid sequence of PPARγ2 is identical to that of PPARγ1 except that it contains an N-terminal extension of 30 amino acids. In contrast to the ubiquitous expression of PPARγ1,9 PPARγ2 is expressed almost exclusively in adipose tissue.6 Consistent with their tissue distribution patterns, PPARγ2, but not PPARγ1, plays a predominant role in adipogenesis.
Given the key role of PPARγ2 in adipogenesis, adipose tissue function and whole body energy homeostasis, this protein has been investigated extensively as a drug target for the treatment of metabolic disorders. One such class of drugs is the potent insulin sensitizers thiazolidinediones (TZDs), which are highly effective in treating type 2 diabetes. However, the side effects caused by this class of compounds, and their eventual withdrawal from the market, point to the urgent need for a comprehensive understanding of the regulatory networks that control PPARγ expression, activity and downstream effects. Unfortunately, despite the continuous efforts in the 20 y since the identification of PPARγ as the master adipogenic factor, the gene regulation mechanism during the early stage of adipogenesis is still not completely understood.
In principle, eukaryotic genes are regulated by the concerted action of both cis-elements, such as promoters, enhancers and repressors, and trans-factors. Many of the trans-factors regulating the Pparg2 gene have been identified. These factors include the protein lysine acetyl-transferases (KAT), CBP/p300, the PPARγ coactivator-1 (PGC-1α) and the CCAAT/enhancer binding proteins (C/EBPβ and C/EBPδ at the early stage and C/EBPα at a late stage during adipogenesis).4 The regulatory function of CBP/p300 was thought to be exerted by direct interaction with PPARγ2 and the activation of its downstream targets, including the gene encoding PPARγ2 itself.10,11 In contrast to the level of understanding about trans-regulation, much less is known about the role of cis-elements in Pparg2 gene transcription. Two recent studies identified regulatory regions upstream of the Pparg2 promoter by either sequence conservation analysis 12 or chromatin marker examination.13 Although convincing evidence was provided for the enhancer activities in both studies, there was not a detailed analysis of the enhancer sequences. For example, the essential transcription factor binding sites within the enhancers remained elusive.
To refine our understanding of Pparg2 regulation, we sought to delineate the cis-regulatory regions within the Pparg2 gene body. The rationale is that the Pparg2 gene spans across approximately 70 kb (mouse), and more than 97% of the sequences are within the introns. Thus, we hypothesized that there might be cis-regulatory elements residing inside these non-coding sequences. The identification and characterization of cis-elements that relate to key enhancer regions have gained recognition over the last couple of decades as a legitimate mode of transcriptional regulation. There are numerous methods that have been employed to identify cis-regulatory regions within the genome. Among these methods, genomic sequence conservation analysis has been widely used. The principle of this methodology is that sequence conservation could be indicative of sequence significance. There have been a number of studies showing that regions spanning a conserved non-coding sequence (CNS) can act as enhancer elements.14-20
However, reliance on sequence conservation alone can have drawbacks. Within a single organism, the genomic sequence is the same for all cell types, but the gene expression patterns and enhancer identities vary significantly. This difference argues that enhancers are determined by cell type or developmental stage-specific signals and not simply by sequence conservation levels. For example, a study by Gordon and Ruvinsky 21 showed that lineage-specific changes in cis or trans-regulation could alter the ability of conserved non-coding sequences to drive gene expression. The use of histone modifications can be an effective way to discern the identity of enhancers.22-25 To date, a number of histone marks, including H3 K4 mono-methylation,22 K27 acetylation 23-25 and K9 acetylation,13 have been used to systematically identify enhancer elements. In addition, most CNS regions are known to function as regulatory factor binding sites.26 As such, it is important to identify the trans-regulatory factor that binds and potentiates the ability of the CNS region to function as an enhancer.
To identify cis-regulatory regions within the murine Pparg2 gene, we used the combined techniques of sequence conservation analysis and chromatin marker examination. Using the VISTA algorithm,27 we first identified 11 highly conserved non-coding sequences within the introns of the Pparg2 gene. Subsequent histone modification ChIP analysis revealed that the CNS10 region was enriched for the enhancer marker H3 K27 acetylation. Further luciferase assays confirmed that the CNS10 region augmented the expression of a reporter gene under the control of the Pparg2 promoter by 20-fold. Detailed sequence analysis and mutagenesis experiments identified a binding site for p300 as the essential enhancer element in CNS10. Moreover, p300 physically bound to CNS10, and the depletion of p300 significantly impaired the enhancer activity of CNS10 on the Pparg2 promoter. In summary, our study identified a potent enhancer element for Pparg2 gene expression and provided a novel mechanism for regulation in which p300 binds to this enhancer to promote Pparg2 gene expression at the early stage of adipogenesis.
Results
Sequence conservation analysis revealed 11 highly conserved non-coding sequences in the murine adipogenic master regulator gene Pparg2
Considering the central role of PPARγ2 in adipogenesis and adipocyte functions, we sought to determine the cis-regulatory elements that contribute to Pparg2 gene regulation. We first compared the mouse PPARγ2 genomic sequence spanning from 2 kb upstream of the transcription start site (TSS) to the 3’-end to the sequences from human, dog, ox and rat using the VISTA algorithm. This analysis yielded a total of 28 conserved regions (data not shown), including the promoter, all 7 exons and 20 CNS located in the introns of the Pparg2 gene. We arbitrarily set the cut-off p-value at 0.0001 and selected the top 11 highly conserved non-coding sequences for further studies. As shown in Figure 1, the 11 CNS were distributed across the whole gene body of Pparg2, and their lengths range from 206 bp (CNS10) to 1812 bp (CNS7). The relative positions of these CNS to the Pparg2 gene transcription start site and their statistical significance (p-value) are listed in Table 1.
Figure 1.
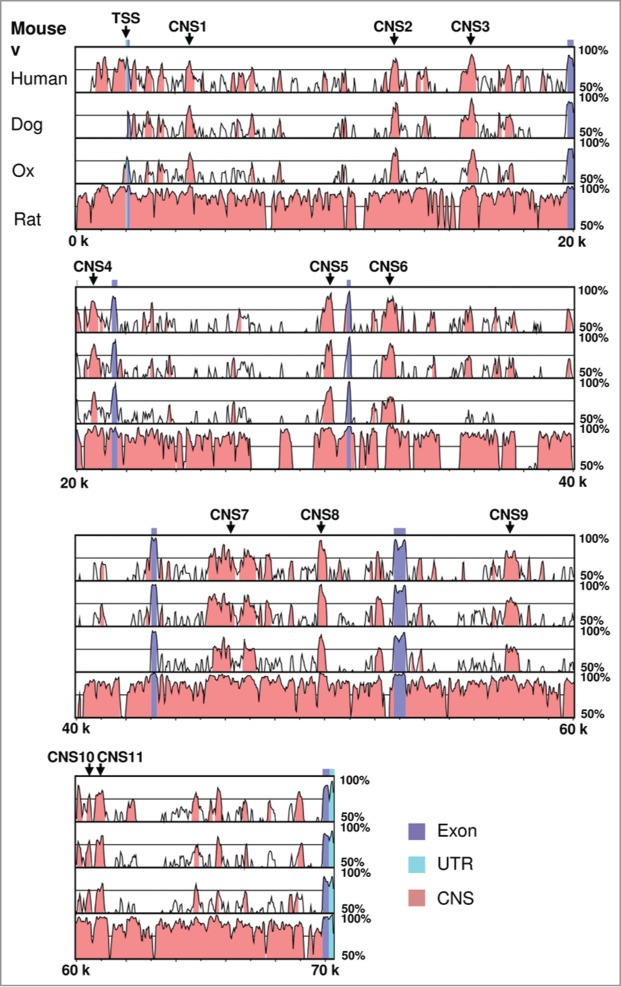
Sequence conservation analysis of the murine adipogenic master regulator gene Pparg2. Mouse PPARγ2 genomic sequence spanning from 2 kb upstream of the transcription start site (TSS) to the 3′-end was compared with human, dog, ox and rat sequences using the VISTA algorithm. The Pparg2 TSS and the 11 highly conserved non-coding sequences (CNS) are indicated in the gene.
Table 1.
List of the conserved non-coding sequences identified by VISTA in the murine Pparg2 gene
| Name | Start* | End* | Length | p-value |
|---|---|---|---|---|
| CNS1 | 2,392 | 2,720 | 329 bp | 3.9e-09 |
| CNS2 | 10,621 | 10,966 | 346 bp | 7.7e-10 |
| CNS3 | 13,721 | 14,092 | 372 bp | 4.3e-09 |
| CNS4 | 18,571 | 19,024 | 454 bp | 1.2e-07 |
| CNS5 | 27,988 | 28,313 | 326 bp | 7.8e-12 |
| CNS6 | 30,469 | 30,928 | 460 bp | 1.7e-09 |
| CNS7 | 43,384 | 45,195 | 1812 bp | 2.1e-24 |
| CNS8 | 47,704 | 48,064 | 361 bp | 1.2e-15 |
| CNS9 | 55,254 | 55,747 | 494 bp | 2.3e-08 |
| CNS10 | 58,379 | 58,584 | 206 bp | 7.3e-05 |
| CNS11 | 58,761 | 59,103 | 343 bp | 4.7e-06 |
Relative to the Pparg2 gene transcription start site.
Histone H3 K27 acetylation marks CNS10 in the murine Pparg2 gene in 2 independent adipocyte progenitor cell lines
Histone H3 K4 mono-methylation (me1) and K27 acetylation (ac) are known markers of enhancer elements in the mammalian system.22-25 Thus, we examined the enrichment of these 2 chromatin modifications at the 11 CNS with the aim of identifying the potential enhancers for the Pparg2 gene. Our histone H3K4me1 ChIP analysis revealed that this modification was generally abundant at all CNS regions in 3T3-L1 cells at the preadipocyte stage (Fig. 2A). Although there were variations in the K4Me1 level among these CNS regions, no CNS was specifically enriched for this modification over the others. By contrast, we observed a marked enrichment of H3K27ac at CNS10 in our ChIP assay (Fig. 2B). CNS10 had a level of H3K27ac that was at least three-fold higher than those of other CNS regions. To verify these observations in another adipocyte progenitor cell line, we used C3H 10T1/2 cells, which are a mouse mesenchymal stem cell line. Consistent with the ChIP results obtained in 3T3-L1 cells, the H3K4me1 levels did not differ significantly among the 11 CNS regions (Fig. S1A). However, H3K27ac was clearly enriched in CNS10, albeit to a lesser extent than in the 3T3-L1 cells (Fig. S1B). In control experiments, we found that both H3K4me1 and K27ac were much more abundant at the actively transcribed locus GAPDH than at the silent region Chr. Fifteen (Fig. 2; Fig. S2). This result concurred with the known role of these 2 chromatin modifications in gene activation.
Figure 2.
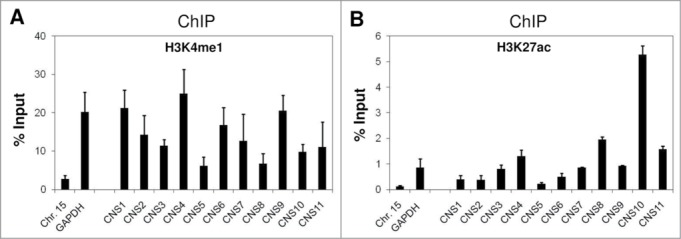
Enrichment of the chromatin markers for enhancer elements at the CNS regions in the murine Pparg2 gene in 3T3-L1 cells. Levels of histone H3 (A), K4 mono-methylation (H3K4me1) and (B) K27 acetylation (H3K27ac) at the CNS regions in the Pparg2 gene in 3T3-L1 cells were examined by ChIP analysis using specific antibodies. The ChIP-qPCR primers used in this study are described in detail in the MATERIALS AND METHODS section. Chr. Fifteen targets a transcriptionally silent region, and GAPDH represents an actively transcribed locus. These results are the averages of 3 to 4 independent ChIP-qPCR assays, and the error bars indicate standard deviations.
CNS10 is a strong enhancer element for the Pparg2 gene
The specific enrichment of H3K27ac at CNS10 suggested that this conserved region could act as an enhancer for Pparg2 gene expression. To test this idea, we used a dual luciferase reporter assay (Promega). We first inserted a 0.6 kb Pparg2 promoter 28 upstream of the firefly luciferase gene in the pGL3 vector. Then, the 11 CNS regions were inserted individually at the 3′-end of the luciferase gene (Fig. 3A). Subsequently, the firefly luciferase reporter vectors containing no CNS (negative control) or CNS1 - 11 were transfected into 10T1/2 cells along with a renilla luciferase vector (pRLTK) as an internal control. As shown in Figure 3B, CNS10 significantly enhanced the promoter activity of Pparg2 by nearly 20-fold.
Figure 3.
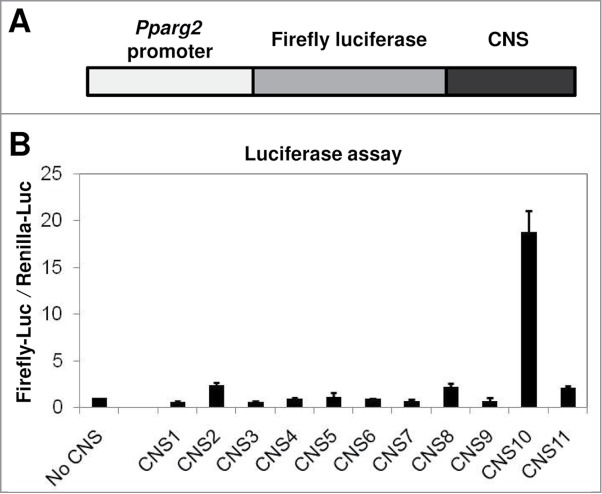
The H3 K27 acetylation-enriched CNS10 is a strong enhancer element for the Pparg2 gene. (A) A schematic of the constructed firefly luciferase reporter plasmid. The 0.6 kb Pparg2 promoter was amplified and inserted upstream of the firefly luciferase gene in the pGL3 vector. The 11 CNS were inserted individually at the 3′-end of the luciferase gene. (B) The firefly luciferase reporter vectors containing no CNS (negative control) or CNS1 - 11 were transfected into C3H 10T1/2 cells along with a renilla luciferase vector (pRLTK) as an internal control. The activities of both the firefly and renilla luciferases were measured 48 hours after transfection. These results are the averages of 3 to 6 independent luciferase assays, and the error bars represent standard deviations.
After identifying CNS10 as a potent enhancer for the Pparg2 gene, we asked whether there were additional chromatin modifications that serve as markers for this DNA element. To address this question, we examined the state of a number of key histone modifications at all CNS regions using ChIP. These histone modifications included both permissive (H3 K4 di- and tri-methylation; H4 K20 mono-methylation; H3 K9/K14 acetylation and H4 K12 acetylation) and non-permissive (H3 K9 tri-methylation and H3 K27 tri-methylation) marks on either H3 or H4. As shown in Figure S2, none of these examined histone modifications were specifically enriched at CNS10 in 3T3-L1 cells, which suggested that H3K27ac is the only identified marker for this enhancer element (Fig. 2B). To confirm our findings in another independent adipocyte progenitor cell line, we performed the same ChIP analysis in 10T1/2 cells. Again, we failed to detect additional chromatin markers for CNS10 (Fig. S3). To rule out the possibility that the elevated level of H3K27ac at CNS10 was due to a higher occupancy of histone H3, we examined the distribution pattern of histone H3 across the 11 CNS regions by ChIP using a specific antibody against the C-terminus of this protein. We found that the histone H3 level was actually relatively low at CNS10 compared with other CNS regions (Figs. S2B and S3B). This result indicated that the enrichment of H3K27ac at this region was not due to a higher occupancy of histone H3. In control experiments, we found that the levels of permissive histone modifications were much higher at the actively transcribed GAPDH promoter than at the silent region Chr. Fifteen, while the non-permissive modifications were enriched at the silent region but not at the GAPDH locus (Figs. S2 and S3).
Enhancer activity of CNS10 is dependent upon a p300 binding site
To identify the transcription factor that binds to CNS10 and promotes its enhancer activity, we sought to determine the crucial transcription factor binding site within the CNS10 sequence. In the luciferase assay, we inserted a 275 bp CNS10 fragment into the firefly luciferase reporter vector. This fragment contains the 206 bp highly conserved CNS10 sequence identified through VISTA analysis (Table 1) and its flanking sequences that are required for PCR amplification. To narrow down the range for the transcription factor binding site search, we divided the 275 bp CNS10 fragment into 4 sub-regions of equal length (CNS10A - 10D) and inserted these fragments individually downstream of the firefly luciferase gene (Fig. 4A). We found that the action of CNS10A largely resembled the enhancer activity of CNS10 on the Pparg2 promoter (Fig. 4A), which indicated that the crucial transcription factor binding site is mainly located within the first 69 bp of the CNS10 fragment. Interestingly, we also noticed that there was a slight decrease in the enhancer activity for CNS10A compared with the full length CNS10, which suggested that the crucial transcription factor binding site might be partially lost in CNS10A. Thus, we took the first 80 bp DNA sequence from the CNS10 fragment for further in silico analysis to avoid losing the potential binding site at the boundary of CNS10A and 10B. To search for transcription factor binding sites within the 80 bp CNS10 sequence (Fig. 4B), we used the online sequence analysis tools TESS and TFSEARCH. Through TESS analysis, we found a glucocorticoid receptor (GR) binding site in CNS10A. TFSEARCH identified C/EBPβ and p300 binding sites within CNS10A or at the boundary between CNS10A and 10B, respectively (Fig. 4B).
Figure 4.
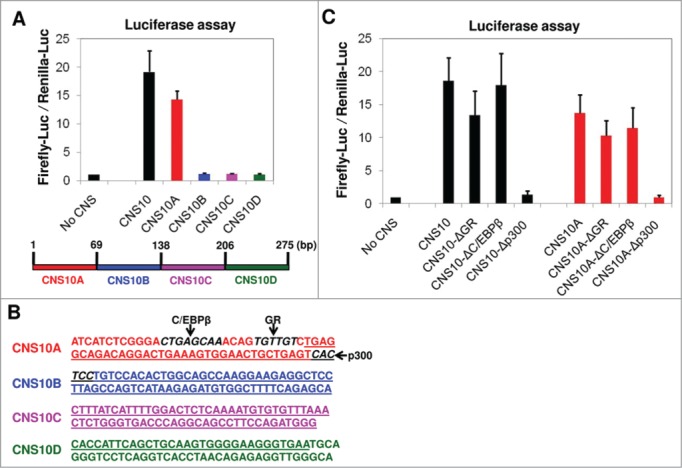
The enhancer activity of CNS10 is dependent upon a p300 binding site within its sequence. (A) The 275 bp inserted CNS10 fragment contains the 206 bp highly conserved CNS10 sequence (Table 1) and its flanking sequences that are required for PCR amplification. To narrow down the range for the transcription factor binding site search, the 275 bp CNS10 fragment was divided into 4 sub-regions (CNS10A - 10D) and inserted downstream of the firefly luciferase gene. The firefly luciferase reporter vectors containing no CNS, CNS10 or CNS10A - 10D were transfected into 10T1/2 cells along with a renilla luciferase vector (pRLTK) as an internal control. The activities of both the firefly and renilla luciferases were measured 48 hours after transfection. (B) The DNA sequence of the 275 bp inserted CNS10 fragment. The sequences of the CNS10 sub-regions are colored in red (CNS10A), blue (CNS10B), magenta (CNS10C) and green (CNS10D). The 206 bp highly conserved CNS10 sequence is underlined. The binding sites of C/EBPβ, glucocorticoid receptor (GR) and p300 are highlighted in bold and italic letters and indicated in the figure. (C) To test the effects of C/EBPβ, GR and p300 binding on CNS10 enhancer activity, their binding sites were deleted from the CNS10 and 10A sequences. The firefly luciferase reporter vectors containing CNS10 and 10A with or without the indicated transcription factor binding site deletions were transfected into 10T1/2 cells along with a renilla luciferase vector (pRLTK) as an internal control. The activities of both the firefly and renilla luciferases were measured 48 hours after transfection. These results are the averages of 3 to 6 independent luciferase assays, and the error bars indicate standard deviations.
To test the effects of C/EBPβ, GR and p300 binding on CNS10 enhancer activity, we first deleted their binding sites from the CNS10 and CNS10A sequences and then examined the firefly luciferase activity. As shown in Figure 4C, the C/EBPβ binding site deletion had virtually no effect on either CNS10 or CNS10A activity. The GR binding site deletion decreased both the CNS10 and CNS10A activities slightly, but the decreases were not of statistical significance. By contrast, the deletion of the p300 binding site completely abolished the activities of both CNS10 and CNS10A, suggesting that the p300 binding was crucial for the enhancer activity of CNS10.
p300 binds to CNS10 and is required for the enhancer activity of CNS10
To directly assess the impact of the p300 depletion on the enhancer activity of CNS10, we depleted p300 using a specific targeting siRNA in 10T1/2 cells. Both the mRNA and protein levels of p300 were reduced to approximately 40% of the level in the scrambled siRNA-treated cells or in the negative control (Fig. 5A). In the subsequent luciferase assay, we found that the knock-down of p300 resulted in a similar level of decrease in the enhancer activity for CNS10 as in mRNA and protein levels (Fig. 5A and B). To confirm that p300 was physically present at the CNS10 region, we designed a pair of ChIP-qPCR primers flanking the p300 binding site in CNS10 and examined p300 binding using ChIP. As shown in Figure 5C, p300 binds specifically to CNS10 but not to the Chr. Fifteen silent region or CNS1 in the Pparg2 gene. At the endogenous level, Pparg2 gene activation during the early stages of 10T1/2 adipogenesis was also significantly impaired by p300-targeting siRNA treatment. Compared with the scrambled siRNA-treated cells, Pparg2 gene expression was reduced to 30–40% in the p300 knock-down cells on days 0, 1 and 2 of adipogenic induction (Fig. 5D). Based on these results, we concluded that p300 bound to CNS10 directly and was essential for its enhancer activity and for Pparg2 gene activation.
Figure 5.
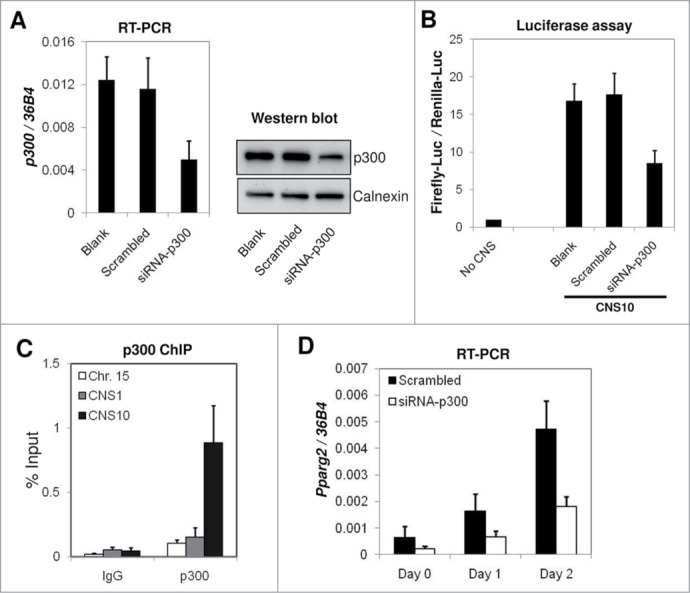
p300 is required for the enhancer activity of CNS10 and directly binds to CNS10. (A) Scrambled siRNA or p300-targeting siRNA were transfected into C3H 10T1/2 cells, and the cells were collected after 2 d to examine p300 mRNA and protein levels by RT-qPCR and western blot, respectively. The mRNA levels of p300 were normalized to the riboprotein gene 36B4. Calnexin was used as a loading control in the protein gel blot analysis. (B) The firefly luciferase reporter vector containing CNS10 was transfected into C3H 10T1/2 cells treated with scrambled siRNA or p300-targeting siRNA. Luciferase activities were measured 48 hours after transfection. (C) p300 binding at the Chr. Fifteen silent region, CNS1 and CNS10 in the Pparg2 gene were examined by ChIP using a p300 specific antibody. Rabbit IgG was included in the ChIP assay as a negative control. (D) C3H 10T1/2 cells treated with scrambled siRNA or p300-targeting siRNA were subjected to adipogenic induction for 2 d Pparg2 gene expression at the early stage of adipogenesis was examined by RT-qPCR and then normalized to 36B4. These results are the averages of 3 to 5 independent experiments, and the error bars indicate standard deviations.
The role of p300 in the enhancer activity of CNS10 is independent of PPARγ2 and RXRα
p300 binds to PPARγ2 directly and functions as a co-activator of the PPARγ2 regulation of its target genes, including Pparg2.10,11 To examine the involvement of PPARγ2 in p300-activated CNS10 enhancer activity, we overexpressed PPARγ2 and its binding partner RXRα either individually or in combination to examine the effects of these regulators on the enhancer activity of CNS10. As shown in Figure S4, in the absence of CNS10 (Pparg2 promoter), PPARγ2 and RXRα expressed together promoted the expression of the luciferase gene. This result can be explained by the presence of PPAR-Response Elements (PPRE) in the promoter sequence of Pparg2. When CNS10 was included, the firefly luciferase activity was dramatically increased (Pparg2 promoter+CNS10), which was consistent with our previous observations (Figs. 3B, 4 and 5B). Additionally, the simultaneous overexpression of PPARγ2 and RXRα only modestly promoted firefly luciferase gene expression, which mainly reflected their effects on promoters. A similar observation was made when CNS10A was included (Pparg2 promoter+CNS10A). Upon the deletion of the p300 binding site from both CNS10 and CNS10A, the level of firefly luciferase gene activation was similar to when only the Pparg2 promoter was present in the firefly luciferase reporter vector. Thus, we concluded that the effects of PPARγ2 and RXRα overexpression on reporter gene activation were exerted through Pparg2 promoter binding and not through CNS10 or p300 binding.
Discussion
PPARγ2, the master regulator of adipogenesis, has been the target of extensive studies over the last 2 decades. Given its pivotal role in regulating mammalian adipose tissue development and function, the discovery of novel mechanisms of Pparg2 gene regulation may contribute substantially to the understanding of human obesity and related metabolic disorders. In this study, we identified a potent novel enhancer element for Pparg2 gene expression and subsequently demonstrated that p300 binding is essential for its activity. Our results elucidate a novel mechanism by which p300 promotes Pparg2 gene expression through enhancer interaction.
Transcriptional enhancers play predominant roles in controlling tissue-specific gene expression patterns.29 Before the implementation of the epigenomic mapping of enhancers, sequence conservation analysis was widely used to identify these functional cis-elements.14-21 Because genes are expressed in a cell-type-selective manner even when the DNA sequences are the same, sequence conservation is not likely to be the primary determinant for enhancer activity in all cell types.21 Therefore, sequence conservation analysis must be complemented by additional measures to accurately predict the cell-type-specific enhancers. Based on this consideration, several epigenomic methodologies have been recently developed for enhancer mapping in the mammalian system. These mapping methodologies are based on open chromatin,30 transcription factor binding,31,32 co-activator binding 24,33 or histone modification enrichment.22-25 In particular, both histone H3 K27 acetylation and the major acetylase p30034 are epigenetic signatures of transcriptional enhancers that fall into the categories of histone modification enrichment and co-activator binding, respectively. In our study, we identified 11 highly conserved non-coding sequences (Fig. 1 and Table 1) using the VISTA algorithm,27 and only CNS10 was enriched with the enhancer marker H3 K27 acetylation (Fig. 2B). Subsequently, CNS10 was shown to be the only enhancer element for the Pparg2 promoter (Fig. 3), which argues for the accuracy of our combined techniques of sequence conservation analysis and chromatin marker examination. Although the other CNS regions were not identified as enhancer elements for Pparg2 in the adipocyte lineage, it is formally possible that they are involved in gene regulation in other lineages. Indeed, when searched against the enhancer lists from the ENCODE project and a recent report,35 many of the CNS regions were identified as potential enhancers in various lineages (Table S5).
Histone H3 K27 is mainly acetylated by CBP and p300,34 which suggests that these 2 KATs may bind to CNS10 in pre-adipocytes. Interestingly, our sequence analysis and mutagenesis experiments revealed that the binding site for p300 is absolutely required for CNS10 activity (Fig. 4). Moreover, p300 binding is evident on CNS10, which is consistent with the role of enhancer binding for this co-activator. p300 is essential for adipogenesis.11 The adipogenic effect of p300 was thought to be exerted by direct interaction with PPARγ2 and the activation of its downstream targets.10 Here, we provided a novel mechanism by which p300 binds to the transcriptional enhancer of the Pparg2 gene and promotes its expression. This function of p300 is apparently independent of PPARγ2 (Fig. S4).
CNS10 is located 58 kb downstream of the transcription start site of the Pparg2 gene (Table 1). Although it is evident that p300 is involved in CNS10 function, the detailed mechanism by which CNS10 activates the Pparg2 promoter remains elusive. One intriguing possibility is that CNS10 physically interacts with the Pparg2 promoter through a long-range interaction36 and brings CNS10 binding factors to the promoter to activate Pparg2 transcription. Further investigation could shed light on the mechanism of CNS10 enhancer function and contribute to the understanding of Pparg2 gene regulation.
Materials and Methods
Cell culture, differentiation and treatment
The mouse pre-adipocyte cell line 3T3-L1 and the mesenchymal stem cell line C3H 10T1/2 were purchased from the ATCC. Both 3T3-L1 and 10T1/2 cells were cultured in DMEM containing 10% fetal bovine serum (FBS). The adipogenic induction of 10T1/2 cells was performed by treating the cells with 1 μM DEX, 0.5 mM IBMX, 10 μg/ml insulin and 1 μM rosiglitazone (Rosi) for 2 d. Knock-down of p300 was carried out in 10T1/2 cells using siGENOME SMARTpool siRNA targeting mouse EP300 (328572) (Thermo Scientific).
Bioinformatic analysis
Sequence conservation analysis of the mouse adipogenic regulatory gene Pparg2 was conducted using the VISTA (visualization tool for alignment, http://genome.lbl.gov/vista/index.shtml) algorithm.27,37 Murine Pparg2 genomic sequences starting 2 kb upstream of the transcription start site (TSS) were compared with human, dog, ox and rat sequences.
To search for transcription factor binding sites within the inserted CNS10 sequence, the online sequence analysis tools Transcription Element Search System (TESS, http://www.cbil.upenn.edu/cgi-bin/tess/tess?RQ = WELCOME) and Searching Transcription Factor Binding Sites (TFSEARCH, http://www.cbrc.jp/research/db/TFSEARCH.html) were used.
Chromatin immunoprecipitation (ChIP)
The ChIP assay was performed using confluent 3T3-L1 or 10T1/2 cells as previously described.38 Briefly, cells were cross-linked with 1% formaldehyde for 10 minutes at 37°C. Then, crude nuclei were purified as previously described.39 Subsequently, the crude nuclei were sonicated using a Bioruptor UCD-300 (Diagenode) to obtain chromatin fragments of approximately 500 bp in length. For each ChIP assay, 2–5 μg of antibodies were added and incubated overnight at 4°C. ChIP and input DNA were quantified by real-time PCR analysis using SYBR green and a 7900HT Fast Real-Time PCR System (Applied Biosystems). The antibodies used in the ChIP assay were as follows: histone H3 K4 mono-methylation (Abcam, ab8895); H3 K27 acetylation (Abcam, ab4729); H3 K4 di-methylation (Upstate, 07–030); H3 K4 tri-methylation (Upstate, 07–473); H3 K9/K14 acetylation (Upstate, 06–599); H4 K12 acetylation (Upstate, 07–595); H3 K9 tri-methylation (Abcam, ab8898); H3 K27 tri-methylation (Upstate, 07–449); H4 K20 mono-methylation (Abcam, ab9051); histone H3 (Abcam, ab1791) and p300 (Santa Cruz, sc-585). Rabbit IgG (Sigma, I-5006) was included as a negative control. ChIP-qPCR primers were designed to target the 11 CNS regions with melting temperatures (Tms) near 60°C. Primers Chr. Fifteen and GAPDH were described previously.40 The primer sequences for ChIP-qPCR are listed in Table S1.
Plasmid construction
Using genomic DNA isolated from 10T1/2 cells as a template, the 0.6 kb murine Pparg2 promoter sequence28 was amplified using primers Pparg2 P-F/P-R and inserted upstream of the firefly luciferase gene in pKR0140 between the KpnI and BglII sites to yield plasmid pKR11. Eleven highly conserved Pparg2 CNS sequences were obtained through VISTA sequence conservation analysis (Table 1) and amplified using the corresponding forward and reverse primers (Table S2). Subsequently, these CNS sequences were inserted downstream of the luciferase gene between the AatII and SalI sites in pKR11 to yield plasmids pKR12 to pKR22 (Table S3).
To obtain the fragmented regions of CNS10, the following pairs of primers (as described in Table S2) were used to PCR amplify the desired sequences: CNS10-F and CNS10A-R (CNS10A, 1–69 bp); CNS10B-F and CNS10B-R (CNS10B, 70–138 bp); CNS10C-F and CNS10C-R (CNS10C, 139–206 bp); CNS10D-F and CNS10-R (CNS10D, 207–275 bp). All PCRs were performed using KAPA HiFi DNA Polymerase (Kapa Biosystems).
Plasmids containing CNS10 or CNS10A with deleted GR binding site TGTTGT (pKR40 and pKR43), C/EBPβ binding site CTGAGCAA (pKR41 and pKR44) or p300 binding site CACTCC41 (pKR42 and pKR45 (CAC deletion in CNS10A)) were constructed using QuikChange Site-Directed Mutagenesis Kit (Stratagene) and respective primers as listed in Table S2.
Luciferase assay
10T1/2 cells were seeded onto 24-well plates in DMEM containing 10% FBS. When the cells were 80–90% confluent, they were transfected with 1 μg Firefly luciferase plasmid (pKR11 or its derivative) and 200 ng renilla luciferase plasmid (pRL-TK) using Lipofectamine 2000 transfection reagent (Invitrogen). Culture media was changed to fresh complete media (DMEM containing 10% FBS and 1% penicillin-streptomycin) at 6 hours and 24 hours after transfection. Two days after transfection, the cells were subjected to a luciferase activity assay using the Dual-Luciferase Reporter 1000 Assay System (Promega) following the manufacturer's protocol.
For the p300-knock-down experiment, cells were transfected 2 d prior to the luciferase assay with either the targeting or scrambled siRNA using DharmaFECT 3 transfection reagent (Thermo Scientific) according to the manufacturer's instructions.
For the PPARγ2 and RXRα overexpression experiment, cells were transfected with the plasmid overexpressing PPARγ2 (Addgene #8895,42) or RXRα (Addgene #8882,6) either individually or in combination with the Lipofectamine 2000 transfection reagent (Invitrogen) together with the luciferase plasmids.
RNA extraction and RT-qPCR
Total RNA was extracted from either scrambled or targeting siRNA-treated cells at the indicated time points using TRIzol reagent (Invitrogen). The RNA (1 μg) was then treated with Amplification Grade DNase I (Invitrogen) to remove DNA contamination and reverse transcribed with random 9-mer and M-MLV Reverse Transcriptase (Invitrogen). The expression levels of the p300 and Pparg2 genes were determined by real-time quantitative PCR analysis using SYBR green and a 7900HT Fast Real-Time PCR System (Applied Biosystems). The primer sequences for RT-qPCR are listed in Table S4. All gene expression data in this study were normalized to the riboprotein gene 36B4.43
Western blot analysis
Whole cell lysates were prepared from 10T1/2 cells transfected with scrambled or p300 targeting siRNAs and quantitated using the NanoDrop 1000 Spectrophotometer (Thermo Scientific). An equal amount of whole cell lysates was resolved by 10% SDS-PAGE, transferred onto PVDF membranes and probed with antibodies recognizing p300 (Santa Cruz, sc-585) and calnexin (Santa Cruz, sc-11397).
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful to the members of the Feng Xu laboratory for their critical comments and discussion throughout this work.
Funding
This work was supported by intramural funding from the Agency for Science, Technology and Research (A*STAR) of Singapore to F.X.
References
- 1. Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 1994; 79:1147-56; PMID:8001151; http://dx.doi.org/ 10.1016/0092-8674(94)90006-X [DOI] [PubMed] [Google Scholar]
- 2. Tontonoz P, Hu E, Spiegelman BM. Regulation of adipocyte gene expression and differentiation by peroxisome proliferator activated receptor gamma. Curr Opin Genet Dev 1995; 5:571-6; PMID:8664544; http://dx.doi.org/ 10.1016/0959-437X(95)80025-5 [DOI] [PubMed] [Google Scholar]
- 3. Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem 2008; 77:289-312; PMID:18518822; http://dx.doi.org/ 10.1146/annurev.biochem.77.061307.091829 [DOI] [PubMed] [Google Scholar]
- 4. Farmer SR. Transcriptional control of adipocyte formation. Cell Metab 2006; 4:263-73; PMID:17011499; http://dx.doi.org/ 10.1016/j.cmet.2006.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, Tallquist MD, Graff JM. White fat progenitor cells reside in the adipose vasculature. Science 2008; 322:583-6; PMID:18801968; http://dx.doi.org/ 10.1126/science.1156232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 1994; 8:1224-34; PMID:7926726; http://dx.doi.org/ 10.1101/gad.8.10.1224 [DOI] [PubMed] [Google Scholar]
- 7. Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, Najib J, Laville M, Fruchart JC, Deeb S, et al. . The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem 1997; 272:18779-89; PMID:9228052; http://dx.doi.org/ 10.1074/jbc.272.30.18779 [DOI] [PubMed] [Google Scholar]
- 8. Zhu Y, Qi C, Korenberg JR, Chen XN, Noya D, Rao MS, Reddy JK. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR gamma) gene: alternative promoter use and different splicing yield two mPPAR gamma isoforms. Proc Natl Acad Sci U S A 1995; 92:7921-5; PMID:7644514; http://dx.doi.org/ 10.1073/pnas.92.17.7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology 1996; 137:354-66; PMID:8536636 [DOI] [PubMed] [Google Scholar]
- 10. Gelman L, Zhou G, Fajas L, Raspe E, Fruchart JC, Auwerx J. p300 interacts with the N- and C-terminal part of PPARgamma2 in a ligand-independent and -dependent manner, respectively. J Biol Chem 1999; 274:7681-8; PMID:10075656; http://dx.doi.org/ 10.1074/jbc.274.12.7681 [DOI] [PubMed] [Google Scholar]
- 11. Takahashi N, Kawada T, Yamamoto T, Goto T, Taimatsu A, Aoki N, Kawasaki H, Taira K, Yokoyama KK, Kamei Y, et al. . Overexpression and ribozyme-mediated targeting of transcriptional coactivators CREB-binding protein and p300 revealed their indispensable roles in adipocyte differentiation through the regulation of peroxisome proliferator-activated receptor gamma. J Biol Chem 2002; 277:16906-12; PMID:11884404; http://dx.doi.org/ 10.1074/jbc.M200585200 [DOI] [PubMed] [Google Scholar]
- 12. Chou WL, Galmozzi A, Partida D, Kwan K, Yeung H, Su AI, Saez E. Identification of regulatory elements that control PPARgamma expression in adipocyte progenitors. PloS one 2013; 8:e72511; PMID:24009687; http://dx.doi.org/ 10.1371/journal.pone.0072511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Jr, Lazar MA. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 2011; 24:1035-44; PMID:20478996; http://dx.doi.org/ 10.1101/gad.1907110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johnson DS, Davidson B, Brown CD, Smith WC, Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res 2004; 14:2448-56; PMID:15545496; http://dx.doi.org/ 10.1101/gr.2964504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li X, Tan L, Wang L, Hu S, Sun C. Isolation and characterization of conserved non-coding sequences among rice (Oryza sativa L.) paralogous regions. Mol Genet Genomics 2009; 281:11-8; PMID:18825415; http://dx.doi.org/ 10.1007/s00438-008-0388-4 [DOI] [PubMed] [Google Scholar]
- 16. Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science 2003; 302:413; PMID:14563999; http://dx.doi.org/ 10.1126/science.1088328 [DOI] [PubMed] [Google Scholar]
- 17. Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. . In vivo enhancer analysis of human conserved non-coding sequences. Nature 2006; 444:499-502; PMID:17086198; http://dx.doi.org/ 10.1038/nature05295 [DOI] [PubMed] [Google Scholar]
- 18. Teng Y, Girard L, Ferreira HB, Sternberg PW, Emmons SW. Dissection of cis-regulatory elements in the C. elegans Hox gene egl-5 promoter. Dev Biol 2004; 276:476-92; PMID:15581880; http://dx.doi.org/ 10.1016/j.ydbio.2004.09.012 [DOI] [PubMed] [Google Scholar]
- 19. Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al. . Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 2005; 3:e7; PMID:15630479; http://dx.doi.org/ 10.1371/journal.pbio.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature 2010; 463:808-12; PMID:20072126; http://dx.doi.org/ 10.1038/nature08750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gordon KL, Ruvinsky I. Tempo and mode in evolution of transcriptional regulation. PLoS Genet 2012; 8:e1002432; PMID:22291600; http://dx.doi.org/ 10.1371/journal.pgen.1002432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. . Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007; 39:311-8; PMID:17277777; http://dx.doi.org/ 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- 23. Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. . Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009; 459:108-12; PMID:19295514; http://dx.doi.org/ 10.1038/nature07829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011; 470:279-83; PMID:21160473; http://dx.doi.org/ 10.1038/nature09692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. . Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 2010; 107:21931-6; PMID:21106759; http://dx.doi.org/ 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hemberg M, Gray JM, Cloonan N, Kuersten S, Grimmond S, Greenberg ME, Kreiman G. Integrated genome analysis suggests that most conserved non-coding sequences are regulatory factor binding sites. Nucleic Acids Res 2012; 40:7858-69; PMID:22684627; http://dx.doi.org/ 10.1093/nar/gks477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dubchak I, Ryaboy DV. VISTA family of computational tools for comparative analysis of DNA sequences and whole genomes. Method Mol Biol 2006; 338:69-89; PMID:16888351 [DOI] [PubMed] [Google Scholar]
- 28. Pei H, Yao Y, Yang Y, Liao K, Wu JR. Kruppel-like factor KLF9 regulates PPARgamma transactivation at the middle stage of adipogenesis. Cell Death Differ 2011; 18:315-27; PMID:20725087; http://dx.doi.org/ 10.1038/cdd.2010.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet: TIG 2012; 28:276-84; PMID:22487374; http://dx.doi.org/ 10.1016/j.tig.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Waki H, Nakamura M, Yamauchi T, Wakabayashi K, Yu J, Hirose-Yotsuya L, Take K, Sun W, Iwabu M, Okada-Iwabu M, et al. . Global mapping of cell type-specific open chromatin by FAIRE-seq reveals the regulatory role of the NFI family in adipocyte differentiation. PLoS Genet 2011; 7:e1002311; PMID:22028663; http://dx.doi.org/ 10.1371/journal.pgen.1002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. . Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008; 133:1106-17; PMID:18555785; http://dx.doi.org/ 10.1016/j.cell.2008.04.043 [DOI] [PubMed] [Google Scholar]
- 32. Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 2009; 462:65-70; PMID:19890324; http://dx.doi.org/ 10.1038/nature08531 [DOI] [PubMed] [Google Scholar]
- 33. Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. . ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 2009; 457:854-8; PMID:19212405; http://dx.doi.org/ 10.1038/nature07730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5PCAF-mediated H3K9ac and CBPp300-mediated H3K1827ac in nuclear receptor transactivation. EMBO J 2011; 30:249-62; PMID:21131905; http://dx.doi.org/ 10.1038/emboj.2010.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2011; 143:156-69; PMID:20887899; http://dx.doi.org/ 10.1016/j.cell.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet 2011; 12:283-93; PMID:21358745; http://dx.doi.org/ 10.1038/nrg2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Visel A, Minovitsky S, Dubchak I, Pennacchio LA. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res 2007; 35:D88-92; PMID:17130149; http://dx.doi.org/ 10.1093/nar/gkl822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xie W, Song C, Young NL, Sperling AS, Xu F, Sridharan R, Conway AE, Garcia BA, Plath K, Clark AT, et al. . Histone h3 lysine 56 acetylation is linked to the core transcriptional network in human embryonic stem cells. Mol Cell 2009; 33:417-27; PMID:19250903; http://dx.doi.org/ 10.1016/j.molcel.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc 2007; 2:1445-57; PMID:17545981; http://dx.doi.org/ 10.1038/nprot.2007.202 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Q, Ramlee MK, Brunmeir R, Villanueva CJ, Halperin D, Xu F. Dynamic and distinct histone modifications modulate the expression of key adipogenesis regulatory genes. Cell Cycle 2012; 11:4310-22; PMID:23085542; http://dx.doi.org/ 10.4161/cc.22224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rikitake Y, Moran E. DNA-binding properties of the E1A-associated 300-kilodalton protein. Mol Cell Biol 1992; 12:2826-36; PMID:1534143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hauser S, Adelmant G, Sarraf P, Wright HM, Mueller E, Spiegelman BM. Degradation of the peroxisome proliferator-activated receptor gamma is linked to ligand-dependent activation. J Biol Chem 2000; 275:18527-33; PMID:10748014; http://dx.doi.org/ 10.1074/jbc.M001297200 [DOI] [PubMed] [Google Scholar]
- 43. Laborda J. 36B4 cDNA used as an estradiol-independent mRNA control is the cDNA for human acidic ribosomal phosphoprotein PO. Nucleic Acids Res 1991; 19:3998; PMID:1861990; http://dx.doi.org/ 10.1093/nar/19.14.3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


