Abstract
The MAGE (Melanoma-associated antigen) protein family members are structurally related to each other by a MAGE-homology domain comprised of 2 winged helix motifs WH/A and WH/B. This family specifically evolved in placental mammals although single homologs designated NSE3 (non-SMC element) exist in most eukaryotes. NSE3, together with its partner proteins NSE1 and NSE4 form a tight subcomplex of the structural maintenance of chromosomes SMC5–6 complex. Previously, we showed that interactions of the WH/B motif of the MAGE proteins with their NSE4/EID partners are evolutionarily conserved (including the MAGEA1-NSE4 interaction). In contrast, the interaction of the WH/A motif of NSE3 with NSE1 diverged in the MAGE paralogs. We hypothesized that the MAGE paralogs acquired new RING-finger-containing partners through their evolution and form MAGE complexes reminiscent of NSE1-NSE3-NSE4 trimers. In this work, we employed the yeast 2-hybrid system to screen a human RING-finger protein library against several MAGE baits. We identified a number of potential MAGE-RING interactions and confirmed several of them (MDM4, PCGF6, RNF166, TRAF6, TRIM8, TRIM31, TRIM41) in co-immunoprecipitation experiments. Among these MAGE-RING pairs, we chose to examine MAGEA1-TRIM31 in detail and showed that both WH/A and WH/B motifs of MAGEA1 bind to the coiled-coil domain of TRIM31 and that MAGEA1 interaction stimulates TRIM31 ubiquitin-ligase activity. In addition, TRIM31 directly binds to NSE4, suggesting the existence of a TRIM31-MAGEA1-NSE4 complex reminiscent of the NSE1-NSE3-NSE4 trimer. These results suggest that MAGEA1 functions as a co-factor of TRIM31 ubiquitin-ligase and that the TRIM31-MAGEA1-NSE4 complex may have evolved from an ancestral NSE1-NSE3-NSE4 complex.
Keywords: E3 ubiquitin ligase, MAGEA1, melanoma-associated antigen family, MDM4, NSE1-NSE3-NSE4 complex, NSE4/EID family, protein evolution, PCGF6, RING-finger proteins, RNF166, TRIM31, TRIM family, TRAF6, TRIM8, TRIM41, ubiquitination
Abbreviations
- MAGE
melanoma-associated antigen
- MHD
MAGE homology domain
- NSE
non-SMC element
- SMC
structure maintenance of chromosomes
- TRIM
tripartite motif
- WH
winged helix
- Y2H
yeast 2-hybrid.
Introduction
The SMC (Structural maintenance of chromosomes) complexes are present from bacteria to humans, demonstrating a high degree of evolutionary conservation.1 In eukaryotes, there are 3 SMC complexes in most genomes and the SMC5–6 protein complex is the most diverged among them.2 The SMC5–6 complex subunits are involved in chromatin dynamics including DNA damage repair, stabilization of stalled replication forks and transcription regulation.3-7 The core of the complex is formed by the NSE2-SMC5-SMC6 subcomplex, which associates with the NSE1-NSE3-NSE4 subcomplex (NSE, non-SMC element;8,9).
The NSE3 subunit is related to the MAGE (Melanoma-associated antigen) superfamily of proteins.10,11 In placental mammals, the MAGE superfamily contains tens of protein-coding genes (37 proteins in humans) while other organisms contain only one NSE3-coding gene.11 Most mammalian MAGE superfamily members share a MAGE homology domain (MHD) composed of 2 winged-helix (WH/A and WH/B) subdomains.12 According to their sequence conservation and tissue-specific expression, they are subdivided into 2 types which contain 10 subfamilies.13 Genes encoding type I MAGEs (A, B and C subfamilies) evolved relatively recently and they are expressed only in specific tissues (mostly in placenta, embryo, testis) and in cancer cells.14–16 Due to their specific expression in tumors and significant immunogenicity (cancer-testis specific antigens), type I MAGEs are implicated in tumorigenesis and cancer progression.17-22 Based on their immunogenicity, several cancer vaccine trials against the type I MAGE antigens are currently ongoing.23–26 The type II MAGE proteins are relatively conserved, the MAGEG1/NSE3 subunit of the SMC5–6 complex being functionally the most conserved. They are expressed ubiquitously and have roles in cell cycle withdrawal, neuronal differentiation and apoptosis.27
Despite the sequence similarities between the MAGE proteins, only MAGEG1/NSE3 is able to form the human NSE1-NSE3-NSE4 subcomplex of the SMC5–6 complex.7 Within the NSE1-NSE3-NSE4 subcomplexes, the MAGEG1/NSE3 protein binds to both NSE1 and NSE4.8,9,28 The composition and function of the other MAGE complexes is relatively poorly understood, although, we previously showed that human MAGE proteins contain a conserved hydrophobic pocket within the WH/B subdomain, which interacts with NSE4/EID proteins.7,29 The other interacting surfaces of the MAGE proteins, including the WH/A subdomain, which in MAGEG1/NSE3 binds to NSE1, diverged significantly. As the NSE1 sequence encodes a RING-finger domain28,30 and several RING-finger proteins have already been reported to interact with MAGE proteins,12,31-34 we hypothesized that the diverged WH/A surfaces may accommodate binding of RING-finger-containing partners.
To support this hypothesis, we screened a yeast 2-hybrid (Y2H) library of human RING-finger genes with representatives of MAGE genes and identified several potential RING-finger-containing partners of MAGE proteins. Among the MAGE-RING pairs, the MAGEA1-TRIM31 interaction exhibits properties seen in NSE1-NSE3-NSE4 complexes. First, MAGEA1 employs its WH/A subdomain surface for its binding to TRIM31. Second, MAGEA1 stimulates TRIM31 E3 ubiquitin-ligase activity (similar to human MAGEG1-driven stimulation of NSE1). In addition, TRIM31 binds directly to the NSE4 protein forming a tripartite TRIM31-MAGEA1-NSE4 complex, suggesting its evolutionary relationship to the ancestral NSE1-NSE3-NSE4 trimer.
Results
Yeast two-hybrid screen for RING-finger-containing partners of MAGE proteins
In our previous work, we showed that yeast as well as human NSE3/MAGEG1 proteins bind to both NSE1 and NSE4 partners within SMC5–6 complexes.7,8,35 In mammals, NSE3/MAGEG1 paralogs evolved differentially and acquired new functions within new MAGE complexes. We demonstrated that the NSE4 interactions are conserved in most human MAGE complexes while the interactions of NSE1 RING-finger subunits diverged.7 We hypothesized that the diverged MAGE complexes acquired new RING-finger-containing partners. To test this hypothesis, a Y2H library of 181 human RING-finger Gal4/AD prey clones, representing 149 distinct E3-ligase RING genes,36 were systematically combined with representative MAGE type I (MAGEA1, -B1 and -C2) and type II (MAGEE2, -F1 and -L2) Gal4/BD bait clones (Table S1). Most Gal4/BD-MAGE fusion constructs contained only the MHD domains, in order to target the screen specifically toward WH/A- and WH/B-binding partners, thereby eliminating partners that may bind to diverse N- and/or C-terminal extensions. Interactions were scored following growth of diploid yeast on media lacking histidine (+2.5 mM 3-aminotriazol, 3-AT) or adenine.
Table 1.
Summary of the MAGE-RING analysis. Most of the RING-finger proteins bind through their diverse C-termini to primarily WH/B subdomains of their MAGE partners. Significant contribution of the WH/A subdomain was visible only for MAGEG1-NSE1 and MAGEA1-TRIM31. Stimulation of ubiquitin ligase activity was observed only for MAGEG1-NSE1 and MAGEA1-TRIM31.
| binding domain | MHD subdomain | Stimulation of ubiquitination | |
|---|---|---|---|
| NSE1 | WH | WH/A | Yes |
| MDM4 | ND | WH/B | No |
| PCGF6 | C-term. | WH/B>WH/A | No |
| RNF166 | C-term. | WH/B>>WH/A | No |
| TRAF6 | C-term. | ND | ND |
| TRIM8 | C-term. | WH/B>WH/A | No |
| TRIM31 | coiled-coil | WH/A∼WH/B | Yes |
| TRIM41 | C-term. | ND | No |
Summary of binding domain analysis of the RING-finger proteins is based on Fig. 2C and Fig. S4. WH/A and WH/B (MHD subdomain) data are from Fig. 2A and Fig. S3. Ubiquitination of RING and MAGE proteins was followed during co-immunoprecipitation (Fig. 1, Figs. S1 and S2) and ubiquitination (Fig. 4 and Fig. S5) experiments. Specifically, data for MAGEG1-NSE1 are from Fig. S3 and Doyle et al.12 ND – not determined.
These targeted matrix screens identified a number of potential MAGE-RING interactions (Fig. 1A and Table S1). MAGEA1(1–309) showed high-confidence positive interactions with LRSAM1, MDM4, PCGF6, RNF166 and TRIM31. Weaker positive interactions were also identified with TRAF4, TRIM8 and UBR3. The only interacting partner found for MAGEB1(63–315) was TRIM41, while the MAGEC2(129–339) fragment showed high-confidence interactions with MNAT1, PCGF6, RNF166, TRAF4, TRIM8 and UBR3 and weaker interactions with CHFR, MARCH7, TRAF6 and UBOX5. Interestingly, the MAGEE2(87–505) fragment interacted with similar partners to MAGEC2, showing strong interactions with MNAT1, PCGF6, RNF166, TRAF6. MAGEL2(420–630) only showed high-confidence binding to PCGF6. Finally, the MAGEF1(74–260) bait was screened against only half of the RING-finger library resulting in discovery of interactions with PHF7 and TRIP. Altogether, targeted Y2H screens identified >10 new potential high-confidence RING-finger-containing partners of the MAGE proteins and >20 lower-confidence putative E3-ligase RING partners (Table S1). These results suggest that particular MAGE proteins may bind to several different RING-finger-containing proteins, thereby providing the potential to selectively regulate a range of biological processes.
Figure 1.
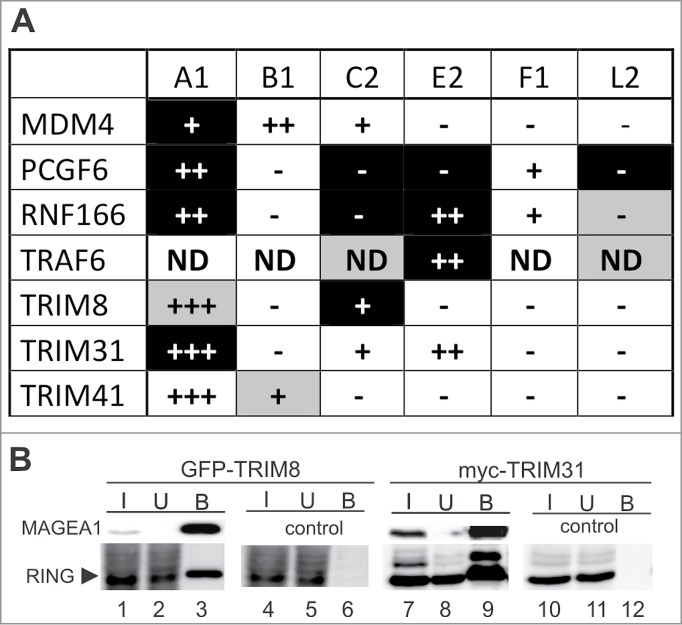
MAGE proteins interact with RING-containing proteins. (A) Summary of the MAGE-RING interaction results. MAGEA1(1–309) (A1), MAGEB1(63–315) (B1), MAGEC2(129–339) (C2), MAGEE2(87–505) (E2), MAGEF1(74–260) (F1) and/or MAGEL2(420–630) (L2) baits were used to screen Y2H library of RING-finger preys. The Y2H interactions are shown: black - high confidence Y2H interactions observed on both -His and -Ade selection; gray - low confidence interactions (from Table S1). Then, selected MAGE-RING interactions were tested using co-immunoprecipitation and pull-down assays: +++ means strong interaction; ++ means intermediate strength of interaction; + means weak interaction; - means no interaction (based on results shown in Figs. S1, S2 and S4); ND – not determined. Nine out of 14 MAGE-RING interactions detected in Y2H were confirmed. (B) Two examples of the MAGE-RING co-immunoprecipitation results. His-S-tag-MAGEA1 (lanes 1–3 and 7–9) was co-transfected with either GFP-TRIM8 (lanes 1–6) or myc-TRIM31 (lanes 7–12) into HEK293T cells. The HEK293T cell extracts were incubated with protein-S-beads and immunoprecipitated proteins were analyzed on western blots. MAGEA1 was detected using protein-S-HRP conjugate and the RING-finger-containing proteins were visualized using anti-GFP and/or anti-myc antibody, respectively. In control experiments, the pTriEx4 vector was co-transfected with either GFP-TRIM8 (lanes 4–6) or myc-TRIM31 (lanes 10–12). The input (I, 1% of lysate), unbound (U, 1% of lysate) and bound (B, 10% of lysate) fractions were separated by 12% SDS-PAGE.
Co-immunoprecipitation analysis of MAGE-RING interactions
To verify several of our highest confidence Y2H interactions, we used our panel of His-S-tagged MAGE proteins against selected GFP- or myc-tagged RING-finger proteins in systematic co-immunoprecipitation assays (2 examples in Fig. 1B; Figs. S1 and S2). Figure 1B shows that the S-tag-MAGEA1(1–309) construct is able to precipitate GFP-TRIM8 (lanes 1–3) and myc-TRIM31 (lanes 7–9) confirming our Y2H data (Fig. 1A). Furthermore, MAGEA1 was also found to bind to GFP-RNF166 (Fig. S1B, lanes 1–3), GFP-PCGF6 (Fig. S2A, lanes 1–3), GFP-TRIM41α (Fig. S2B, lanes 1–3) and weakly to myc-MDM4 (Fig. S1A, lanes 1–3). Further analysis showed that 9 out of 14 interactions identified by Y2H were also detected by co-immunoprecipitation assays (Fig. 1A; Figs. S1 and S2). Following up selected interactions (of MDM4, PCGF6, RNF166, TRAF6, TRIM8, TRIM31, TRIM41) we further analyzed and classified them according to: (1) which subdomain of the MAGE protein was involved in the interaction (Fig. 2; Fig. S3), (2) which binding domain of the RING-finger protein was involved in the interaction (Fig. 2; Fig. S4) and (3) whether MAGE binding stimulated RING-finger E3 ubiquitin-ligase activity (Fig. 4; Fig. S5). The results are summarized in Table 1 (for details see next). As the parameters of MAGEA1-TRIM31 corresponded best to the features of the conserved NSE3/MAGEG1-NSE1 interactions, we focused our detailed analysis on the MAGEA1-TRIM31 complex (see next).
Figure 2.
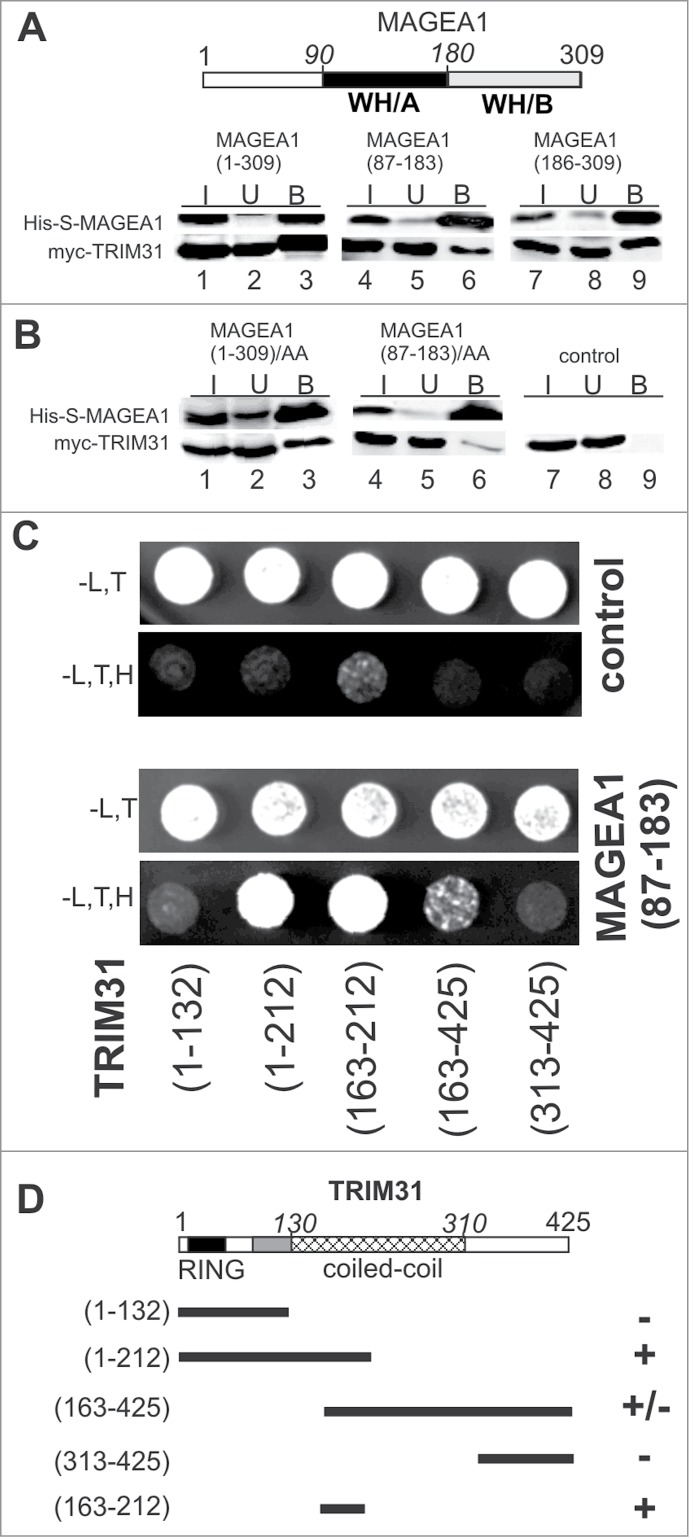
Winged-helix (WH) subdomains of MAGEA1 homology domain (MHD) interact with coiled-coil domain of TRIM31. (A) Full-length (lanes 1–3), WH/A subdomain (lanes 4–6) and WH/B subdomain (lanes 7–9) of the His-S-tag-MAGEA1 construct bind myc-TRIM31 protein in co-immunoprecipitation experiments (further details as in Fig. 1), suggesting a role of whole MHD in the MAGEA1-TRIM31 interaction. The evolutionarily conserved WH/A (black) and WH/B (gray) subdomains of MAGEA1 are indicated. (B) Mutated form (L114A, L115A; AA) of the full-length MAGEA1 (lanes 1–3) and/or its WH/A subdomain fragment (lanes 4–6) was used in co-immunoprecipitation experiments. The leucine motif mutations in the WH/A subdomain fragment almost completely abolish the interaction with TRIM31, suggesting an important role of the leucine motif of the WH/A subdomain in the MAGEA1-TRIM31 interaction. (C) Fragments of TRIM31 were fused to Gal4/BD domain, co-transformed along with Gal4/AD-MAGEA1(83–187) and analyzed by Y2H. Growth of the transformants was verified on plates without Leu and Trp (-L,T panels). Interactions were scored by growth of yeast transformants on plates without Leu, Trp and His (-L,T,H panels). The TRIM31(1–212), TRIM31(163–425) and TRIM31(163–212) coiled-coil-containing fragments bind to MAGEA1 (bottom panels). In contrast, neither the RING-B/box motif alone (aa 1–132) nor the very C-terminal fragment (aa 312–425) of TRIM31 is able to interact with MAGEA1. In control experiments (top panels), each Gal4/BD-TRIM31 construct was co-transformed with the Gal4/AD (pACT2) vector. (D) Schematic representation of the TRIM31 fragments used for the Y2H. The Y2H interactions from panel C are presented as follows: + means Y2H interaction observed on the plate without histidine; - means no interaction. These results suggest that the coiled-coil aa163–212 region of TRIM31 is both essential and sufficient for the TRIM31-MAGEA1 interaction. The subdomains of TRIM31 tripartite motif are indicated: RING-finger domain (black), B-box (gray) and coiled-coil (crosshatched).
Figure 4.
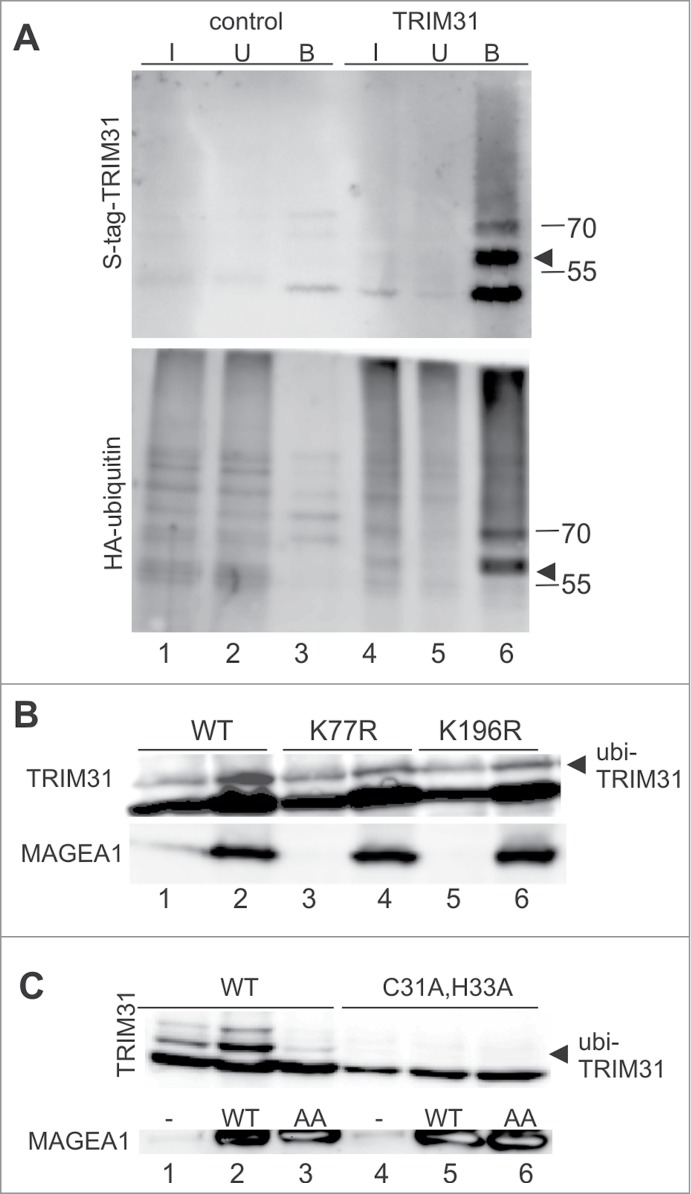
MAGEA1 enhances auto-ubiquitination activity of TRIM31 protein. (A) His-S-tag-TRIM31 (top panel, lanes 4–6) and HA-ubiquitin (bottom panel, lanes 1–6) constructs were co-transfected into HEK293T cells. The cell extracts were denatured with 0.5% SDS, 5-times diluted and precipitated on protein-S-beads (further details in Materials and Methods and in Fig. 1). Several bands are visible in the TRIM31 panel. The major TRIM31 band (∼50kDa) corresponds to His-S-tag-TRIM31 (theoretical molecular weight 53kDa) while the band above (∼60kDa, arrowhead) corresponds to the mono-ubiquitinated TRIM31 protein (bottom panel, arrowhead). The higher molecular weight species correspond to multi-ubiquitinated forms of TRIM31. (B and C) The co-expression of wild-type His-S-tag-MAGEA1 (WT) and myc-TRIM31 (WT) results in higher levels of ubiquitin-TRIM31 conjugates (top panels; compare lanes 1 and 2). (B) The K77R (lane 4) and K196R (lane 6) mutations of TRIM31 reduce MAGEA1-stimulated ubi-TRIM31 levels by 50%, suggesting that MAGEA1 stimulates TRIM31 ubiquitination on the specific lysines. (C) While coexpression of the wild-type MAGEA1 protein (WT) results in higher ubi-TRIM31 levels, the MAGEA1/L114A,L115A mutant (AA) does not increase ubi-TRIM31 levels (top panel, lane 3), suggesting WH/A-mediated MAGEA1-binding dependent mechanism of TRIM31 ubiquitination. In addition, the myc-TRIM31 mutations in the RING-finger domain (C31A, H33A) abrogate ubi-TRIM31 levels (top panel, lanes 4–6), suggesting that the ubi-TRIM31 conjugates result from TRIM31 auto-ubiquitination activity which can be stimulated by MAGEA1 binding. Arrowheads point to ubi-TRIM31 conjugates.
Analysis of MAGEA1-TRIM31 interaction
First, we compared the contribution of the MHD subdomains of the MAGE proteins with the conserved NSE3-NSE1 binding mode. In NSE3, the N-terminal winged-helix domain (WH/A subdomain) of the fission yeast Nse3 protein binds to Nse17 and similarly the WH/A surface of the human NSE3/MAGEG1 protein interacts with NSE1 (Fig. S3A, lanes 4–6;12). In our co-immunoprecipitation experiments, both MAGEA1(87–183) (WH/A) and MAGEA1(186–309) (WH/B) fragments bind TRIM31 (Fig. 2A), suggesting a contribution of both subdomains to the TRIM31 interaction. Note that the contribution of the WH/A subdomains to the other MAGE-RING interactions is much lower or missing (Table 1; Fig. S3, sections C-E, lanes 1–3).
Second, based on our fission yeast Nse3 analysis (7; Palecek, unpublished data) and the human MAGEG1-NSE1 interaction data (Fig. S3B; Ref.12 and PDB: 3NW0) we generated WH/A-specific mutations (L114A and L115A) in MAGEA1 and analyzed their binding to TRIM31 (Fig. 2B). Consistent with the above findings that both WH/A and WH/B mediate the MAGEA1-TRIM31 interaction, the L114A and L115A mutations do not significantly decrease the full-length MAGEA1 interaction (Fig. 2B, lane 3). In contrast, these mutations almost eliminate binding of the MAGEA1(87–183) fragment to TRIM31 (Fig. 2B, lane 6), suggesting that the WH/A amino acids L114 and L115 form part of the MAGEA1 interacting surface with TRIM31.
Third, an extensive analysis to find out which motif(s) of RING-finger-containing proteins bind to MHD domains reveals no conserved MAGE-binding motif but suggests binding of their variable C-terminal parts (Table 1; Fig. S4; L. Kozakova, data not shown). For the MAGEA1-TRIM31 interaction, we observe binding of the MAGEA1(87–183) construct to the TRIM31(1–212) fragment containing RING-B/box (RB) and part of the coiled-coil domain (Fig. 2C). However, the RB motif alone (aa 1–132) is not able to bind MAGEA1 in Y2H assays. The C-terminal (aa 163–425) fragment of TRIM31 weakly binds MAGEA1, while the TRIM31(313–425) fragment, missing the coiled-coil domain, does not bind MAGEA1, suggesting that the coiled-coil region of TRIM31 is essential for its interaction with MAGEA1. As the TRIM31(1–212) and TRIM31(163–425) constructs overlap in the coiled-coil region (aa 163–212), we suggest that this region may mediate the MAGEA1 interaction. Indeed, the TRIM31(163–212) construct interacts with MAGEA1 in the Y2H assays, suggesting that the coiled-coil region aa 163–212 is essential and sufficient for the binding of TRIM31 to the WH/A subdomain of MAGEA1 (Fig. 2C and D).
Human MAGEG1/NSE3 enhances ubiquitin-ligase activity of the NSE1 protein
The NSE1 proteins contain RING-finger domain homologous to typical E3 ubiquitin-ligases30,37 and are able to catalyze ubiquitin-chain formation in vitro.12,38 In our His-S-tag-MAGEG1 immunoprecipitation experiments we observed a weak band above the major MAGEG1 band when the protein was co-expressed and co-precipitated with NSE1 (Fig. 3A, lane 3). Using LC-MS/MS analysis we confirmed the presence of ubi-MAGEG1 conjugates in the band and identified ubiquitination of lysines K220 and K221 (Fig. S5A; Table S2). This band was missing when the NSE1 was co-expressed with the MAGEG1/L97A, I98A mutant protein (Fig. 3A, lane 6). As these mutations diminish the MAGEG1-NSE1 interaction (Fig. S3B;12), these results suggest that the ubiquitination of MAGEG1 is dependent on NSE1 binding. Given that human NSE1 has ubiquitin-ligase activity in vitro (12 and P. Kolesar, unpublished data), we conclude that NSE1 may ubiquitinate its MAGEG1 partner in HEK293 cells.
Figure 3.
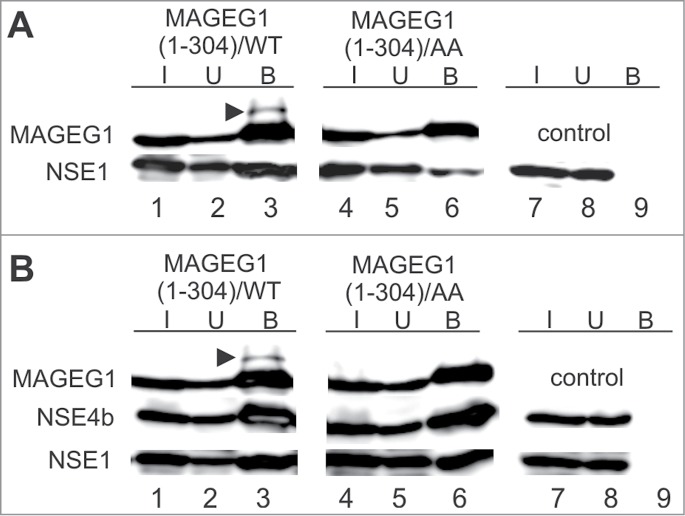
The WH/A-mediated interaction of MAGEG1 enhances the NSE1 E3-ligase activity. Either wild-type (wt, lanes 1–3) or L97A, I98A mutant (AA, lanes 4–6) of the His-S-tag-MAGEG1 construct (top panels) was co-transfected with FLAG-NSE1 alone (section A) or FLAG-NSE1 together with FLAG-NSE4b (section B) into HEK293T cells (and treated with MG132). The HEK293T cell extracts were immunoprecipitated with protein-S-beads and proteins were analyzed on western blots. In control experiments, the pTriEx4 vector (lanes 7–9) was co-transfected with NSE1 and NSE4b constructs. (A) MAGEG1 is ubiquitinated (arrowhead) in the presence of NSE1 (lane 3, top panel). The L97A, I98A (AA) mutations diminish the MAGEG1 binding to NSE1 (lane 6, bottom panel; see also Fig. S3B) and result in the disappearance of the ubi-MAGEG1 band (lane 6, top panel), suggesting the MAGEG1-NSE1 interaction-dependent ubiquitination of MAGEG1. (B) Similarly, MAGEG1 is ubiquitinated within the wt MAGEG1-NSE1-NSE4b complex (lane 3, top panel). In the presence of NSE4b, the NSE1 levels incorporated into the wild-type (lanes 1–3) and/or L97A, I98A mutant (lanes 4–6) MAGEG1-NSE1-NSE4b complexes are similar, suggesting a role of NSE4b for the stability of the complex. However, the MAGEG1/L97A, I98A protein is not ubiquitinated (lane 6, top panel) in the mutant MAGEG1-NSE1-NSE4b complex, suggesting key role of the MAGEG1 (WH/A) binding to NSE1 for its ubiquitination. Further details are in Fig. 1.
Figure 5.
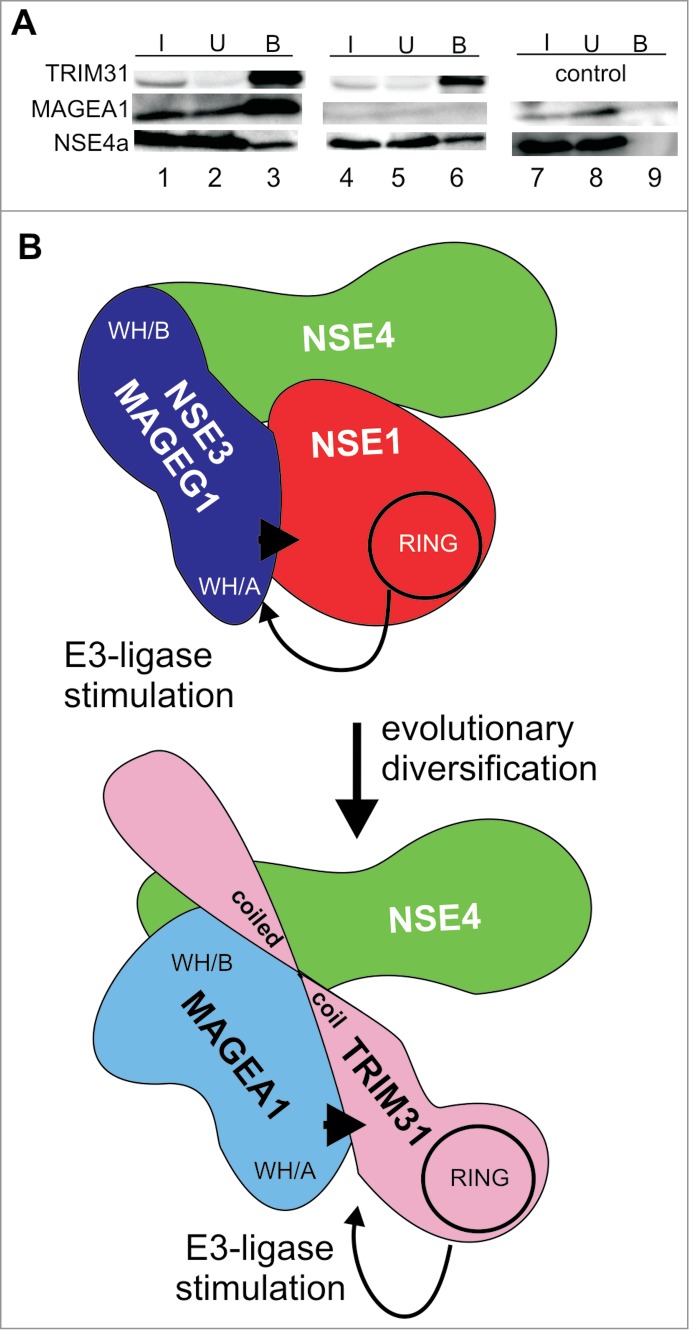
TRIM31 can form TRIM31-MAGEA1-NSE4 complexes reminiscent of NSE1-NSE3-NSE4 trimers. (A) myc-FLAG-TRIM31 (top panel, lanes 1–6), MAGEA1 (middle panel, lanes 1–3 and 7–9) and His-S-tag-NSE4a (bottom panel, lanes 1–9) constructs were co-transfected into HEK293T cells. The cell extracts were precipitated using the anti-FLAG-beads and immunoprecipitated proteins were analyzed on western blots. TRIM31 was detected using anti-myc, NSE4a was visualized using protein-S-HRP conjugate and the MAGEA1 protein was detected using anti-MAGEA1 antibody. Note that the endogenous levels of MAGEA1 in HEK293T cells are very low (lanes 4–6) compared to the levels of ectopically expressed MAGEA1 (lanes 1–3). These data suggest that TRIM31 binds directly to NSE4a and forms TRIM31-MAGEA1-NSE4a complex. (B) Cartoon comparing common features of the NSE1-NSE3-NSE4 and the TRIM31-MAGEA1-NSE4 complexes. The NSE3/MAGE proteins share conserved hydrophobic pockets (within the WH/B subdomain) interacting with NSE4/EID proteins. NSE3/MAGEG1 (WH/A) binds to NSE1 and stimulates (arrow) its RING-finger (RING) E3 ubiquitin-ligase activity. Similarly, MAGEA1 binds to TRIM31 and stimulates (arrow) its RING-finger (RING) E3 ubiquitin-ligase activity. Both, NSE3/MAGEG1 and MAGEA1, require their WH/A binding motifs for their stimulatory effects (arrowheads). As the TRIM31-MAGEA1-NSE4 complex shares common features with the yeast and human NSE1-NSE3-NSE4 complexes we propose that they may have evolved from a common ancestral NSE1-NSE3-NSE4 complex.
The ubi-MAGEG1 band is also visible when the wt NSE1-MAGEG1-NSE4b trimer is co-precipitated (Fig. 3B, lane 3). Consistent with our previous yeast trimer results,7,8 the co-expression of NSE4b with mutant MAGEG1/L97A, I98A stabilizes the human NSE1-MAGEG1-NSE4b trimer and increases NSE1 incorporation back to wild-type levels (compare lanes 3 and 6 of Fig. 3B with those of Fig. 3A). However, the NSE4b co-expression does not restore the ubi-MAGEG1 levels in the MAGEG1/L97A, I98A mutant trimer (Fig. 3B, lane 6), suggesting that even though all 3 components are present in the MAGEG1 mutant complex, specific WH/A-mediated NSE1-MAGEG1 interaction is required for the ubiquitination reaction. Consistent with published findings,12 these results suggest that MAGEG1 functions as co-factor of the NSE1 ubiquitin-ligase in vitro.
MAGEA1 enhances ubiquitin-ligase activity of the TRIM31 protein
Similarly, we looked for post-translational modifications of the RING-containing proteins during our systematic co-immunoprecipitation experiments (Fig. 1; Fig. S1). However, we were only able to detect higher molecular weight bands above TRIM31 (Fig. 1B, lane 9). To identify TRIM31 post-translational modifications, we used different analytical methods. First, we co-transfected an HA-ubiquitin construct along with His-S-tag-TRIM31 plasmid into HEK293T cells. HEK293T cell extracts were denatured (to disrupt non-covalent interactions) and then TRIM31 was precipitated using protein-S beads.Figure 4A shows that the HA-ubiquitin conjugates precipitate specifically with the TRIM31 protein and the size of the major 60kDa-band corresponds to the mono-ubiquitinated form of the TRIM31 protein (Fig. 4A, lane 6, compare top and bottom panels). Second, using LC-MS/MS analysis we identified ubiquitin-TRIM31 conjugates in His-S-tag-MAGEA1 precipitates (Table 2; Fig. S5B). In addition, we found several phosphorylation sites (Table S3) in TRIM31 and identified ubiquitin-MAGEA1 conjugates (Table 2; Table S2; Fig. S5B).
Table 2.
List of LC-MS/MS identified (TRIM31 and MAGEA1) peptides which carried the GG residues of the ubiquitin modification.60
| TRIM31 (GG position) | peptide |
|---|---|
| K77 | R.NLVEKIQALQASEVQSK.R |
| K196 | R.ILTEFELLHQVLEEEK.N |
| K230 | K.KLVDSLK.T |
| K236 | K.LVDSLK.T |
| K275 |
R.SEEFQFLNPTPVPLELEKK.L |
|
MAGEA1 (GG position) |
|
| K186 | K.AEMLESVIKNYK.H |
| K189 | K.AEMLESVIKNYK.H |
| K289 | R.KLLTQDLVQEK.K |
| K299 | K.LLTQDLVQEKYLEYR.Q |
| K337 | K.VLEYVIKVSA.R |
For detail results see Table S2.
Interestingly, the levels of mono- and di-ubiquitin-TRIM31 conjugates are enhanced upon the co-expression of MAGEA1 (Fig. 4B and C, compare lanes 1 and 2). Note that co-expression of MAGE proteins with the other RING-finger partners did not stimulate their ubiquitination (Fig. S5C; data not shown). As our mass spectrometric analysis identified several ubiquitinated TRIM31 lysine residues (Table 2), we changed these lysine residues one by one to arginine and tested these TRIM31 mutants for their MAGEA1-stimulated ubiquitination. The K77R and K196R mutations reduce the MAGEA1-stimulated TRIM31 ubiquitination levels by ∼50% while the other mutations have no effect (Fig. 4B, lanes 4 and 6; Fig. S5D), suggesting that TRIM31 is ubiquitinated specifically on K77 and K196 residues (and possibly on other as yet unidentified residues).
To test the possibility that MAGEA1 stimulates ubiquitination of TRIM31 through direct interaction, we co-expressed full-length MAGEA1/L114A, L115A mutant together with wt TRIM31 in HEK293T cells. Although these MAGEA1 mutations disturb only the binding of its WH/A subdomain, but not its WH/B binding interface, the MAGEA1/L114A, L115A mutant is not able to stimulate ubiquitination of TRIM31 (Fig. 4C, lane 3). These data suggest that the MAGEA1 interaction enhances ubiquitination of TRIM31 through its WH/A-mediated interaction.
To distinguish between direct TRIM31 auto-ubiquitination and TRIM31-ligase-independent ubiquitination, we also prepared a TRIM31 RING-finger mutant (C31A, H33A) and used it in our MAGEA1-stimulation assays.39 TRIM31 ubiquitination was abolished in the RING-finger mutant and no MAGEA1 stimulation was observed (Fig. 4C, lanes 4–6, top panel). Altogether, our data suggest that MAGEA1 stimulates TRIM31 E3 ubiquitin-ligase activity in a similar way to MAGEG1 stimulation of NSE1 i.e. through its WH/A subdomain interaction (Fig. 3 and Ref.12).
MAGEA1 forms a heterotrimer with TRIM31 and NSE4 proteins
Yeast and human NSE3/MAGEG1 proteins interact with NSE1 and NSE4 subunits forming heterotrimeric subcomplexes within the SMC5–6 complexes (Fig. 3B;7,9,35,40). We showed that the NSE3-NSE4 interaction is evolutionarily conserved in both NSE3/MAGE and NSE4/EID families.7,29 Consistent with these data, we detected endogenous NSE4a (using LC-MS/MS) in our His-S-tag-MAGEA1 precipitates from HEK293T cells (Table S2). To test if TRIM31 can form a stable trimer with NSE4a and MAGEA1, we employed a FLAG-TRIM31 construct in our co-immunoprecipitation assays. The FLAG-TRIM31 construct was co-transfected together with His-S-tag-NSE4a and untagged MAGEA1. Both NSE4a and MAGEA1 proteins were detected in bound fractions (Fig. 5A, lanes 1–3), suggesting that TRIM31 may form a TRIM31-MAGEA1-NSE4a complex.
To analyze the NSE4a binding mode, we co-transfected FLAG-TRIM31 with the NSE4a construct alone. In our co-immunoprecipitation experiment, the FLAG-TRIM31 protein precipitates NSE4a despite the absence of detectable endogenous MAGEA1, suggesting that the interaction between TRIM31 and NSE4a is direct (Fig. 5A, lanes 4–6). Indeed, we confirmed direct interactions between TRIM31 and either of the human NSE4 proteins (NSE4a and NSE4b/EID3) using in vitro pull-down experiments (Fig. S6). Altogether, human MAGEA1 is able to form TRIM31-MAGEA1-NSE4 complexes (reminiscent of the human NSE1-MAGEG1-NSE4 trimers) and stimulates the E3 ubiquitin-ligase activity of the TRIM31 subunit.
Discussion
In our previous studies, we showed that NSE1, NSE3 and NSE4 form a subcomplex within the highly conserved SMC5–6 complex and that NSE3 is homologous to the mammalian MAGE protein superfamily.7,35 Although several human MAGE family members interact with NSE4/EID proteins, they neither bind NSE1 nor form SMC5–6 complexes.7,12 As several RING-finger proteins have already been reported to interact with MAGE proteins12,31-34 we hypothesized that the evolutionarily diverged MAGE complexes may acquired alternative RING-finger-containing components, which substitute the NSE1 subunit. Therefore we screened a collection of human E3-RING-finger Y2H prey clones36 with several MAGE baits and found a number of new MAGE partners.
Because the conserved WH subdomains in NSE1 bind to human NSE3/MAGEG1,12 we looked for such structural elements in the newly identified MAGE partners. However, we found neither WH domains nor any other common MAGE-binding motif (Fig. S4). Although several MAGE partners contain coiled-coil motifs in addition to their RING-finger domains (PCGF6, TRAF6, TRIM8, TRIM31, TRIM41), most of them employ their variable C-terminal regions for binding to MAGE proteins (Figs. S2 and S4). The only exception is TRIM31, which binds to MAGEA1 protein through the coiled-coil domain (Fig. 2C and D). These results are consistent with the recent findings of Potts and collaborators,12 who found no common motif within their various MAGE-RING complexes except for the MAGEC2 interaction with the coiled-coil domain of TRIM28.
The above-mentioned variability of MAGE-partner binding modes suggests no evolutionary relation to the ancestral NSE3-NSE1 interaction for the most of our MAGE-RING interactions. In addition, most of the RING-finger proteins bind preferentially to the WH/B subdomains of the MAGE proteins, while NSE1 binds to the WH/A subdomain of NSE3/MAGEG1 (Fig. S3; L. Kozakova, unpublished data;7,12), further supporting the suggestion that their evolution is independent from that of the NSE1-NSE3-NSE4 complex. However, binding of TRIM31 to the WH/A subdomain of MAGEA1 suggests a similarity to NSE1-NSE3/MAGEG1 interaction. Similar to MAGEG1, the MAGEA1 WH/A surface (LL) motif mediates binding to TRIM31 (Fig. 2B; Refs.7,12). The additional interaction through the WH/B-subdomain might be a consequence of a different conformation of type I MAGE proteins, as already suggested for the MAGEC2-TRIM28 complex, which also employs both WH/A surface motif and WH/B subdomain for TRIM28 binding.12
Interestingly, TRIM31 binding to the WH/B subdomain of MAGEA1 could position it close to the NSE4 protein, as NSE4 also binds to the WH/B subdomain of MAGEA1.7,29 Consistent with this idea, the coiled-coil domain of TRIM31 binds to both the WH/B subdomain of MAGEA1 and the NSE4 protein (L. Vondrova, unpublished data). Importantly, binding of NSE4 to the MAGEA1-TRIM31 complex further supports our idea that the TRIM31-MAGEA1-NSE4 complexes may have evolved from the ancestral NSE1-NSE3-NSE4 complexes (Fig. 5B). Interestingly, as MAGEC2 interacts with NSE4/EID proteins, particularly with EID2,29 it is tempting to speculate that a TRIM28-MAGEC2-EID2 complex may have evolved in a similar way. In addition, TRIM27-MAGEL2-EID1 complex may also exist as EID1 associates with TRIM27/RFP.41,42 These latter assumptions are supported by the fact that the MAGEA1 MHD sequence is relatively more diverged than the MHD sequence of MAGEL2 and/or MAGEC2, respectively.13 Altogether, several findings suggest co-evolution of the TRIM, NSE3/MAGE and NSE4/EID protein families.
In the NSE1-NSE3-NSE4 complexes, binding of NSE3 (WH/A) to NSE1 enhances its ubiquitin-ligase activity (Figs. 3 and 5B; P. Kolesar, unpublished data;12). Consistent with the proposed evolution of the TRIM31-MAGEA1-NSE4 complexes, MAGEA1 stimulates the ubiquitin-ligase activity of TRIM31 (Fig. 4). Similar to the NSE3/MAGEG1 proteins, the stimulatory effect of the MAGEA1 protein is dependent on the WH/A subdomain interaction. Interestingly, the MAGEC2 binding (via WH/A) to TRIM28 enhances ubiquitination of p53,12,43 while the MAGEC2 binding (via WH/B; Fig. S3E) to MDM4/MDM2 does not increase the ubi-p53 levels (L. Kozakova and S. Uldrijan, to be published elsewhere). Similarly, MAGEA1 binding to the C-terminal part of TRIM41 does not increase its ubiquitination levels, while its binding to the coiled-coil domain of TRIM31 stimulates its auto-ubiquitination (Fig. 4; Fig. S5C). These results suggest co-evolution of the particular protein-protein interaction features of the MAGE proteins and their functional co-evolution as specific co-factors of RING-finger ligases.44
It was proposed that MAGEC2 binding to both TRIM28 E3-ligase and UbcH2 E2-conjugating enzyme facilitates increased substrate ubiquitination by enhancing physical E2-E3-substrate connections.12,45 As we were unable to detect MAGEA1 interaction with the UbcH5b E2-conjugating enzyme (39; P. Kolesar, unpublished data), we propose an alternative model for the MAGE-TRIM stimulatory effect based on the tight structural and functional relationship of the RING-finger and coiled-coil domains in TRIM proteins.46,47 Possibly, binding of MAGE to the TRIM coiled-coil domain may initiate conformational change(s), resulting in stimulation of RING-finger mediated ubiquitination activity. Consistent with this notion, MAGEA1 stimulates only TRIM31 E3-ligase while there is no detectable stimulation of TRIM8 or TRIM41, which bind MAGEA1 via their C-termini (Fig. 1B; Fig. S5C). The latter interactions may represent different interaction types. For example, MAGED1-Praja1 interaction, which directs MAGED1 to proteasome-dependent degradation,31,48 represents a substrate-ligase type of interaction rather than the stimulation-of-ligase type of interaction proposed for MAGEA1-TRIM31. In other words, different MAGE-RING interactions may have different natures, may serve different purposes and may have different evolutionary origins. Based on the MAGEA1-TRIM31 and MAGEC2-TRIM28 data, we propose the existence of a class of MAGE-TRIM complexes, in which MAGE proteins bind to coiled-coil domains of TRIM proteins and stimulate their RING-finger ligase activities. This speculative hypothesis will need to be tested in future work.
Functions of such MAGE-TRIM complexes will differ depending (at least partially) on the substrate specificities of their diverse RING-finger subunits. The NSE1-NSE3-NSE4 complexes and the MAGEC2-TRIM28 complexes are involved in DNA repair and transcription regulation5,7,12,40,43,49 while the MAGEL2-TRIM27 complexes regulate endocytosis.42 Interestingly, all 3 subunits of the TRIM31-MAGEA1-NSE4 complex are implicated in transcription regulation.7,50-52 In particular, TRIM31 overexpression inhibits HIV entry and induces AP-1 transcription activity in a RING-dependent manner, suggesting a possible role in the immune response.52,53 Given the role of MAGEA1 in immunity, we speculate that the TRIM31-MAGEA1-NSE4 complexes may function in cellular immune pathways.54
Altogether, our results provide new insights into composition and function of the TRIM31-MAGEA1-NSE4 complex and suggest its evolutionary relation to the NSE1-NSE3-NSE4 complexes.
Materials and Methods
Plasmids
All pTriEx4 plasmids described in this study were generated by PCR and ligase-independent cloning (Merck Millipore, http://www.merckmillipore.com/czech-republic/chemicals/ptriex-4-ek-lic-vector-kit/EMD_BIO-70905). The PCR primers used in this study are listed in Table S4. The other plasmids were described already in Ref.7
To prepare Y2H clones, the pTriEx4-MAGE constructs were digested with the following restriction enzymes: MAGEA1(1–309) NcoI/EcoRI, MAGEA1(87–183) NcoI/XhoI, MAGEB1(63–315) NcoI/EcoRI, MAGEC2(129–339) NcoI/XhoI, MAGEE2(87–505) NcoI/EcoRI, MAGEF1(74–260) NcoI/XhoI and MAGEL2(420–630) NcoI/EcoRI. These MAGE fragments were inserted into pGBKT7 and/or pACT2 vectors, which were predigested with the corresponding restriction enzymes.
The pCDNA3-GFP-TRIM8 plasmid was kindly provided by Dr. Germana Meroni.55 The pCMV6-entry-myc-FLAG-TRIM31 TrueORF Gold plasmid was purchased from OriGene company (http://www.origene.com/orf_clone/trueclone/NM_007028/RC201930/TRIM31.aspx, RC201930). The untagged version of the MAGEA1 was prepared by NcoI digestion of pTriEx4-MAGEA1(1–309) construct and its relegation. The TRIM31(312–425) NcoI/XhoI fragment was inserted into predigested pGBKT7 vector. The other TRIM31 fragments were amplified by PCR (fragments aa 1–212, aa 1–132, aa 163–212 and aa 163–425; see Table S4 for primers) and cloned directly into pGBKT7 vector using in-fusion system (Clontech, http://www.clontech.com/CZ/Products/Cloning_and_Competent_Cells/Cloning_Kits/Cloning_Kits-HD-Liquid).
The QuikChange Lightning Site-Directed Mutagenesis system (Agilent Technologies, http://www.chem.agilent.com/search/?Ntt=site +direct+mutagenesis+kit) was used to generate point mutations in the pTriEx4-MAGEA1, pTriEx4-MAGEG1 and pCMV6-Entry-myc-FLAG-TRIM31 plasmids. The primers are listed in Table S5. All the new clones were sequence verified.
Yeast two-hybrid assays
Construction of the human E3-RING prey Y2H clone collection in pACTBD-B or pACTBD(/E)-B was performed as previously described in.36,56 All clones were sequence verified following auto-activation tests. Targeted matrix interaction studies were performed as previously described.57 Diploid colonies were replicated onto selective plates lacking adenine or histidine (+2.5 mM 3AT) to independently test for activation of the ADE2 and HIS3 reporters.
The pGBKT7/pACT2-based Y2H system was used for further analysis of particular MAGE interaction with its RING-finger-containing partner (Clontech, http://www.clontech.com/CZ/Products/Protein_Interactions_and_Profiling/Yeast_Two-Hybrid). Each Gal4/BD-TRIM31 construct was co-transformed with Gal4/AD-MAGEA1(87–183) plasmid. In control experiments, the Gal4/BD-TRIM31 constructs were co-transformed with empty pACT2 vector. Colonies were inoculated into YPD media and cultivated overnight. 10-fold dilutions were dropped onto the SD-Leu, -Trp (control), SD-Leu, -Trp, -Ade, and/or SD-Leu, -Trp, -His (with 0, 1, 2, 5, 10, 20, 30 mM 3-aminotriazole) plates. Each combination was co-transformed at least 3 times into S. cerevisiae PJ69–4a and at least 2 independent tests were carried out from each transformation.
Co-immunoprecipitation experiments
We used co-immunoprecipitation assays based on S-tag affinity to protein-S-beads (Merck Millipore, http://www.emdmillipore.com/life-science-research/s-protein-agarose/EMD_BIO-69704). Corresponding plasmid mixes were co-transfected into HEK293T (DSMZ, https://www.dsmz.de/catalogs/details/culture/ACC-305) cells using Lipofectamine2000 (Invitrogen, http://www.lifetechnologies.com/order/catalog/product/11668019?ICID=search-product). Cells were cultivated overnight and treated with 20 μM MG132 inhibitor (Merck Millipore, http://www.millipore.com/catalog/item/474790–1mg) for 4 hours. Alternatively, cells were cultivated for 22–24 hrs without MG132 treatment. Then cells were washed with 1x PBS and lysed by scraping in lysis buffer (50 mM Tris-HCl pH7.7, 0.5% NP40, 150 mM NaCl, 1mM DTT, 1x protease inhibitor cocktail; Roche) and further incubated on ice for 15 minutes. Lysates were sonicated and cleared by centrifugation at 21,000 × g for 15 minutes/4°C.29 Alternatively, cells were lysed in 0.5% SDS, boiled for 20 minutes to denature proteins (to disrupt non-covalent interactions) and then the lysate was 5-times diluted with the lysis buffer (to get 0.1% final concentration of SDS).58 The agarose beads conjugated to protein-S were mixed with lysates and incubated for 4 hours at 4°C (alternatively, the anti-FLAG M2 antibodies conjugated to magnetic beads were used; http://www.sigmaaldrich.com/catalog/product/sigma/m8823). Beads were washed 3 times for 10 min with lysis buffer and eluted in 50μl SDS-loading buffer. The input (1% of lysate), unbound (1% of lysate) and bound (10% of lysate) fractions were analyzed by 12–15% SDS–PAGE gel electrophoresis and western blotting. The amount of S-tagged proteins was analyzed on western blots using protein-S-HRP conjugate (Merck Millipore, http://www.emdmillipore.com/life-science-research/s-protein-hrp-conjugate/EMD_BIO-69047). The other proteins were detected using either mouse anti-GFP (Roche, http://www.roche-applied-science.com/shop/en/global/products/anti-gfp), mouse anti-FLAG (Sigma, http://www.sigmaaldrich.com/catalog/product/sigma/f3165), mouse anti-myc (Cell Signaling, http://www.cellsignal.com/products/2276.html) or rabbit anti-MAGEA1 (AbCam, http://www.abcam.com/mage-1-antibody-ab21472.html) antibody. The secondary anti-mouse (Sigma, http://www.sigmaaldrich.com/catalog/search?interface=All andterm=A0168) or anti-rabbit (Abcam, http://www.abcam.com/products?keywords=97051) antibodies conjugated to HRP were applied and visualized using ECL systems (either Cell Signaling Technology, http://www.cellsignal.com/products/7003.html or Thermo Scientific, http://www.piercenet.com/product/supersignal-west-femto-chemiluminescent-substrate).
Ubiquitin-ligase stimulation assays
To follow the MAGEA1-dependent stimulation of TRIM31 ubiquitination, we co-transfected the His-S-tag-MAGEA1 and myc-TRIM31 constructs into HEK293T cells (for details see co-immunoprecipitation section above). Cell extracts were directly mixed with SDS-loading buffer. The reaction mixtures were analyzed by 12% SDS–PAGE gel electrophoresis and western blotting. The western blot signals were scanned by LAS-3000 system (Fujifilm) and the ubi-TRIM31 levels were analyzed using Multi Gauge software (Fujifilm).
Mass spectrometry analysis
Samples from co-immunoprecipitation experiments were separated by 15% SDS-PAGE gel electrophoresis. Either selected protein bands or gel regions were excised manually. After destaining and alkylation, each piece of gel was incubated with trypsin (Promega, http://worldwide.promega.com/search-results/?q=V5111) and extracted as described in.59 Resulting peptides were injected to LC-MS/MS system (RSLCnano connected on line to Orbitrap Elite; Thermo Fisher Scientific). MS and HCD MS/MS data were acquired under high resolution mode (60000 and 15000, respectively). The mass spectrometric data were processed by the Proteome Discoverer software (version 1.4, Thermo Fisher Scientific) with in-house Mascot (version 2.4.1, Matrix Science). Ubiquitination (as GlyGly or LeuArgGlyGly remnants on K;60), phosphorylation (S, T and Y), oxidation (M) and deamidation (N) as optional modifications, carbamidomethylation of C as fixed modification and 3 enzyme miscleavages were set for all searches. Post-processing of database-search results was done by Percolator and only peptides with false discovery rate < 1% (q-value 0.01), rank 1 and with at least 6 amino acids were considered (further details in Supplementary Material section).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. R. Hanai and Dr. G. Meroni for providing us with their constructs. We are grateful to Prof. K. Vousden, Dr. B. Vojtesek and Dr. P. Muller for plasmids, anti-HA antibody and advices on ubiquitination assays.
Funding
Construction of E3-RING prey clones and the initial targeted Y2H studies were performed in CS lab (Wellcome Trust, GRANT: 080911/2/06/2). Early MAGE studies have been funded in ARL lab (MRC Grant G0501450). The major part of this work was supported by a grant to JP from the Grant Agency of the Czech Academy of Sciences (IAA501630902). The final steps were accomplished with support from the Czech Science Foundation (project GA13–00774S, GA14–12166S and GAP206–12-G151). The following institutional support is acknowledged: CEITEC (Central European Institute of Technology, CZ.1.05/1.1.00/02.0068) funded from the European Regional Development Fund and funding from the European Social Funds (CZ.1.07/2.3.00/20.0189 and CZ.1.07/2.3.00/30.0037).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 2006; 7:311-22; PMID:16633335; http://dx.doi.org/ 10.1038/nrm1909 [DOI] [PubMed] [Google Scholar]
- 2. Cobbe N, Heck MM. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol Biol Evol 2004; 21:332-47; PMID:14660695; http://dx.doi.org/ 10.1093/molbev/msh023 [DOI] [PubMed] [Google Scholar]
- 3. Lehmann AR. The role of SMC proteins in the responses to DNA damage. DNA Repair 2005; 4:309-14; PMID:15661654; http://dx.doi.org/ 10.1016/j.dnarep.2004.07.009 [DOI] [PubMed] [Google Scholar]
- 4. Murray JM, Carr AM. Smc5/6: a link between DNA repair and unidirectional replication? Nat Rev Mol Cell Biol 2007; 9:177-82; PMID:18059412; http://dx.doi.org/ 10.1038/nrm2309 [DOI] [PubMed] [Google Scholar]
- 5. Lopez-Sanchez N, Gonzalez-Fernandez Z, Niinobe M, Yoshikawa K, Frade JM. A single mage gene in the chicken genome encodes CMAGE, a protein with functional similarities to mammalian type II MAGE proteins. Physiol Genomics 2007; 30:156-71; PMID:17374844; http://dx.doi.org/ 10.1152/physiolgenomics.00249.2006 [DOI] [PubMed] [Google Scholar]
- 6. De Piccoli G, Torres-Rosell J, Aragon L. The unnamed complex: what do we know about smc5-smc6? Chromosome Res 2009; 17:251-63; PMID:19308705; http://dx.doi.org/ 10.1007/s10577-008-9016-8 [DOI] [PubMed] [Google Scholar]
- 7. Hudson JJR, Bednarova K, Kozakova L, Liao C, Guerineau M, Colnaghi R, Vidot S, Marke J, Bathula SR, Lehmann AR, et al. Interactions between the Nse3 and Nse4 components of the SMC5-6 complex identify evolutionarily conserved interactions between MAGE and EID families. PLoS One 2011; 6:e17270; PMID:21364888; http://dx.doi.org/ 10.1371/journal.pone.0017270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Palecek J, Vidot S, Feng M, Doherty AJ, Lehmann AR. The SMC5-6 DNA repair complex: Bridging of the SMC5-6 heads by the kleisin, NSE4, and non-kleisin subunits. J Biol Chem 2006; 281:36952-9; PMID:17005570; http://dx.doi.org/ 10.1074/jbc.M608004200 [DOI] [PubMed] [Google Scholar]
- 9. Duan X, Yang Y, Chen YH, Arenz J, Rangi GK, Zhao X, Ye H. Architecture of the smc5/6 complex of saccharomyces cerevisiae reveals a unique interaction between the nse5-6 subcomplex and the hinge regions of smc5 and smc6. J Biol Chem 2009; 284:8507-15; PMID:19141609; http://dx.doi.org/ 10.1074/jbc.M809139200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res 2001; 61:5544-51; PMID:11454705 [PubMed] [Google Scholar]
- 11. Zhao Q, Caballero OL, Simpson AJ, Strausberg RL. Differential evolution of MAGE genes based on expression pattern and selection pressure. PLoS One 2012; 7:e48240; PMID:23133577; http://dx.doi.org/ 10.1371/journal.pone.0048240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell 2010; 39:963-74; PMID:20864041; http://dx.doi.org/ 10.1016/j.molcel.2010.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katsura Y, Satta Y. Evolutionary history of the cancer immunity antigen MAGE gene family. PLoS One 2011; 6:e20365; PMID:21695252; http://dx.doi.org/ 10.1371/journal.pone.0020365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 2005; 5:615-25; PMID:16034368; http://dx.doi.org/ 10.1038/nrc1669 [DOI] [PubMed] [Google Scholar]
- 15. Hofmann O, Caballero OL, Stevenson BJ, Chen YT, Cohen T, Chua R, Maher CA, Panji S, Schaefer U, Kruger A, et al. Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci U S A 2008; 105:20422-7; PMID:19088187; http://dx.doi.org/ 10.1073/pnas.0810777105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lifantseva N, Koltsova A, Krylova T, Yakovleva T, Poljanskaya G, Gordeeva O. Expression patterns of cancer-testis antigens in human embryonic stem cells and their cell derivatives indicate lineage tracks. Stem Cells Int 2011; -; PMID:21785609; http://dx.doi.org/ 10.4061/2011/795239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim J, Reber HA, Hines OJ, Kazanjian KK, Tran A, Ye X, Amersi FF, Martinez SR, Dry SM, Bilchik AJ, et al. The clinical significance of MAGEA3 expression in pancreatic cancer. Int J Cancer 2006; 118:2269-75; PMID:16331618; http://dx.doi.org/ 10.1002/ijc.21656 [DOI] [PubMed] [Google Scholar]
- 18. Gure AO, Chua R, Williamson B, Gonen M, Ferrera CA, Gnjatic S, Ritter G, Simpson AJ, Chen YT, Old LJ, et al. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin Cancer Res 2005; 11:8055-62; PMID:16299236; http://dx.doi.org/ 10.1158/1078-0432.CCR-05-1203 [DOI] [PubMed] [Google Scholar]
- 19. Atanackovic D, Luetkens T, Hildebrandt Y, Arfsten J, Bartels K, Horn C, Stahl T, Cao Y, Zander AR, Bokemeyer C, et al. Longitudinal analysis and prognostic effect of cancer-testis antigen expression in multiple myeloma. Clin Cancer Res 2009; 15:1343-52; PMID:19190130; http://dx.doi.org/ 10.1158/1078-0432.CCR-08-0989 [DOI] [PubMed] [Google Scholar]
- 20. Shantha Kumara HM, Grieco MJ, Caballero OL, Su T, Ahmed A, Ritter E, Gnjatic S, Cekic V, Old LJ, Simpson AJ, et al. MAGE-A3 is highly expressed in a subset of colorectal cancer patients. Cancer Immun 2012; 12:16-24; PMID:23390371 [PMC free article] [PubMed] [Google Scholar]
- 21. Svobodova S, Browning J, MacGregor D, Pollara G, Scolyer RA, Murali R, Thompson JF, Deb S, Azad A, Davis ID, et al. Cancer-testis antigen expression in primary cutaneous melanoma has independent prognostic value comparable to that of breslow thickness, ulceration and mitotic rate. Europ J Cancer 2011; 47:460-9; PMID:21115342; http://dx.doi.org/ 10.1016/j.ejca.2010.09.042 [DOI] [PubMed] [Google Scholar]
- 22. Su S, Minges JT, Grossman G, Blackwelder AJ, Mohler JL, Wilson EM. Proto-oncogene activity of melanoma antigen-A11 (MAGE-A11) regulates retinoblastoma-related p107 and E2F1 proteins. J Biol Chem 2013; 288:24809-24; PMID:23853093; http://dx.doi.org/ 10.1074/jbc.M113.468579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lonchay C, van der Bruggen P, Connerotte T, Hanagiri T, Coulie P, Colau D, Lucas S, Van Pel A, Thielemans K, van Baren N, et al. Correlation between tumor regression and T cell responses in melanoma patients vaccinated with a MAGE antigen. Proc Natl Acad Sci U S A 2004; 101:14631-8; PMID:15452345; http://dx.doi.org/ 10.1073/pnas.0405743101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci 2009; 100:2014-21; PMID:19719775; http://dx.doi.org/ 10.1111/j.1349-7006.2009.01303.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sang M, Wang L, Ding C, Zhou X, Wang B, Lian Y, Shan B. Melanoma-associated antigen genes - an update. Cancer Lett 2011; 302:85-90; PMID:21093980; http://dx.doi.org/ 10.1016/j.canlet.2010.10.021 [DOI] [PubMed] [Google Scholar]
- 26. Caballero OL, Simpson AJ, Neville AM. Vaccines and early breast cancer. Cell Cycle 2014; 13;PMID:24699824; http://dx.doi.org/ 10.4161/cc.28747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barker PA, Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J Neurosci Res 2002; 67:705-12; PMID:11891783; http://dx.doi.org/ 10.1002/jnr.10160 [DOI] [PubMed] [Google Scholar]
- 28. Pebernard S, Perry JJ, Tainer JA, Boddy MN. Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability. Mol Biol Cell 2008; 19:4099-109; PMID:18667531; http://dx.doi.org/ 10.1091/mbc.E08-02-0226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guerineau M, Kriz Z, Kozakova L, Bednarova K, Janos P, Palecek J. Analysis of the nse3/MAGE-binding domain of the nse4/EID family proteins. PLoS One 2012; 7:e35813; PMID:22536443; http://dx.doi.org/ 10.1371/journal.pone.0035813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fujioka Y, Kimata Y, Nomaguchi K, Watanabe K, Kohno K. Identification of a novel non-SMC component of the SMC5/SMC6 complex involved in DNA repair. J Biol Chem 2002; 277:21585-91; PMID:11927594; http://dx.doi.org/ 10.1074/jbc.M201523200 [DOI] [PubMed] [Google Scholar]
- 31. Sasaki A, Masuda Y, Iwai K, Ikeda K, Watanabe K. A RING finger protein praja1 regulates dlx5-dependent transcription through its ubiquitin ligase activity for the dlx/Msx-interacting MAGE/necdin family protein, dlxin-1. J Biol Chem 2002; 277:22541-6; PMID:11959851; http://dx.doi.org/ 10.1074/jbc.M109728200 [DOI] [PubMed] [Google Scholar]
- 32. Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 2005; 437:1173-8; PMID:16189514; http://dx.doi.org/ 10.1038/nature04209 [DOI] [PubMed] [Google Scholar]
- 33. Yang B, O'Herrin SM, Wu J, Reagan-Shaw S, Ma Y, Bhat KM, Gravekamp C, Setaluri V, Peters N, Hoffmann FM, et al. MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res 2007; 67:9954-62; PMID:17942928; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-1478 [DOI] [PubMed] [Google Scholar]
- 34. Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 2007; 3:PMID:17353931; http://dx.doi.org/ 10.1038/msb4100134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sergeant J, Taylor E, Palecek J, Fousteri M, Andrews E, Sweeney S, Shinagawa H, Watts FZ, Lehmann AR. Composition and architecture of the schizosaccharomyces pombe rad18 (smc5-6) complex. Mol Cell Biol 2005; 25:172-84; PMID:15601840; http://dx.doi.org/ 10.1128/MCB.25.1.172-184.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Markson G, Kiel C, Hyde R, Brown S, Charalabous P, Bremm A, Semple J, Woodsmith J, Duley S, Salehi-Ashtiani K, et al. Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res 2009; 19:1905-11; PMID:19549727; http://dx.doi.org/ 10.1101/gr.093963.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McDonald WH, Pavlova Y, Yates JRJ, Boddy MN. Novel essential DNA repair proteins nse1 and nse2 are subunits of the fission yeast smc5-smc6 complex. J Biol Chem 2003; 278:45460-7; PMID:12966087; http://dx.doi.org/ 10.1074/jbc.M308828200 [DOI] [PubMed] [Google Scholar]
- 38. Hou X, Zhang W, Xiao Z, Gan H, Lin X, Liao S, Han C. Mining and characterization of ubiquitin E3 ligases expressed in the mouse testis. BMC Genomics 2012; 13:495; PMID:22992278; http://dx.doi.org/ 10.1186/1471-2164-13-495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugiura T, Miyamoto K. Characterization of TRIM31, upregulated in gastric adenocarcinoma, as a novel RBCC protein. J Cell Biochem 2008; 105:1081-91; PMID:18773414; http://dx.doi.org/ 10.1002/jcb.21908 [DOI] [PubMed] [Google Scholar]
- 40. Taylor EM, Copsey AC, Hudson JJ, Vidot S, Lehmann AR. Identification of the proteins, including MAGEG1, that make up the human SMC5-6 protein complex. Mol Cell Biol 2008; 28:1197-206; PMID:18086888; http://dx.doi.org/ 10.1128/MCB.00767-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krutzfeldt M, Ellis M, Weekes DB, Bull JJ, Eilers M, Vivanco MD, Sellers WR, Mittnacht S. Selective ablation of retinoblastoma protein function by the RET finger protein. Mol Cell 2005; 18:213-24; PMID:15837424; http://dx.doi.org/ 10.1016/j.molcel.2005.03.009 [DOI] [PubMed] [Google Scholar]
- 42. Hao YH, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK, Potts PR. Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell 2013; 152:1051-64; PMID:23452853; http://dx.doi.org/ 10.1016/j.cell.2013.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiao TZ, Bhatia N, Urrutia R, Lomberk GA, Simpson A, Longley BJ. MAGE I transcription factors regulate KAP1 and KRAB domain zinc finger transcription factor mediated gene repression. PLoS One 2011; 6:e23747; PMID:21876767; http://dx.doi.org/ 10.1371/journal.pone.0023747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Espantman KC, O'Shea CC. aMAGEing new players enter the RING to promote ubiquitylation. Mol Cell 2010; 39:835-7; PMID:20864031; http://dx.doi.org/ 10.1016/j.molcel.2010.09.006 [DOI] [PubMed] [Google Scholar]
- 45. Feng Y, Gao J, Yang M. When MAGE meets RING: insights into biological functions of MAGE proteins. Protein Cell 2011; 2:7-12; PMID:21337005; http://dx.doi.org/ 10.1007/s13238-011-1002-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol 2008; 8:PMID:18673550; http://dx.doi.org/ 10.1186/1471-2148-8-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Streich FC, Jr., Ronchi VP, Connick JP, Haas AL. Tripartite motif ligases catalyze polyubiquitin chain formation through a cooperative allosteric mechanism. J Biol Chem 2013; 288:8209-21; PMID:23408431; http://dx.doi.org/ 10.1074/jbc.M113.451567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Teuber J, Mueller B, Fukabori R, Lang D, Albrecht A, Stork O. The ubiquitin ligase praja1 reduces NRAGE expression and inhibits neuronal differentiation of PC12 cells. PLoS One 2013; 8:e63067; PMID:23717400; http://dx.doi.org/ 10.1371/journal.pone.0063067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bhatia N, Xiao TZ, Rosenthal KA, Siddiqui IA, Thiyagarajan S, Smart B, Meng Q, Zuleger CL, Mukhtar H, Kenney SC, et al. MAGE-C2 promotes growth and tumorigenicity of melanoma cells, phosphorylation of KAP1, and DNA damage repair. The J Inves Dermatol 2013; 133:759-67; PMID:23096706; http://dx.doi.org/ 10.1038/jid.2012.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bavner A, Matthews J, Sanyal S, Gustafsson JA, Treuter E. EID3 is a novel EID family member and an inhibitor of CBP-dependent co-activation. Nucleic Acids Res 2005; 33:3561-9; PMID:15987788; http://dx.doi.org/ 10.1093/nar/gki667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Laduron S, Deplus R, Zhou S, Kholmanskikh O, Godelaine D, De Smet C, Hayward SD, Fuks F, Boon T, De Plaen E. MAGE-A1 interacts with adaptor SKIP and the deacetylase HDAC1 to repress transcription. Nucleic Acids Res 2004; 32:4340-50; PMID:15316101; http://dx.doi.org/ 10.1093/nar/gkh735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, Luban J, Mothes W. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J Virol 2013; 87:257-72; PMID:23077300; http://dx.doi.org/ 10.1128/JVI.01804-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Uchil PD, Quinlan BD, Chan WT, Luna JM, Mothes W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog 2008; 4:e16; PMID:18248090; http://dx.doi.org/ 10.1371/journal.ppat.0040016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 1991; 254:1643-7; PMID:1840703; http://dx.doi.org/ 10.1126/science.1840703 [DOI] [PubMed] [Google Scholar]
- 55. Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. EMBO J 2001; 20:2140-51; PMID:11331580; http://dx.doi.org/ 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Semple JI, Prime G, Wallis LJ, Sanderson CM, Markie D. Two-hybrid reporter vectors for gap repair cloning. Biotechniques 2005; 38:927-34; PMID:16018554; http://dx.doi.org/ 10.2144/05386RR03 [DOI] [PubMed] [Google Scholar]
- 57. Woodsmith J, Jenn RC, Sanderson CM. Systematic analysis of dimeric E3-RING interactions reveals increased combinatorial complexity in human ubiquitination networks. Mol Cell Proteomics 2012; 11:PMID:22493164; http://dx.doi.org/ 10.1074/mcp.M111.016162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J 2007; 26:102-12; PMID:17159902; http://dx.doi.org/ 10.1038/sj.emboj.7601469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stejskal K, Potesil D, Zdrahal Z. Suppression of peptide sample losses in autosampler vials. J Proteome Res 2013; 12:3057-62; PMID:23590590; http://dx.doi.org/ 10.1021/pr400183v [DOI] [PubMed] [Google Scholar]
- 60. Jeram SM, Srikumar T, Pedrioli PG, Raught B. Using mass spectrometry to identify ubiquitin and ubiquitin-like protein conjugation sites. Proteomics 2009; 9:922-34; PMID:19180541; http://dx.doi.org/ 10.1002/pmic.200800666 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


