Key Clinical Message
Intrachromosomal amplification of chromosome 21 (iAMP21) defines a distinct cytogenetic subgroup of B-cell precursor acute lymphoblastic leukemia (BCP-ALL) with poor prognosis that should be investigated in routine practice. Single-nucleotide polymorphism (SNP)-array provides a useful method to detect such cases showing a highly characteristic profile.
Keywords: BCP-ALL, cytogenetics, iAMP21, RUNX1, SNP-array
Images in Hematology
We report the case of a 7-year-old girl with no previous medical history, presenting in our department with fever, fatigue, weakness, bruising, and tumor syndrome (lymphadenopathy and hepatosplenomegaly). A total blood count showed: leukocytes 29 × 109/L (including neutrophils 3%, lymphocytes 17%, and blasts 80%), hemoglobin concentration 82 g/L, and platelet count 63 × 109/L. The bone marrow aspirate was of high cellularity with a decreased number of megakaryocytes and 97% peroxidase-negative staining blast cells. Immunophenotyping confirmed the diagnosis of B-cell precursor acute lymphoblastic leukemia (BCP-ALL) with expression of CD34, HLA-DR, and B-lymphoid markers CD19, CD10, CD20, and CD22. T-lymphoid markers were absents. Blasts also aberrantly expressed the myeloid markers CD33 and CD117. Molecular studies did not show any rearrangement usually observed in BCP-ALL (ETV6-RUNX1 TCF3-PBX1 BCR-ABL1, or KMT2A fusions). Cytogenetics analyses revealed a female karyotype with 46 chromosomes including a grossly abnormal chromosome supposed to derive from chromosome 21 in all metaphases (Fig.1A). All other chromosomes appeared normal. Interphase fluorescent in situ hybridization (FISH) using RUNX1/RUNX1T1 probe (Vysis®) was performed (Fig.1B) and revealed extra signals of RUNX1 clustered together. The same analysis performed on metaphases (Fig.1C) showed RUNX1 extra copies all along the abnormal chromosome 21, leading to the identification of intrachromosomal amplification of chromosome 21 (iAMP21). Single-nucleotide polymorphism (SNP)-array was performed on the same sample using Affymetrix® Cytoscan High Density Array. Analysis revealed only few abnormalities. Leukemic cells were characterized by small deletions targeting ETV6 (location: 12p13, size: 242 kb), RB1 (location: 13q14; size: 98 kb), and BTLA genes (location: 3q13; size: 164 kb) without IKZF1 deletion, and a highly rearranged chromosome 21 with multiple regions of gain, amplification, and deletion (Fig.1D). Notably, the highest level of amplification spanned the RUNX1 locus with a copy number of eight in accordance with the FISH analysis. The patient was enrolled in the very high-risk (VHR) group of the EORTC 58081 protocol and achieved complete remission with complete molecular response.
Figure 1.
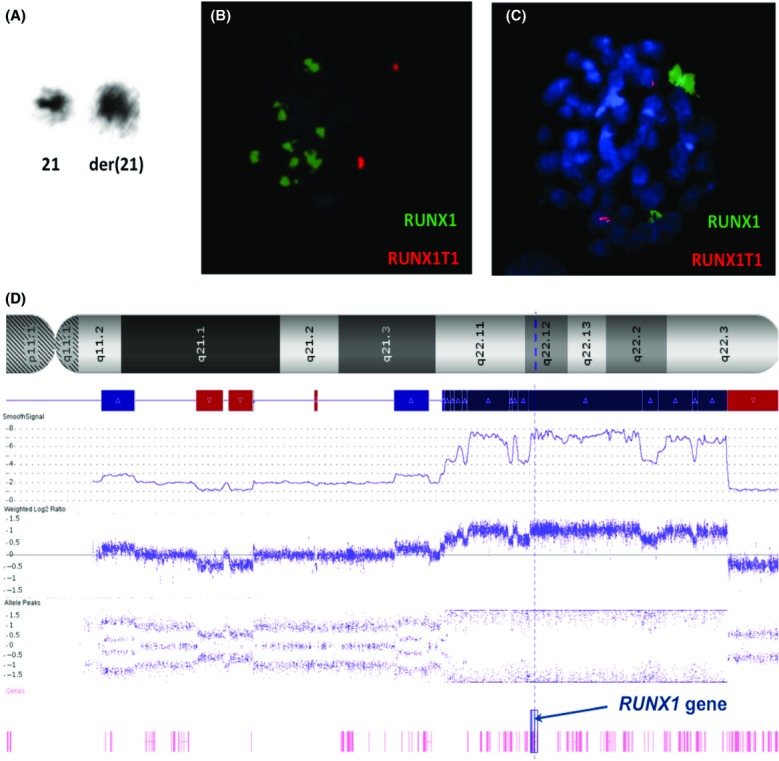
Cytogenetics study of bone marrow sample at BCP-ALL diagnosis. Karyotype was: 46,XX,der(21) [20] and displayed a grossly abnormal derivative chromosome 21 (A). FISH analysis on interphase (B) and metaphase (C) using a probe specific to RUNX1 (green signals) on chromosome 21q22 confirmed the amplification of RUNX1 with a median of 6 extra copies and implication of the derivative chromosome 21. SNP-array revealed complex copy number variations with the highest level of amplification located within the RUNX1 locus (screenshot from Chromosome Analysis® suite). Copy number (referred to SmoothSignal) is 2 for normal situation, <2 for loss (red boxes), and >2 for gain/amplification (blue/dark blue boxes) (D).
iAMP21 is now considered as a distinct cytogenetic subgroup of BCP-ALL which has been confirmed to be a primary genetic event 1,2. This abnormality has been reported in about 2% of childhood BCP-ALL with a poor event-free survival rate when treated with standard chemotherapy, which justifies to assign such patients in the very high-risk group and treat them intensively 3. Despite its strong prognostic association, iAMP21 is not systematically investigated in routine practice and its detection may be difficult by conventional cytogenetics. Especially, the morphology of the abnormal chromosome 21 is highly heterogeneous between patients. FISH analysis provides an efficient detection method to evaluate the number of copies of RUNX1. Finding three or more extra copies of RUNX1 is very suggestive of iAMP21. However, interpretation of cases with FISH analysis on interphase alone should be made with caution, since RUNX1 extra copies could also be associated with hyperdiploid BCP-ALL. In this context, SNP-array is of great interest as no cell culture is needed, and it provides a distinctive profile of iAMP21, with the highest level amplification of a region spanning RUNX1 and the lowest level of copy number at the telomeric region of the chromosome 21. Recent findings have highlighted the mechanism giving rise to iAMP21. The abnormality occurs nonrandomly as its formation starts by telomere attrition initiating breakage-fusion-bridge cycles that explains highly amplified regions juxtaposed to a deleted telomeric region 4,5. Overall, looking for iAMP21 should be performed in routine practice in BCP-ALL patients, especially in cases without recurrent cytogenetic abnormalities, or when an abnormal chromosome 21 is suspected or when a small marker chromosome of undetermined origin is found at karyotype, particularly in older children (median age of 9 years) 4 or poor responders to standard chemotherapy. In this context, SNP-array is a useful method to confirm the accuracy of iAMP21 diagnosis with its pathognomonic profile. Moreover, its use is of increasing importance to refine risk stratification in pediatric BCP-ALL 6.
Conflict of Interest
None declared.
References
- Soulier J, Trakhtenbrot L, Najfeld V, Lipton JM, Mathew S, Avet-Loiseau H, et al. Amplification of band q22 of chromosome 21, including AML1, in older children with acute lymphoblastic leukemia: an emerging molecular cytogenetic subgroup. Leukemia. 2003;17:1679–1682. doi: 10.1038/sj.leu.2403000. [DOI] [PubMed] [Google Scholar]
- Rand V, Parker H, Russell LJ, Schwab C, Ensor H, Irving J, et al. Genomic characterization implicates iAMP21 as a likely primary genetic event in childhood B-cell precursor acute lymphoblastic leukemia. Blood. 2011;117:6848–6855. doi: 10.1182/blood-2011-01-329961. [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Moorman AV, Schwab C, Carroll AJ, Raetz EA, Devidas M, et al. An international study of intrachromosomal amplification of chromosome 21 (iAMP21): cytogenetic characterization and outcome. Leukemia. 2014;28:1015–1021. doi: 10.1038/leu.2013.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ. Blood Spotlight on iAMP21 acute lymphoblastic leukemia (ALL), a high-risk pediatric disease. Blood. 2015;125:1383–1386. doi: 10.1182/blood-2014-08-569228. [DOI] [PubMed] [Google Scholar]
- Li Y, Schwab C, Ryan SL, Papaemmanuil E, Robinson HM, Jacobs P, et al. Constitutional and somatic rearrangement of chromosome 21 in acute lymphoblastic leukaemia. Nature. 2014;508:98–102. doi: 10.1038/nature13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman AV, Enshaei A, Schwab C, Wade R, Chilton L, Elliott A, et al. A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood. 2014;124:1434–1444. doi: 10.1182/blood-2014-03-562918. [DOI] [PubMed] [Google Scholar]


