Abstract
Aims
Citrullination, the post-translational conversion of arginine to citrulline by the enzyme family of peptidylarginine deiminases (PADs), is associated with several diseases, and specific citrullinated proteins have been shown to alter function while others act as auto-antigens. In this study, we identified citrullinated proteins in human myocardial samples, from healthy and heart failure patients, and determined several potential functional consequences. Further we investigated PAD isoform cell-specific expression in the heart.
Methods and results
A citrullination-targeted proteomic strategy using data-independent (SWATH) acquisition method was used to identify the modified cardiac proteins. Citrullinated-induced sarcomeric proteins were validated using two-dimensional gel electrophoresis and investigated using biochemical and functional assays. Myocardial PAD isoforms were confirmed by RT-PCR with PAD2 being the major isoform in myocytes. In total, 304 citrullinated sites were identified that map to 145 proteins among the three study groups: normal, ischaemia, and dilated cardiomyopathy. Citrullination of myosin (using HMM fragment) decreased its intrinsic ATPase activity and inhibited the acto-HMM-ATPase activity. Citrullinated TM resulted in stronger F-actin binding and inhibited the acto-HMM-ATPase activity. Citrullinated TnI did not alter the binding to F-actin or acto-HMM-ATPase activity. Overall, citrullination of sarcomeric proteins caused a decrease in Ca2+ sensitivity in skinned cardiomyocytes, with no change in maximal calcium-activated force or hill coefficient.
Conclusion
Citrullination unique to the cardiac proteome was identified. Our data indicate important structural and functional alterations to the cardiac sarcomere and the contribution of protein citrullination to this process.
Keywords: Myofilament, Citrullination, Peptidylarginine deiminases, Heart failure, Sarcomere
1. Introduction
Citrullination, the irreversible post-translational modification (PTM) involving the conversion of arginine to citrulline by the family of enzymes peptidylarginine deiminase (PAD), is associated with several diseases.1–5 Citrullination appears to be a generalized process; autoantibodies targeting citrullinated proteins are relatively specific for rheumatoid arthritis (RA) and, although occasionally observed in other autoimmune conditions, are uncommonly observed in healthy individuals.6 In our previous study, elevated levels of citrullination were found in the myocardium of RA patients.7 PADs were detected in cardiomyocytes, resident inflammatory cells, endothelial cells and vascular smooth muscle cells.7 However, the actual proteins that are citrullinated in myocardium are unknown as is whether (i) myocardial citrullinated proteins are immune targets for circulating autoantibodies, (ii) myocardial citrullinated proteins can themselves induce an autoimmune response, and (iii) citrullinated proteins directly mediate phenotypic modifications to cardiac structure or function. There is precedent for PTMs of myocardial proteins leading to changes in cardiac contractility and structure in heart failure (HF). Phosphorylation, oxidation, and acetylation of sarcomeric proteins cause morphologic changes to proteins that lead to decreased contractile performance and adverse cardiac remodelling with HF.8 However, it is unknown whether citrullination of myocardial proteins plays a similar role.
Citrullination results in a small increase in molecular mass (+0.984 Da) but converts the positively charged guanidine group on an arginine residue into the neutrally charged ureido group on the citrulline amino acid. The loss of charge from an arginine to a citrulline can have dramatic consequences on protein structure,9 proteolytic susceptibility,10 protein–protein interactions,11,12 and intracellular signalling.12 Since citrullination can lead to profound changes in protein structure and function, it is not surprising that increased citrullination and the PAD enzymes are found in numerous chronic diseases.13 Furthermore, the conversion of arginine to citrulline is catalyzed in a Ca2+-dependent manner with relatively high intracellular concentration of calcium. Because the cytosolic and nucleoplasmic calcium concentrations are relatively low, PADs should be inactive under normal conditions. However, PADs become activated in injured and dying cells, when calcium concentrations increase because of the influx of calcium ions from the extracellular environment and release from intracellular calcium stores.14 HF is a chronic maladaptive state in which complex transcriptional, proteomic, and morphological changes result in profound perturbations in intracellular Ca2+ cycling that drive the progressive deterioration in cardiac function.15 Hallmark features of HF pathogenesis are mechanical dysfunction and Ca2+ handling dysfunction,16–19 suggesting a potential for PAD activation.
There have been challenges in identification of citrullinated proteins and the modified amino acid residues.20 Here for the first time, we use a mass spectrometry (MS) approach, SWATH, to define the citrullinated proteome. In this application, SWATH is a two-step process involving the development of a citrullination-specific peptide ion library that is used to subsequently compare the individual sample MS ion data allowing quantification of site-specific differences between control and ischaemic and dilated cardiomyopathy HF subjects. Citrullination of the high abundant sarcomeric proteins was further analysed using 1 and 2 DE analysis. In addition, we investigate the biochemical and physiological effects of citrullinated sarcomeric proteins, providing the initial evidence supporting the hypothesis that citrullination may play a role in the reduced contractility observed in HF.
2. Methods
An expanded Methods section can be found in the Supplementary material online.
2.1. Reagents and materials
The following reagents were obtained: rabbit skeletal muscle PAD2 (Sigma); PAD cocktail (SignalChem); PAD2, heavy meromyosin (HMM), tropomyosin (TM) (Sigma); F-actin (Cytoskeleton Inc.), cardiac troponin (TnI) (Abcam), anti-modified citrulline antibody (Millipore); sequencing grade Lys-C protease and protease inhibitor cocktail (Roche).
2.2. Human heart tissue
Left ventricular tissue samples were obtained from Cris Dos Remedios, University of Sydney, Australia after informed consent and with approval of the local Ethical Committee. The samples were acquired during heart transplantation surgery, from patients with HF [ischemic heart disease (ISHD) and idiopathic cardiomyopathy (IDCM), n = 10 each] and non-failing donor hearts (n = 10) as previously described.21
2.3. Mouse heart tissue and neonatal myocytes
Male C57BL/6 mice (n = 3) (5-day-old neonatal mice, Jackson Laboratories) were obtained. Animal study was approved by The Johns Hopkins University Animal Care and Use Committee and followed established NIH guidelines. Briefly, primary cultured ventricular myocytes isolated from neonatal mice were a kind gift from Dr Koitabashi.22 Collagenase-digested isolated myocytes were incubated in buffer with increasing concentrations of Ca2+, achieving a final concentration of 1.2 mM Ca2+ as in the MEM culture media. Cells were seeded at 25 000 rod-shaped myocytes/mL on 6-well plates or 60 mm dishes coated with laminin. After 1 h incubation in 37°C, 5% CO2, the culture media were replaced to remove unattached cells.
2.4. Protein extraction and SWATH-MS
Hearts were fractionated into myofilament- and cytosolic-enriched fractions using the IN Sequence protocol.23 Protein extraction and generation of LysC peptides from subfractions was performed using a filter-aided sample preparation (FASP) protocol.24 When needed, recombinant proteins or the In Sequence fractions were incubated with PAD's cocktail at a ratio of 1:20 for 2 h at 37°C in 100 mM Tris, pH 7.6, 5 mM DTT, 10 mM CaCl2. The reaction was stopped by addition of 5 mM EDTA prior to digestion. A TripleTOF 6600 mass spectrometer (Sciex) was used for both data-dependent acquisition to build peptide spectral ion library and SWATH-MS (data-independent acquisition) for each individual sample analysis. The raw data were searched with ProteinPilot™ Software 5.0 to create a spectral ion library. Individual SWATH-MS runs were matched against the spectral library created in the presence or absence of PAD (plus and minus PAD) for both the myofilament- and cytosolic-enriched protein fractions (see Supplementary material online).
2.5. Preparation of citrullinated samples
Recombinant proteins and the fractions obtained from IN Sequence were incubated with PAD2 at a ratio of 1:20 for 2 h at 37°C in working buffer (100 mM Tris, pH 7.6, 5 mM DTT, 10 mM CaCl2). The reaction was stopped by addition of 5 mM EDTA.
2.6. Statistical validation peptides, proteins and citrullination residues: acceptance criteria
Bioinformatics analysis was performed with the workflow described in data supplement. The peptide normalization used in this study was based on the iRT peptide retention time,25 and normalized values were used for downstream analysis. Ensemble protein ID accession numbers were mapped back to their associated encoding Ensemble gene entries. Data analysis and mining were performed using iProXpress (http://proteininformationresource.org/iproxpress2)26 and Cytoscape.27 The Kruskal–Wallis test (non-parametric one-way ANOVA) for each peptide was used to calculate P-values. The significance of the biochemical changes was determined by performing a t-test (P ≤ 0.05) on the differences for all paired data. Unless otherwise stated all biochemical assays were replicated three times.
2.7. SDS–PAGE immunoblot for citrullination
A 1:2000 diluted of the anti-citrulline (Modified) antibody was used for the 1 DE western blot (see Supplementary material online).
2.8. Two-dimensional gel electrophoresis
The independent verification of proteomics data was performed with fluorescence two-dimensional (2DE) gel electrophoresis (2D-DIGE, pI range and SDS–PAGE range) as reported previously.28 The treatment of sample with PAD2 enzyme, which removes a guanidino group from specific arginine residues within the modified protein, can be used to identify citrullinated proteins based on the change in charge of the protein after treatment (see Supplementary material online).
2.9. Membrane-permeabilized myocytes
Left ventricular tissue from C57BL/6 mice was flash-frozen in liquid nitrogen and stored at −80°C. For analysis, tissue was homogenized in the presence of 0.3% Triton X-100, and protease and phosphatase inhibitors, as described.29 Myocytes were washed without Triton X-100 to remove the detergent and resuspended in isolation buffer. PAD2 was activated in 10 mM Ca2+ and 50 mM DTT for 60 min at 37°C. PAD2-treated myocytes were then exposed to a 1:10 dilution of activated PAD2 in isolation buffer for 30 min at room temperature. Myocytes were then glued with silicone to the tips of 150 μm diameter minutia pins attached to a force transducer and motor arm (Aurora Scientific Inc., Aurora, ON, Canada). Sarcomere length was monitored by video camera (Imperx, Boca Raton, FL, USA) and calculated by the High-speed Video Sarcomere Length Program (Aurora Scientific Inc.). Myocyte sarcomere length was set at 2.1 μm. A complete activation of the myocyte occurred at the beginning and end of the experiment, and the myocyte discarded if there was >10% rundown, as described.29
2.10. ATPase activity
The HMM-ATPase activity was analysed at three separate experiments described previously.30,31 First experiment was carried out at constant HMM and F-actin concentration with citrullinated or non-citrullinated HMM and/or F-actin. Second, HMM-ATPase activity was determined at increasing TM concentrations (citrullinated or non-citrullinated). Third experiment HMM-ATPase activity was determined at constant HMM, F-actin, and TM concentration with increasing citrullinated or non-citrullinated TnI. Each experiment was done in triplicate and three separate times. For more details, see Supplementary material online.
2.11. Actin-binding experiments
Various concentrations of citrullinated and non-citrullinated HMM, TM, and/or TnI were added to F-actin and centrifuged to determine extent of binding. Pellet and supernatant were analysed using 10% SDS–PAGE, and amount of each protein was quantified by densitometry as previously described.32,33 Each assay was carried out in triplicate. For more details, see Supplementary material online.
2.12. Reverse transcription-PCR
Total mRNA from mouse neonatal cardiac myocytes was extracted using TRI-reagent (Sigma) according to the manufacturer's protocols. cDNA was generated using the SuperScript III First-Standard Synthesis System (Invitrogen) according to the instructions of the manufacturer. Reverse transcription (RT)-PCR was performed using primers specific for the PAD1, PAD2, PAD3, PAD4, and β-actin. The PCR products were separated by electrophoresis on a 1.8% agarose gel and visualized under UV light. Each assay was done in duplicate.
3. Results
3.1. Identification of myocardial citrullinated proteins
To identify citrullinated targets in the heart, we assessed the citrullinome in three groups, ISHD, IDCM, and non-failing donor hearts (n = 10 per group) using SWATH-MS.34 SWATH-MS allowed for the quantification. Fifty-three citrullinated sites were altered with HF compared with the non-failing controls (P < 0.05) and are listed in Table 1 (see Supplementary material online, Table S1 for details of all citrullinated peptides in this study). These were proteins with diverse cellular functions, including the regulators of transcription and chromatin structure, cytoskeletal and contraction, cellular signalling processes, and metabolism (Figure 1). Western blotting of myofilament- and cytosolic-enriched fractions obtained from ISHD, IDCM, and non-failing donor hearts (n = 10 per group) using an anti-modified citrulline antibody confirmed that citrullination occurs in intracellular proteins. Although there was no difference (see Supplementary material online, Figure S1) in the overall immunoreactivity between groups with regard to the number of bands or band density per blot, in-gel digestion and subsequent MS of the immunoreactive bands identified the major sarcomeric proteins at their expected molecular weight (e.g. myosin heavy and light chains and actin, data not shown). It must be noted that quantitative assessment by immuno-1DE was confounded by the presence of other proteins in the gel bands and challenges of direct site-specific assessment using MS.
Table 1.
List of citrullinated protein (P = 0.05) with citrullinated peptide sequence
| Protein | Peptide | UP identifier | p_kw | |
|---|---|---|---|---|
| Cytoplasm | Adenylate kinase isoenzyme 1 | RGETSGR[Dea]VDDNEETIK | Q5T9B7 | 0.03 |
| Cytoplasm | Alcohol dehydrogenase 1B | AAGAAR[Dea]IIAVDINK | ADH1B | 0.04 |
| Cytoplasm | Beta-enolase | SPDDPAR[Dea]HITGEK | ENOB | 0.00 |
| Cytoplasm | Carbonic anhydrase 3 | DIR[Dea]HDPSLQPWSVSYDGGSAK | CAH3 | 0.01 |
| Cytoplasm | Fatty acid-binding protein, heart | LILTLTHGTAVC[CAM]TR[Dea]TYEK | FABPH | 0.04 |
| Cytoplasm | Peptidyl-prolyl cis-trans isomerase A | TAENFR[Dea]ALSTGEK | PPIA | 0.01 |
| Cytoplasm | Serum deprivation-response protein | FQHPGSDMR[Dea]QEK | SDPR | 0.01 |
| Cytoplasm | Serum deprivation-response protein | VSPLTFGR[Dea]K | SDPR | 0.01 |
| Cytoplasm | Heat shock protein beta-7 | RHPHTEHVQQTFR[Dea]TEIK | HSPB7 | 0.05 |
| Cytoplasm | Alpha-crystallin B chain | GLSEMR[Dea]LEK | CRYAB | 0.01 |
| Cytoskeleton | Actina | C[CAM]DIDIR[Dea]K | ACTA1 | >0.05 |
| Cytoskeleton | Actina | QEYDEAGPSIVHR[Dea]K | ACTA2 | >0.05 |
| Cytoskeleton | Filamin-C | SSSSR[Dea]GSSYSSIPK | FLNC | 0.03 |
| Cytoskeleton | LIM domain-binding protein 3 | TSPEGAR[Dea]DLLGPK | LDB3 | 0.00 |
| Cytoskeleton | Myosin-binding protein C, cardiac-type | EPVFIPR[Dea]PGITYEPPNYK | A8MXZ9 | 0.03 |
| Cytoskeleton | Myozenin-2 | R[Dea]VATPFGGFEK | MYOZ2 | 0.01 |
| Cytoskeleton | Myozenin-2 | AELPDYR[Dea]SFNR[Dea]VATPFGGFEK | MYOZ2 | 0.02 |
| Cytoskeleton | Troponin I, cardiac muscle | NIDALSGMEGR[Dea]K | TNNI3 | 0.02 |
| Cytoskeleton | Troponin I, cardiac muscle | ESLDLR[Dea]AHLK | TNNI3 | 0.02 |
| Cytoskeleton | Troponin T, cardiac muscle | PR[Dea]SFMPNLVPPK | E7EPW4 | 0.01 |
| Cytoskeleton | Tropomyosinb | ETR[Dea]AEFAERSVTKLEK | TPM1 | 0.002 |
| Cytoskeleton | Vimentin | FADLSEAAN[Dea]RNNDALR[Dea]QAK | VIME | 0.03 |
| Cytoskeleton | Glyceraldehyde-3-phosphate dehydrogenase | LWR[Dea]D[Dhy]GRGALQN[Oxi]IIPASTGAAK | G3P | 0.01 |
| Cytoskeleton | Myosin-7 | AEETQR[Dea]SVNDLTSQR[Dea]AK | MYH7 | 0.05 |
| Mitochondrion | Aconitate hydratase | SYLR[Dea]LRPDRVAMQDATAQ[Dea]M[Oxi]AMLQFISSGLSK | ACON | 0.01 |
| Mitochondrion | Aconitate hydratase | ANSVR[Dea]NAVTQEFGPVPDTAR[Dea]YYK | ACON | 0.03 |
| Mitochondrion | Aconitate hydratase | IVYGHLDDPASQEIER[Dea]GK | ACON | 0.03 |
| Mitochondrion | Adenylate kinase 4 | LLR[Dea]AVILGPPGSGK | KAD4 | 0.00 |
| Mitochondrion | Alcohol dehydrogenase class-3 | VAGASR[Dea]IIGVDINK | ADHX | 0.05 |
| Mitochondrion | ATP synthase subunit alpha | R[Dea]TGAIVDVPVGEELLGR[Dea]VVDALGNAIDGK | ATPA | 0.00 |
| Mitochondrion | ATP synthase subunit alpha | AIEEQVAVIYAGVR[Dea]GYLDK | ATPA | 0.01 |
| Mitochondrion | ATP synthase subunit alpha | GIRPAINVGLSVSR[Dea]VGSAAQ[Dea]TRAMK | ATPA | 0.01 |
| Mitochondrion | ATP synthase subunit alpha | QGQYSPMAIEEQVAVIYAGVR[Dea]GYLDK | ATPA | 0.03 |
| Mitochondrion | ATP synthase-coupling factor 6 | SGGPVDASSEYQQELER[Dea]ELFK | ATP5J | 0.00 |
| Mitochondrion | Chloride intracellular channel protein 4 | YR[Dea]NFDIPK | CLIC4 | 0.04 |
| Mitochondrion | Cytochrome c (Fragment) | EER[Dea]ADLIAYLK | C9JFR7 | 0.01 |
| Mitochondrion | Delta-1-pyrroline-5-carboxylate dehydrogenase | VLR[Dea]NAAGNFYINDK | AL4A1 | 0.00 |
| Mitochondrion | Delta-1-pyrroline-5-carboxylate dehydrogenase | AADMLSGPR[Dea]R[Dea]AEILAK | AL4A1 | 0.01 |
| Mitochondrion | Delta-1-pyrroline-5-carboxylate dehydrogenase | VANEPVLAFTQGSPER[Dea]DALQK | AL4A1 | 0.03 |
| Mitochondrion | Enoyl-CoA hydratase | EMVLTGDR[Dea]ISAQDAK | ECHM | 0.01 |
| Mitochondrion | ES1 protein homologue | VLR[Dea]GVEVTVGHEQEEGGK | ES1 | 0.00 |
| Mitochondrion | Heat shock protein HSP 90-alpha | HLEINPDHSIIETLR[Dea]QK | HS90A | 0.04 |
| Mitochondrion | NAD(P) transhydrogenase | AATITPFR[Dea]K | NNTM | 0.05 |
| Mitochondrion | Phosphate carrier protein | VYFR[Dea]LPRPPPPEMPESLK | MPCP | 0.00 |
| Mitochondrion | Pyruvate dehydrogenase E1 component subunit alpha, somatic form | C[CAM]DLHR[Dea]LEEGPPVTTVLTR[Dea]EDGLK | ODPA | 0.01 |
| Mitochondrion | Succinate dehydrogenase [ubiquinone] iron-sulfur subunit | FAIYR[Dea]WDPDK | SDHB | 0.05 |
| Mitochondrion | Succinyl-CoA ligase [ADP/GDP-forming] subunit alpha | TR[Dea]LIGPNC[CAM]PGVINPGEC[CAM]K | SUCA | 0.02 |
| Mitochondrion | Succinyl-CoA:3-ketoacid coenzyme A transferase 1 | ADR[Dea]AGNVIFR[Dea]K | SCOT1 | 0.04 |
| Mitochondrion | Voltage-dependent anion-selective channel protein 1 | SR[Dea]VTQSNFAVGYK | VDAC1 | 0.02 |
| Mitochondrion | 60 kDa heat shock protein | FDR[Dea]GYISPYFINTSK | CH60 | 0.04 |
| Mitochondrion | Trifunctional enzyme subunit beta | PNIR[Dea]NVVVVDGVR[Dea]TPFLLSGTSYK | ECHB | 0.02 |
| Mitochondrion | Aconitate hydratase | TGR[Dea]EDIANLADEFK | ACON | 0.00 |
| Mitochondrion | Aconitate hydratase | R[Dea]LQLLEPFDK | ACON | 0.01 |
| Nucleus | Dual specificity protein phosphatase 3 | LGITHVLNAAEGR[Dea]SFMHVNTNANFYK | DUS3 | 0.01 |
| Nucleus | Neuroblast differentiation-associated protein AHNAK | FGVSTGR[Dea]EGQTPK | AHNK | 0.01 |
| Collagen alpha-3(VI) chain | DVVFLLDGSEGVR[Dea]SGFPLLK | E7ENL6 | 0.03 |
Citrullinated proteins were grouped by cellular component.
apkw (P) < 0.05.
bBased on 2DE gel analysis.
Figure 1.
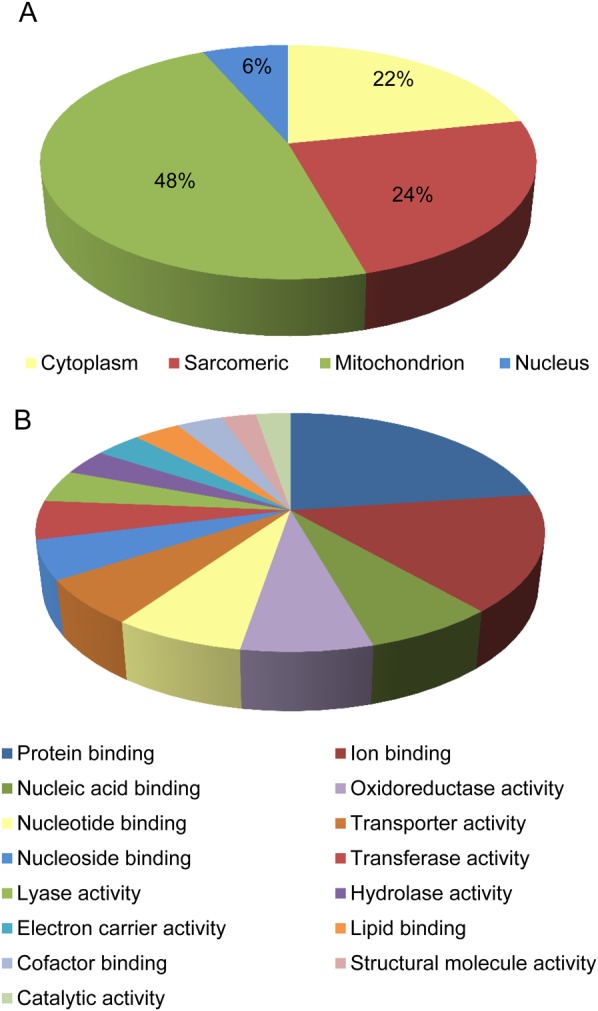
The cardiac citrullinated proteome. Diagrams show citrullinated proteins with significant P-value group by (A) cellular component and (B) molecular function. Details can be found in Table 1.
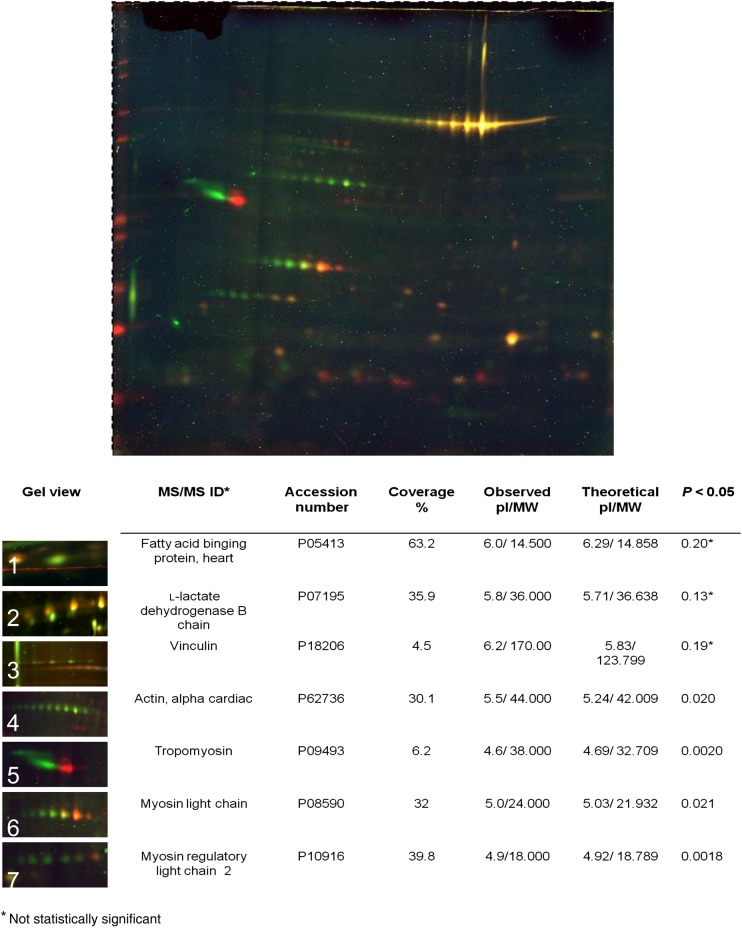
To further validate citrullination of the high abundant sarcomeric proteins, DIGE 2D gel electrophoresis (pH 4–7, 10% SDS–PAGE) was carried out. Myofilament- and cytosolic-enriched fractions of ISHD, IDCM, and non-failing donor hearts (n = 4 per group) were pretreated with PAD2 to induce maximum citrullination, combined at a 1:1 ratio with the matching untreated samples and simultaneously resolved by 2D gel electrophoresis. The sarcomeric proteins, including actin, TM, and myosin light chains were shown to be citrullinated with ectopic treatment of PAD2 (Figure 2).
Figure 2.
2DE DIGE analysis with samples treated with PAD2. Samples labelled with Cy2 (internal control), Cy3 (untreated), and Cy5 (treated) as described in Methods section. Details can be found in insert table.
3.2. Biochemical assessment of modified sarcomeric proteins
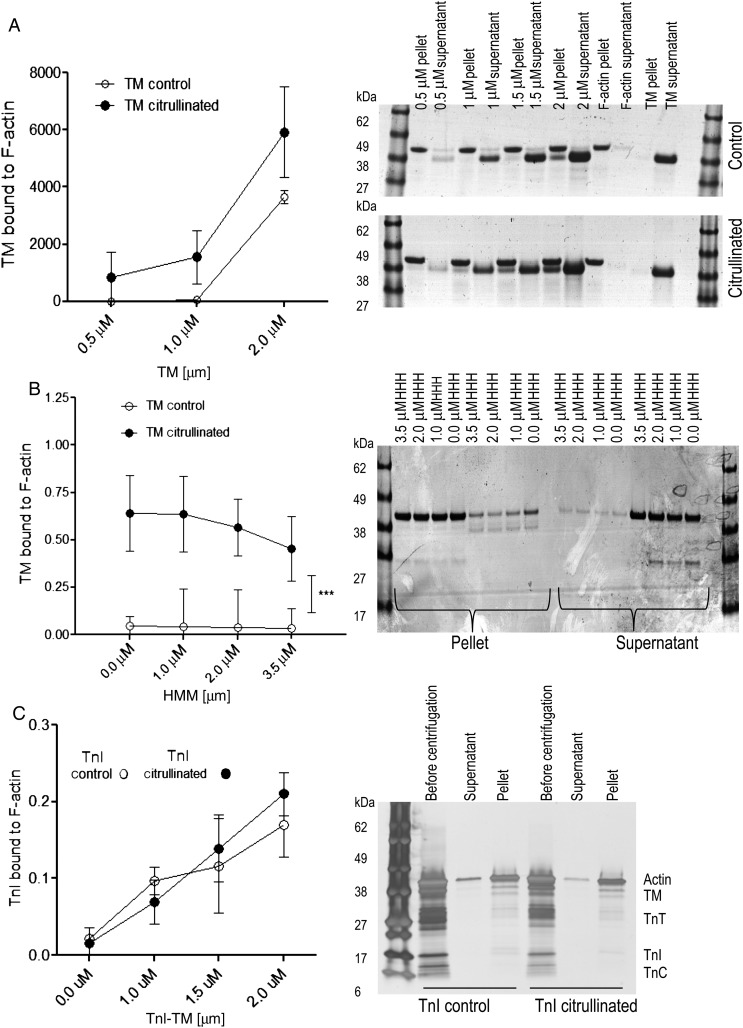
To test whether citrullination can affect sarcomeric protein function, actin, HMM, TM, and troponin were citrullinated by ectopic PAD2 and then compared with respective unmodified recombinant proteins to determine whether citrullination affects their biochemical, structural, or enzymatic properties. The binding of citrullinated or untreated HMM, TM, or TM–TnI to F-actin was determined using classical co-sedimentation assays (Figure 3A–C). Citrullination of actin did not alter F-actin formation and over 95% of the actin was pelleted upon centrifugation (data not shown). Binding of citrullinated TM to F-actin was enhanced compared with unmodified TM (Figure 3A). Since TM and HMM can affect binding of each other to F-actin in a co-operative manner,35 we tested for co-operativity under conditions in which binding of TM alone to F-actin is poor, but increased upon the binding of myosin heads to F-actin.36 As illustrated in Figure 3B, at low salt concentration, citrullinated TM in the presence of HMM was able to bind to F-actin.
Figure 3.
Actin-binding studies. Increasing concentrations of (A) TM (0.5–2 µM) were incubated with F-actin or (B) actin-HMM and (C) TnI (0.5–2 µM) in buffer containing 40 mM Tris–HCl (pH 7.6), 100 mM NaCl, 5 mM MgCl2, and 1 mM DTT. Binding of TM to F-actin was carried out at 25°C for 30 min and ultracentrifuged at 156 565 g for 25 min, 20°C, in a Beckman model TL-100.2. Both pellet and supernatant (unbound protein) were analysed. Representative silver-stained gels show proteins composition of the supernatants and pellets. A triplicate set of gels was analysed by densitometry. Each data point is an average (and range) of the values obtained from the three sets of gels.
The binding of cardiac TnI to F-actin in the presence of TM was performed by co-sedimenting citrullinated TnI in the presence of F-actin and TM. It was found that both citrullinated and non-citrullinated forms of cardiac TnI bound to F-actin equally well (Figure 3C).
3.3. Inhibition of HMM-ATPase activity
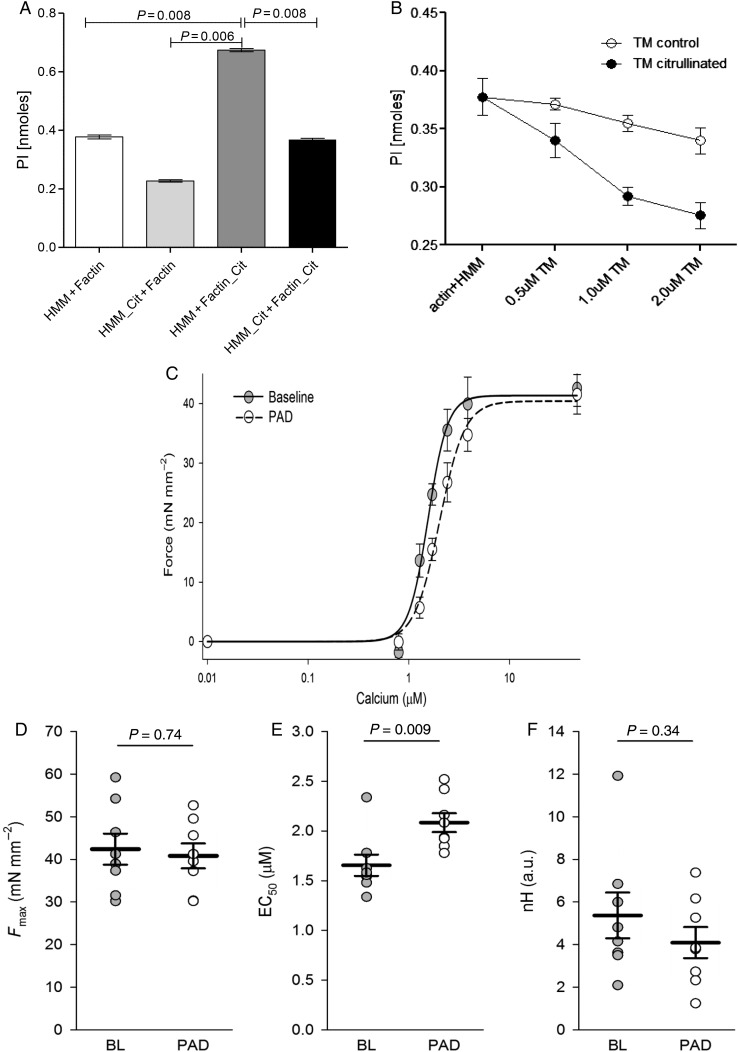
To verify a possible modulation of the actomyosin HMM-ATPase activity by citrullination of sarcomeric proteins, experiments were performed in the presence of citrullinated HMM, F-actin, and TM. The data summarized in Figure 4A show that, in the presence of citrullinated HMM (0.5 µmol/L HMM and 7 µmol/L F-actin), the actomyosin HMM-ATPase activity decreased from 0.32 ± 0.01 to 0.22 ± 0.002 nmol Pi, corresponding to 30% decrease in the enzyme activity. In contrast, citrullination of F-actin enhanced actomyosin HMM-ATPase activity up to 55% on the enzyme activity (Figure 4A). When both F-actin and HMM were citrullinated, the rate of ATP hydrolysis remained greater than control (0.368 ± 0.005 to 0.32 ± 0.01 nmol Pi). Figure 4B showed that actomyosin HMM-ATPase activity was affected by TM citrullination. The inhibition of actomyosin HMM-ATPase conferred by non-citrullinated and citrullinated TnI was compared. Addition of non-citrullinated TnI to HMM-F-actin-TM caused a decrease in the ATPase rate at 37 °C. Citrullinated TnI acted similar to non-citrullinated TnI and caused a decrease in the ATPase with no significant deference to non-citrullinated form (data not show).
Figure 4.
Citrullination of sarcomeric proteins, biochemical and physiological effects. (A) Regulation of the actomyosin HMM-ATPase activity by citrullinated F-actin and/or citrullinated HMM. (B) Inhibition of actomyosin HMM-ATPase activity by TM. ATPase activity was measured as a function of TM concentration. The results are the average of four independent experiments for each protein at each TM concentration. Assay conditions: 0.2 mg/mL F-actin, 0.02 mg/mL HMM, 0–2.0 µM TM in 10 mM Hepes, pH 7.5, 30 mM NaCl, 5 mM MgCl2, 4 mM ATP. PAD2 treatment reduced myofilament calcium sensitivity. (C) Force–calcium relationships for untreated membrane-permeabilized myocytes from untreated control (n = 8 myocytes from three mice, grey circles) and PAD2-treated (n = 8 myocytes from three mice, open circles) groups. (D) There was no difference in maximal calcium-activated force (Fmax) between the two groups. (E) PAD2 treatment caused a significant (P = 0.009) increase in EC50 (calcium required to generate 50% Fmax), indicating a decrease in calcium sensitivity. (F) While the hill coefficient (nH) trended to be decreased by PAD2 treatment (see steepness of curve in C), the difference was not significant (P = 0.34).
3.4. PAD2 reduces calcium sensitivity in skinned myocytes
Chemically skinned cardiomyocytes isolated from the left ventricle of wild-type C57Bl6 male mice were exposed to varying concentrations of calcium (n = 8 myocytes from three mice per group, Figure 4C–F). PAD2 treatment had no effect on either maximal calcium-activated force (Fmax) or hill coefficient (nH). However, PAD2 caused a rightward shift in the force–calcium relationship that indicates an increase in EC50 or a decrease in calcium sensitivity (P = 0.009, Figure 4C). This suggests that citrullination of myofilament proteins causes a loss-of-function phenotype, reducing its ability to generate force in response to intracellular calcium.
3.5. Analysis of PAD mRNA expression
To determine the cell specificity of the PAD isoform expression in the myocardium, mRNA expression level was determined using isoform-specific primers by Nested-PCR on isolated cardiac myocytes and fibroblasts (Figure 5). PAD2 was the primary isoform expressed in both cardiomyocytes and cardiac fibroblasts, although fibroblasts also had significant expression of PAD1 and PAD4 mRNA. In contrast, PAD3 mRNA was not detected in any of the cell types and conditions tested (Figure 5, note no band at 200 bp, see Supplementary material online for more details).
Figure 5.
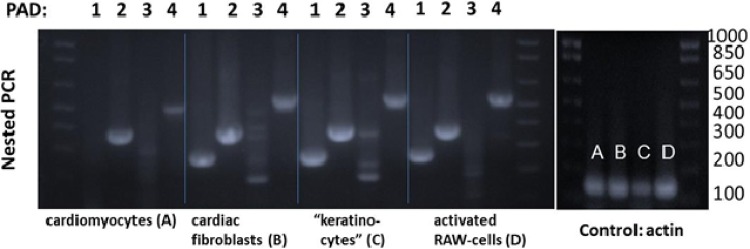
RT-PCR analysis of expression level of PAD isoforms in (A and B) heart from control mouse, (C) mouse keratinocytes, and (D) mouse macrophage cell line activated by lipopolysaccharide. The PCR product PAD2 is seen in all types of samples; PAD4 and PAD1 are seen in cardiac fibroblast, keratinocytes, and macrophage cell line. PAD3 has not been detected. (MW: PAD1, 285 bp; PAD2, 390 bp; PAD3, 200 bp; PAD4, 550 bp).
4. Discussion
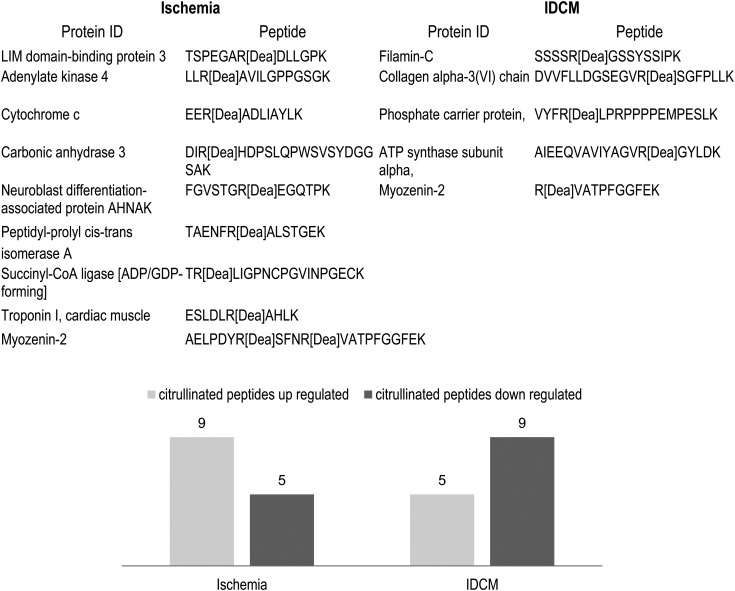
Our experimental findings characterize citrullinated proteins in the normal and HF myocardium. The analysis revealed that protein citrullination has broad cellular distribution (Figure 1) but is highly enriched in the mitochondria and sacromeric subproteome. A close connection between these two subproteomes is not unexpected due to the high energetic requirements of the sarcomere. We speculated that citrullination, similar to phosphorylation and acetylation, could potentially regulate muscle-contractile proteins in a co-ordinated manner. Interestingly, the multiple enzymes involved in metabolic pathways/metabolism were up-regulated in ischaemia but down-regulated in IDCM (Figure 6). To understand the topology and functional annotation of citrullinated protein–protein interaction in the heart system, the protein network was constructed using the STRING database (see Supplementary material online, Figure S2).37 The entire protein network consisted of high scoring interaction partners (STRING relevant confidence score ≥0.5, ischaemia/IDCM vs. control). The visual analysis of this sub-network showed that citrullinated proteins are interacted to each other and are involved in the metabolism and respiratory chain targets, contraction and signal transduction systems.38 As an initial assessment to understand potential functional consequences of the citrullination, recombinant sarcomere proteins HMM, F-actin, TM, and TnI were used as model proteins to study the interactions that govern the thick and thin filament function and by measuring the contractile properties of single skinned myocytes treated with PAD2.
Figure 6.
Citrullinated peptides up-/down-regulated between heart failure groups, ischaemia, and IDCM.
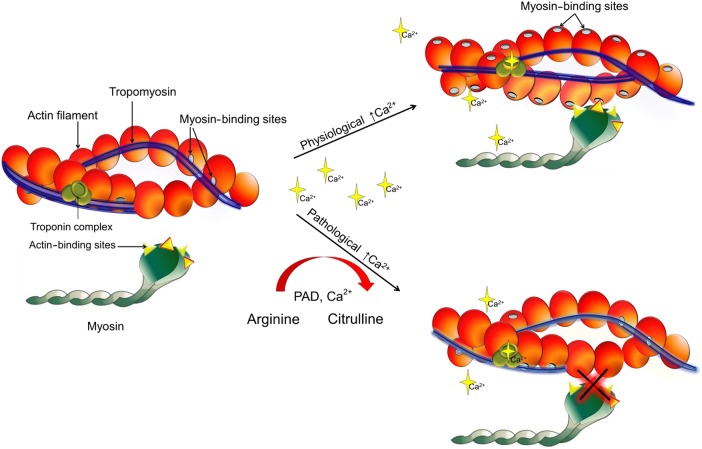
Ca2+-dependent alterations to Tn alter an azimuthal movement of TM on the actin surface, which allows myosin binding and cross-bridge isomerization to strong binding, force-producing states, and muscle contraction. At low intracellular Ca2+, Tn-TM sterically blocks myosin-binding sites on actin (blocked state), while in the presence of elevated Ca2+ Tn-TM moves and partially exposes the myosin-binding sites on F-actin (closed state). Myosin binding in the presence of Ca2+ is required for full activation.39,40 It is a combination of Ca2+-induced TnI conformational change which is, in part, transmitted via TnT to TM (especially the T1 region that binds along TM) as well as myosin binding that influence the exact positioning of TM on the actin filament. Based on our MS data, citrullination sites on myosin, actin, TM, TnI, and TnT are in regions what can influence these interactions and thus the actomyosin HMM-ATPase activity and contraction. Below outlines the potential impact based on the biochemical and physiological experiments presented in this manuscript.
First, intrinsic actomyosin HMM-ATPase was inhibited by citrullination, but this was overcome in the presence of F-actin regardless of whether F-actin is citrullinated or not. Citrullinated F-actin was a more potent modulator of HMM enzymatic activity and increased actomyosin HMM-ATPase rate by 55% compared with the unmodified F-actin. This suggests that citrullination of F-actin changes the confirmation of the actin filament to alter the ease of ATP hydrolysis by the myosin once it is bound. The actin-HMM interaction was also affected by the citrullination of TM. Citrullinated TM displayed enhanced binding to F-actin compared with unmodified TM. This was also observed in the presence of HMM (based on the centrifugation assays). This correlated to an inhibition of the citrullinated TM-actin-HMM-ATPase activity compared with the non-citrullinated TM, suggesting that citrullinated TM altered the ability of HMM to bind to the actin filament (Figure 7). Intriguingly, citrullinated TnI, like the unmodified protein complex, bound tightly to actin-TM based on centrifugation studies. Figure 3C shows that >95% of the unmodified and modified TnI was present in the pellet.
Figure 7.
Citrullination of the contractile proteins could affect different aspects of regulatory function. It could either trigger a structural change or stabilize a conformation that is necessary for actin-activated release of Pi and completion of the ATPase cycle.
Biochemical results were related to our physiological finding and showed that citrullination of the sarcomeric proteins caused a decrease in Ca2+ sensitivity in the skinned cardiac myocyte (Figure 4C). On TnI, C-terminus residues 191–210, which contains the citrullinated residue 203, is primarily responsible for maintaining the TM conformation that prevents cross-bridge cycling.41 Thus, not only does TnI promote the blocked state, but also contributes to the stabilization of TM in the closed state. In mice, cardiac TnI containing cardiomyopathy mutations, R192H or R204H (which are equivalent to the human sequence of 191 and 203 the latter being the same citrullinatable residue as discussed above), increases the binding affinity of Tn for actin-TM.35 Perhaps this is augmented by the citrullination of TnT at residue 77, which is located in the N-terminal tail (TnT1) that lies along TM and has been shown to be involved in co-operativity of the actin-TM-Tn filament. In addition, binding of Ca2+ or myosin to actin-TM-Tn can displace TnI residues 151 and 188, which flank the second citrullinated residue 169, away from the outer domain of F-actin.42 This is consistent with an azimuthal displacement to TnI–TM by Ca2+ that can expose the high-affinity binding site on F-actin for myosin.40 Studies on familial hypertrophic cardiomyopathic mutations occurring on thick filament proteins (TM, MHC, MyBP-C, ELC, and RLC) show that a change in one amino acid side chain can have an enormous effect on cardiac morphology and function.43,44 Furthermore, factors that lead to abnormal contraction and relaxation in the failing heart include metabolic pathway abnormalities that result in decreased energy production, energy transfer, and energy utilization.45
Finally, to better understand the involvement of citrullinated proteins in the heart, it is important to obtain insight about cell specificity. Previous data acquired by the immunohistochemistry showed that PADs 1–3 and, to some degree, PAD6 were detected in cardiomyocytes with PADs 2 and 4 found in endothelial cells and vascular smooth muscle cells.7 In the present study, we relied not on antibodies and tissue slices, but rather examined the mRNA expression of PAD family members in isolated mouse cardiomyocytes and cardiac fibroblasts. The data showed that PAD2 was primarily expressed in both cardiomyocytes and cardiac fibroblasts, whereas PAD1 and PAD4 mRNA were the major forms in the cardiac fibroblasts.
In summary, we have presented previously unexplored roles for citrullination in the heart. Ultimately, identification of citrullination of the majority of the sarcomeric proteins and alterations in their biochemical properties suggest that there is potentially a new PTM regulation of cardiac contractility. Citrullination at some of these residues was increased in the myocardium of individuals with HF compared with controls, suggesting that citrullination could play a role in the decrease in contractile dysfunction in HF.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
This work was supported by Johns Hopkins NHBLI Proteomic Initiative contracts NHLBI-HV-10-05(2) (J.E.V.) and HHSN268201000032C (J.E.V.); the National Institutes of Health grants R21 HL112586-01 (J.E.V. and J.M.B.) and P01-HL077180 (J.E.V.).
Acknowledgements
The authors thank Dr Dos Remidois C for providing the human heart samples and Dr Norimichi Koitabashi for providing the neonatal mice heart tissue.
Conflict of interest: none declared.
References
- 1.Jang B, Kim E, Choi JK, Jin JK, Kim JI, Ishigami A, Maruyama N, Carp RI, Kim YS, Choi EK. Accumulation of citrullinated proteins by up-regulated peptidylargininedeiminase 2 in brains of scrapie-infected mice: a possible role inpathogenesis. Am J Pathol 2008;173:1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang X, Han J. Expression of peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol Carcinog 2006;45:183–196. [DOI] [PubMed] [Google Scholar]
- 3.Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer 2009;9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mastronardi FG, Noor A, Wood DD, Paton T, Moscarello MA. Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J Neurosci Res 2007;85:2006–2016. [DOI] [PubMed] [Google Scholar]
- 5.Ishigami A, Ohsawa T, Hiratsuka M, Taguchi H, Kobayashi S, Saito Y, Murayama S, Asaga H, Toda T, Kimura N, Maruyama N. Abnormal accumulation of citrullinated proteins catalyzed by peptidylarginine deiminase in hippocampal extracts from patients with Alzheimer's disease. J Neurosci Res 2005;80:120–128. [DOI] [PubMed] [Google Scholar]
- 6.Schellekens GA, Visser H, de Jong BA, van den Hoogen FH, Hazes JM, Breedveld FC, van Venrooij WJ. The diagnostic properties of rheumatoid arthritis antibodies recognizing a cyclic citrullinated peptide. Arthritis Rheum 2000;43:155–163. [DOI] [PubMed] [Google Scholar]
- 7.Giles JT, Fert-Bober J, Park JK, Bingham CO III, Andrade F, Fox-Talbot K, Pappas D, Rosen A, van Eyk J, Bathon JM, Halushka MK. Myocardial citrullination in rheumatoid arthritis: a correlative histopathologic study. Arthritis Res Ther 2012;14:R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeWinter MM. Functional consequences of sarcomeric protein abnormalities in failing myocardium. Heart Fail Rev 2005;10:249–257. [DOI] [PubMed] [Google Scholar]
- 9.Borissoff JI, Joosen IA, Versteylen MO, Brill A, Fuchs TA, Savchenko AS, Gallant M, Martinod K, Ten Cate H, Hofstra L, Crijns HJ, Wagner DD, Kietselaer BL. Elevated levels of circulating DNA and chromatin are independently associated with severe coronary atherosclerosis and a prothrombotic state. Arterioscler Thromb Vasc Biol 2013;33:2032–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarcsa E, Marekov LN, Mei G, Melino G, Lee SC, Steinert PM. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J Biol Chem 1996;271:30709–30716. [DOI] [PubMed] [Google Scholar]
- 11.Pritzker LB, Joshi S, Gowan JJ, Harauz G, Moscarello MA. Deimination of myelin basic protein. 1. Effect of deimination of arginyl residues of myelin basic protein on its structure and susceptibility to digestion by cathepsin D. Biochemistry 2000;39:5374–5381. [DOI] [PubMed] [Google Scholar]
- 12.Fan LY, He DY, Wang Q, Zong M, Zhang H, Yang L, Sun LS. Citrullinated vimentin stimulates proliferation, pro-inflammatory cytokine secretion, and PADI4 and RANKL expression of fibroblast-like synoviocytes in rheumatoid arthritis. Scand J Rheumatol 2012;41:354–358. [DOI] [PubMed] [Google Scholar]
- 13.Gyorgy B, Toth E, Tarcsa E, Falus A, Buzas EI. Citrullination: a posttranslational modification in health and disease. Int J Biochem Cell Biol 2006;38:1662–1677. [DOI] [PubMed] [Google Scholar]
- 14.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Protein arginine deiminase 4 (PAD4): current understanding and future therapeutic potential. Curr Opin Drug Discov Devel 2009;12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 15.Bers DM. Cardiac excitation-contraction coupling. Nature 2002;415:198–205. [DOI] [PubMed] [Google Scholar]
- 16.Snaidecki B, Solaro J. When hearts fail so does skeletal muscle. Breaking a vicious cycle. Circ Res 2006;98:1456–1458. [DOI] [PubMed] [Google Scholar]
- 17.Luo M, Anderson ME. Ca2+ cycling in heart failure. Circ Res 2013;6:690–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solaro RJ, Sheehan KA, Lei M, Ke Y. The curious role of sarcomeric proteins in control of diverse processes in cardiac myocytes. J Gen Physiol 2010;136:13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon TR, Pogwizd SM, Bers DM. Elevated sarcoplasmic reticulum Ca2+ leak in intact ventricular myocytes from rabbits in heart failure. Circ Res 2003;93:592–594. [DOI] [PubMed] [Google Scholar]
- 20.De Ceuleneer M, Van Steendam K, Dhaenens M, Deforce D. In vivo relevance of citrullinated proteins and the challenges in their detection. Proteomics 2012;12:752–760. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P, Kirk JA, Ji W, dos Remedios CG, Kass DA, Van Eyk JE, Murphy AM. Multiple reaction monitoring to identify site-specific troponin I phosphorylated residues in the failing human heart. Circulation 2012;126:1828–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koitabashi N, Aiba T, Hesketh GG, Rowell J, Zhang M, Takimoto E, Tomaselli GF, Kass DA. Cyclic GMP/PKG-dependent inhibition of TRPC6 channel activity and expression negatively regulates cardiomyocyte NFAT activation: novel mechanism of cardiac stress modulation by PDE5 inhibition. J Mol Cell Cardiol 2010;48:713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kane LA, Neverova I, Van Eyk JE. Subfractionation of heart tissue: the ‘in sequence’ myofilament protein extraction of myocardial tissue. Methods Mol Biol 2007;357:87–90. [DOI] [PubMed] [Google Scholar]
- 24.Romero V, Fert-Bober J, Nigrovic PA, Darrah E, Haque UJ, Lee DM, van Eyk J, Rosen A, Andrade F. Immune-mediated pore-forming pathways induce cellular hypercitrullination and generate citrullinated autoantigens in rheumatoid arthritis. Sci Transl Med 2013;5:209ra150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Escher C, Reiter L, MacLean B, Ossola R, Herzog F, Chilton J, MacCoss MJ, Rinner O. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 2012;12:1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Natale DA, Arighi CN, Blake JA, Bult CJ, Christie KR, Cowart J, D'Eustachio P, Diehl AD, Drabkin HJ, Helfer O, Huang H, Masci AM, Ren J, Roberts NV, Ross K, Ruttenberg A, Shamovsky V, Smith B, Yerramalla MS, Zhang J, AlJanahi A, Çelen I, Gan C, Lv M, Schuster-Lezell E, Wu CH. Protein Ontology: a controlled structured network of protein entities. Nucleic Acids Res 2014;42:D415–D421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods 2012;9:1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matt P, Fu Z, Fu Q, Van Eyk JE. Biomarker discovery: proteome fractionation and separation in biological samples. Physiol Genomics 2008;33:12–17. [DOI] [PubMed] [Google Scholar]
- 29.Kirk JA, Holewinski RJ, Kooij V, Agnetti G, Tunin RS, Witayavanitkul N, de Tombe PP, Gao WD, Van Eyk J, Kass DA. Cardiac resynchronization sensitizes the sarcomere to calcium by reactivating GSK-3beta. J Clin Invest 2014;1:129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White HD. Special instrumentation and techniques for kinetic studies of contractile systems. Methods Enzymol 1982;85:698–708. [DOI] [PubMed] [Google Scholar]
- 31.Tauhata SB, dos Santos DV, Taylor EW, Mooseker MS, Larson RE. High affinity binding of brain myosin-Va to F-actin induced by calcium in the presence of ATP. J Biol Chem 2001;276:39812–39818. [DOI] [PubMed] [Google Scholar]
- 32.Skorzewski R, Sliwinska M, Borys D, Sobieszek A, Moczarewska J. Effect of actin C-terminal modification on tropomyosin isoforms binding and thin filament regulation. Biochim Biophys Acta 2009;2:237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nascimento AA, Cheney RE, Tauhata SB, Larson RE, Mooseker MS. Enzymatic characterization and functional domain mapping of brain myosin-V. J Biol Chem 1996;271:17561–17569. [DOI] [PubMed] [Google Scholar]
- 34.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 2012;11:O111 016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galinska A, Hatch V, Craig R, Murphy AM, Van Eyk JE, Wang CL, Lehman W, Foster DB. The C terminus of cardiac troponin I stabilizes the Ca2+-activated state of tropomyosin on actin filaments. Circ Res 2010;106:705–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eaton BL. Tropomyosin binding to F-actin induced by myosin heads. Science 1976;192:1337–1339. [DOI] [PubMed] [Google Scholar]
- 37.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 2009;37:D412–D416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehman W, Craig R. Tropomyosin and the steric mechanism of muscle regulation. Adv Exp Med Biol 2008;644:95–109. [DOI] [PubMed] [Google Scholar]
- 40.Mijailovich SM, Kayser-Herold O, Li X, Griffiths H, Geeves MA. Cooperative regulation of myosin-S1 binding to actin filaments by a continuous flexible Tm–Tn chain. Eur Biophys J 2012;12:1015–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer S, Kentish JC. The role of troponin C in modulating the Ca2″ sensitivity of mammalian skinned cardiac and skeletal muscle fibres. J Physiol 1994;480:45–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lehman W, Galinska-Rakoczy A, Hatch V, Tobacman LS, Craig R. Structural basis for the activation of muscle contraction by troponin and tropomyosin. J Mol Biol 2009;388:673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Chalovich JM, Marriott G. Structural dynamics of troponin I during Ca2+-activation of cardiac thin filaments: a multi-site Forster resonance energy transfer study. PLoS ONE 2012;7:e50420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szczesna D, Ghosh D, Li Q, Gomes AV, Guzman G, Arana C, Zhi G, Stull JT, Potter JD. Familial hypertrophic cardiomyopathy mutations in the regulatory light chains of myosin affect their structure, Ca2+ binding, and phosphorylation. J Biol Chem 2001;276:7086–7092. [DOI] [PubMed] [Google Scholar]
- 45.Marston SB. How do mutations in contractile proteins cause the primary familial cardiomyopathies? J Cardiovasc Transl Res 2011;4:245–255. [DOI] [PubMed] [Google Scholar]