Key Clinical message
Case report involving a normal female by NIPT with male external genitalia on routine fetal morphology assessment. QF-PCR, CGH microarray, and FISH revealed an unbalanced translocation, involving the short arms of the X and Y chromosomes. This case demonstrates the possible limitations of correctly identifying sex chromosome abnormalities via NIPT.
Keywords: CGH microarray, noninvasive prenatal testing (NIPT), sex discordance
Introduction
Sex discordance between prenatal cytogenetic results and ultrasound findings is not common, but gender discordance is observed in approximately 1/2500 pregnancies 1. Now with the widespread uptake of noninvasive prenatal testing (NIPT), a larger cohort of women has access to fetal DNA information. Previously this information was typically only available to women undertaking an invasive prenatal procedure 2, so the potential for identifying prenatal sex discordance is likely to increase.
Clinical Report
This case involves a 40-year-old Caucasian woman with an appropriately developed 2-year-old son following an uncomplicated first pregnancy, during which she undertook combined first trimester screening but no invasive testing. There was no family history of sex discordance. In the index (second) pregnancy, she underwent NIPT (Harmony™ Prenatal Test, Ariosa Diagnostics, San Jose, CA, USA) at 10 weeks and 4 days’ gestation on the basis of advanced maternal age. The Harmony test result indicated a risk of <1 in 10,000 for the common trisomies and the fetal sex was reported as female. Sex chromosome analysis indicated a probability of XX above 99%. The fetal fraction was reported at 17.7%.
Fetal anatomy and nuchal translucency assessment in this case showed no abnormality at 12 weeks and 5 days’ gestation, but the protocol does not include imaging the genitalia during this examination. The fetus again showed no structural anomalies at the routine 19-week morphology examination although, male external genitalia was identified. Following a discussion with a maternal-fetal medicine specialist, further investigations were instigated, which included amniocentesis at 20 weeks and 4 days’ gestation, with 25 mL of clear amniotic fluid collected following a single-needle insertion that did not traverse the placenta, which was sent for Quantitative fluorescent polymerase chain reaction (QF-PCR) analysis for chromosomes 13, 18, 21, X and Y and CGH microarray studies.
Materials and Methods
High-molecular weight DNA was extracted from 15 mL of amniotic fluid, using the Promega Wizard Genomic DNA Purification Kit (Madison, WI, USA). A reserve cell culture was established from the remaining sample following standard laboratory protocols. QF-PCR for rapid aneuploidy detection was performed, using the Devyser Compact v3 QF-PCR kit 3. The sample was then processed on a targeted version of the Agilent ISCA 8 × 60k design (SUFWprenatalArray) against a normal male control and slides were scanned using a DNA microarray scanner with Surescan High Resolution Technology (Agilent, Santa Clara, USA) as previously described 4. Aberration detection was performed by Agilent CytoGenomics 2.0.6 software. All results were reported according to GRCh37 (hg19) assembly. Metaphase fluorescence in situ hybridization (FISH) testing was performed on the reserve cultured amniocytes.
Results
QF-PCR showed a profile consistent with a fetus disomic for 13, 18, 21, and X with a partial Y chromosome present. The marker for SRY was present, and the DXY267 (pseudoautosomal marker, present on both X and Y chromosomes) showed a triallelic peak of which only one allele was identified in the maternal profile, as depicted in Fig.1.
Figure 1.
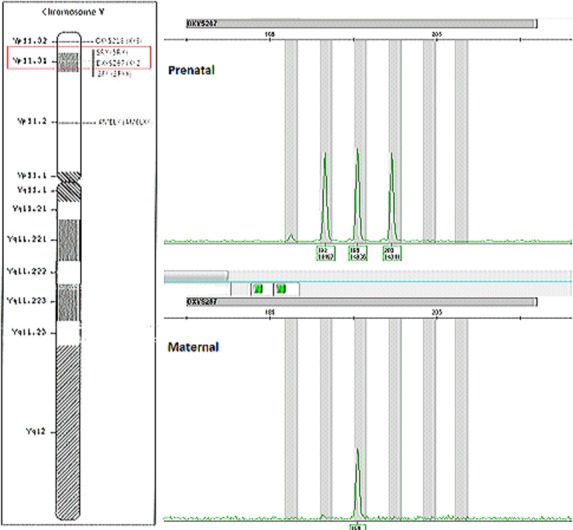
Graphical representation of the Y chromosome using QF-PCR. SRY was present and DXY267 (pseudoautosomal marker XY2) showed three alleles present. DXY218 (pseudoautosomal marker XY3) showed two alleles while amelogenin showed only a single allele and the ZFY allele was not present (not pictured).
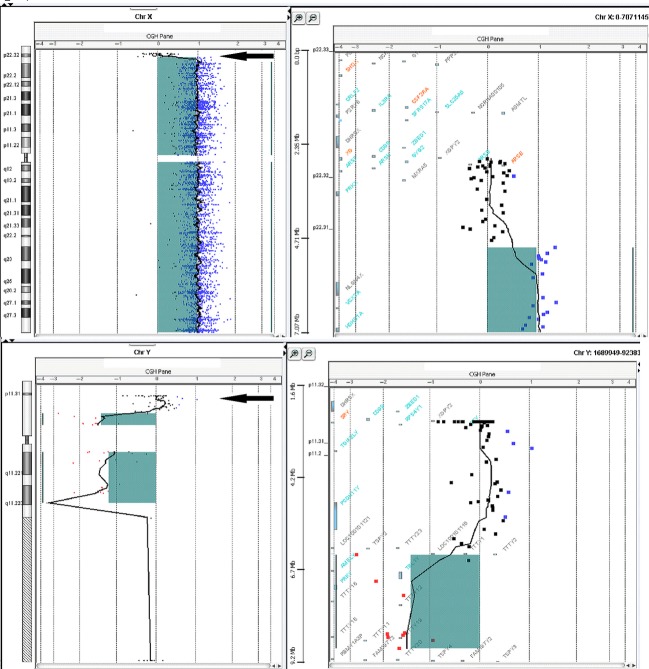
The CGH microarray result showed a normal +1 log ratio along the majority of the X chromosome, but a log ratio of 0 from Xp22.33 to Xp22.32 indicated a copy number loss of 2.2 Mb (from 2 copies to 1). The Y chromosome showed a log ratio of −1 along the majority of the Y chromosome, but a log ratio of 0 from Yp11.31 to Yp11.2, indicated a copy number gain of 3.7 Mb (from 0 copies to 1). The SRY region was included in this gain. Graphical representation of these changes is depicted in Fig.2.
Figure 2.
CGH microarray images of X and Y chromosomes. Log ratio of 0 from Xp22.33 to Xp22.32 indicate a copy number loss of 2.2 Mb. Log ratio of 0 from Yp11.31 to Yp11.2, indicate a copy number gain of 3.7 Mb.
An unbalanced translocation involving the short arms of the X and Y chromosomes was suggested by the CGH microarray and FISH testing confirmed translocation of the additional Y material onto the Xp telomere, as depicted in Fig.3.
Figure 3.
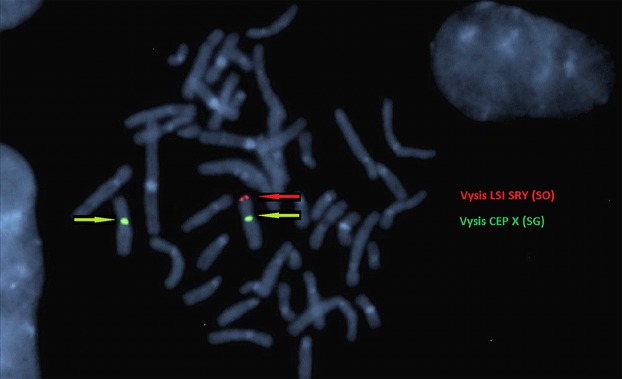
A representative metaphase showing FISH with LSI SRY Spectrum Orange (SO) CEP X Spectrum Green (SG) probes [Vysis, Abbott Molecular, Abbott Park IL, USA]. Positive SRY signal indicated by the red arrow is found at the terminal p arm of the derivative X. The centromeres of both X chromosomes are shown by the green arrows (image courtesy of South Eastern Area Laboratory Services).
Paternal cytogenetic and FISH studies were recommended, but declined, and the pregnancy was ongoing.
Discussion
46,XX males are rare, with a reported incidence of 1:20,000 newborn males. In the majority of cases, the Y chromosome (including the SRY region in 90% of cases) is translocated onto the X chromosome as a result of recombination during paternal meiosis 5. The phenotype of 46,XX males can vary from normal to ambiguous genitalia and is now referred to as testicular disorders of sex development (DSD) 6 The phenotypic variation is thought to be related to the amount of Y chromosome translocated and also to the skewed inactivation of the derivative (X:Y) chromosome 7. A larger fragment of Y translocated material could potentially protect the derivative X from inactivation, allowing a normal male phenotype, whilst those with a smaller fragment of Y material are more likely to exhibit genital ambiguity 8. Most cases of 46,XX SRY positive males are diagnosed in late adolescence or adulthood through cytogenetic studies investigating infertility and/or small testes 5.
Recently, there has been a case reported with a similar unbalanced translocation involving the short arms of the X and Y chromosomes, detected prenatally in a patient who underwent invasive testing for advanced maternal age. Although the cytogenetic result was reported as a normal 46,XX karyotype, the QF-PCR and FISH profiles were concordant with our case 6. Without fortuitous definitive prenatal testing, the sex discordance would have gone unidentified until puberty or later in this instance. In the current case, discordance between the female NIPT gender result and the phenotypically male features on ultrasound prompted invasive testing for clarification. As the uptake of NIPT increases across the obstetric population, more gender discordance will be identified prenatally.
When the SRY gene is present, normal external genitalia and masculinization is usually observed as the SRY is considered to play a crucial role in the differentiation of testes in early embryonic life 9. However, with the absence of the spermatogenic genes on Yq (AZFa, AZFb, and AZFc) azoospermia is possible 8.
Ariosa Diagnostics uses a novel assay (digital analysis of selected chromosome regions: DANSR) to determine trisomy 21, 18 10, 13 11, and sex chromosome aneuploidy 12 risk for each patient. This assay is designed to evaluate specific genomic fragments best able to discriminate aneuploid from euploid samples but only sequences selected loci on the chromosomes of interest, rather than whole-genome sequencing utilized during the massively parallel sequencing (MPS) method 13. The DANSR method decreases the amount of raw sequencing reads per sample by tenfold compared to MPS, which leads to a decrease in cost and improved efficiency 10.
Due to the low guanosine-cytosine (CG) content of the X chromosome, MPS is subject to highly variable amplification. A study of sex chromosome aneuploidy detection by DANSR assay compared to MPS 12 has attributed its higher detection rate to the selective avoidance of these regions. The resulting data are analysed using the fetal-fraction optimised risk of trisomy evaluation (FORTE) algorithm. This model considers the age-related risks and the percentage of fetal cfDNA in the sample to provide an individualized risk score for trisomy. The higher the fetal DNA fraction, the greater the difference in the number of cfDNA fragments originating from trisomic compared with disomic chromosomes 10. A sample with a calculated z-score over 3.6 is considered indicative of an aneuploidy result 14.
The DANSR assay includes 32 selected chromosome regions on the Y chromosome, and although the test does not claim to be able to detect small copy number variations, a recent study correctly identified six out of six cases XXY males 15. With almost two full X chromosomes and 3.7 Mb of additional Y chromosome (including SRY) present in this case, it is interesting that this was not reported as a 47,XXY Klinefelter syndrome result. The SRY gene is considered responsible for the determination of normal male external genitalia and masculinization but its presence alone does not seem to be sufficient for the FORTE algorithm to determine there is at least a partial Y chromosome present.
Two important points are raised by this case. Firstly, the chromosome regions selected as important when reporting on gender may need to be reviewed to reduce the potential for erroneous gender results, which may also positively impact on the rate of false-positive diagnoses of a range of sex chromosome abnormalities. Secondly, this case demonstrates the possible limitations of correctly identifying sex chromosome abnormalities other than the classical syndromes and highlights the necessity of good quality ultrasound, counseling and molecular and cytogenetic techniques, as definitive diagnostic testing is recommended for any abnormal NIPT result 16.
Conflicts of interest
None declared.
References
- Trujillo-Tiebas MJ, Gonzàlez-Gonzàlez C, Lorda-Sànchez I, Querejeta ME, Ayuso C, Ramos C. Prenatal diagnosis of 46, XX male fetus. J. Assist. Reprod. Genet. 2006;23:253–254. doi: 10.1007/s10815-005-9020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey M, Sharma S. Aggarwa S. Prenatal screening methods for aneuploidies. N. Am. J. Med. Sci. 2013;5:182–190. doi: 10.4103/1947-2714.109180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann K, Fox SP, Abbs SJ, Yau SC, Scriven PN, Docherty Z, et al. Development and implementation of a new rapid aneuploidy diagnostic service within the UK National Health Service and implications for the future of prenatal diagnosis. Lancet. 2001;358:1057–1061. doi: 10.1016/S0140-6736(01)06183-9. [DOI] [PubMed] [Google Scholar]
- Scott F, Murphy K, Carey L, Greville W, Mansfield N, Barahona P, et al. Prenatal diagnosis using combined quantitative fluorescent polymerase chain reaction and array comparative genomic hybridization analysis as a first-line test: results from over 1000 consecutive cases. Ultrasound Obstet. Gynecol. 2013;41:500–507. doi: 10.1002/uog.12429. [DOI] [PubMed] [Google Scholar]
- Anık A, Çatlı G, Abacı A. Bober E. 46.XX male disorder of sexual development: a case report. J. Clin. Res. Pediatr. Endocrinol. 2013;5:258–260. doi: 10.4274/Jcrpe.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo F, Cantalupo G, Ciavarella M, Monica MD, Lombardi C, Maioli M, et al. Prenatal diagnosis of 46, XX testicular DSD. Molecular, cytogenetic, molecular-cytogenetic, and ultrasonographic evaluation. Prenat. Diagn. 2009;29:998–1001. doi: 10.1002/pd.2329. [DOI] [PubMed] [Google Scholar]
- Gunes S, Asci R, Okten G, Atac F, Onat OE, Ogur G, et al. Two males with SRY-positive 46, XX testicular disorder of sex development. Syst. Biol. Reprod. Med. 2013;59:42–47. doi: 10.3109/19396368.2012.731624. [DOI] [PubMed] [Google Scholar]
- Wu QY, Li N, Li WW, Li TF, Zhang C, Cui YX, et al. Clinical, molecular and cytogenetic analysis of 46, XX testicular disorder of sex development with SRY-positive. BMC Urol. 2014;14:70. doi: 10.1186/1471-2490-14-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Veerapandian V. Chaudhary I. Halder A. The sertoli cell only syndrome glaucoma in a sex – determining region Y (SRY) positive XX infertile male. J. Clin. Diagn. Res. 2013;7:1457–1459. doi: 10.7860/JCDR/2013/5186.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks AB, Struble CA, Wang ET, Song K. Oliphant A. Noninvasive prenatal detection selective analysis of cell-free DNA obtained from maternal blood: evaluation for trisomy 21 and trisomy 18. Am. J. Obstet. Gynecol. 2012;206:319. doi: 10.1016/j.ajog.2012.01.030. [DOI] [PubMed] [Google Scholar]
- Ashoor G, Syngelaki A, Wang E, Struble C, Olophant A, Song K, et al. Trisomy 13 detection in the first trimester of pregnancy using a chromosome-selective cell-free DNA analysis method. Ultrasound Obstet. Gynecol. 2013;41:21–25. doi: 10.1002/uog.12299. [DOI] [PubMed] [Google Scholar]
- Nicolaides KH, Musci TJ, Struble CA, Syngelaki A. Gil MDM. Assessment of fetal sex chromosome aneuploidy using directed cell-free DNA analysis. Fetal Diagn. Ther. 2014;35:1–6. doi: 10.1159/000357198. [DOI] [PubMed] [Google Scholar]
- Mazloom AR, Zeljko D, Oeth P, Wang H, Jensen T, Tynan J, et al. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell-free DNA from maternal plasma. Prenat. Diagn. 2013;33:591–597. doi: 10.1002/pd.4127. [DOI] [PubMed] [Google Scholar]
- Sparks AB, Wang ET, Struble CA, Barrett W, Stokowski R, McBride C, et al. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat. Diagn. 2012;32:3–9. doi: 10.1002/pd.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks J, Wolfberg AJ, Wang ET, Struble CA, Zhan J, Juneau K, et al. Non-Invasive risk assessment of fetal sex chromosome aneuploidy through directed analysis and incorporation of fetal fraction. Prenat. Diagn. 2014;34:496–499. doi: 10.1002/pd.4338. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Parker L, Wentworth J, Madankumar R, Saffer C, Das AF, et al. DNA sequencing versus standard prenatal aneuploidy screening. N. Engl. J. Med. 2014;370:799–808. doi: 10.1056/NEJMoa1311037. [DOI] [PubMed] [Google Scholar]