Abstract
Wilms' tumor-1 protein (WT1) is a transcription factor that can either activate or repress genes to regulate cell growth, apoptosis and differentiation. WT1 can act as either a tumor suppressor or an oncogene. The cellular functions of WT1 are predominantly regulated by its various interacting partners. Recently we have found that WT1 can regulate the fidelity of chromosome segregation through its interaction with the spindle assembly checkpoint protein, Mitotic arrest deficient-2 (MAD2). WT1 delays anaphase entry by inhibiting the ubiquitination activity of the Anaphase promoting complex/cyclosome (APC/C). Our findings have revealed an important role of WT1 in the regulation of mitotic checkpoint and genomic stability.
Keywords: checkpoint, mitosis, Mad2, WT1
Introduction
WT1 is a multifunctional transcription factor that is essential for embryonic development, including the genito-urinary system, sensory system and heart.1-10 WT1 regulates several cellular processes including apoptosis, proliferation, differentiation and mRNA processing.4,11,12 Very recently we showed that WT1 plays an important role in the process of cell division by regulating the spindle/mitotic checkpoint function.13 WT1 has been reported with contrasting roles of a tumor suppressor and an oncogene. There is a general agreement that wild type WT1 likely functions as tumor suppressor during embryonic development, where loss-of-function mutation can cause pediatric cancer of the kidney (Wilms’ tumor). In contrast, overexpression or mutation of WT1 in adult cells leads to tumorigenesis, including cancers of the lung, brain and breast.1,14-16 Indeed, WT1 peptide therapy has shown promising results in targeting several tumors and is currently under clinical trials.17,18 We still do not fully understand that how these diverse roles of WT1 control organogenesis during embryonic growth or contribute to the process of malignant transformation in adults. Thus, identification of a novel role of WT1 in the fundamental cellular process of cell division contributes to our understanding of how WT1 can regulate genomic integrity and chromosomal stability both in embryonic development and in adult tumors.
Mechanism of Mitotic Checkpoint Regulation: Emerging Players
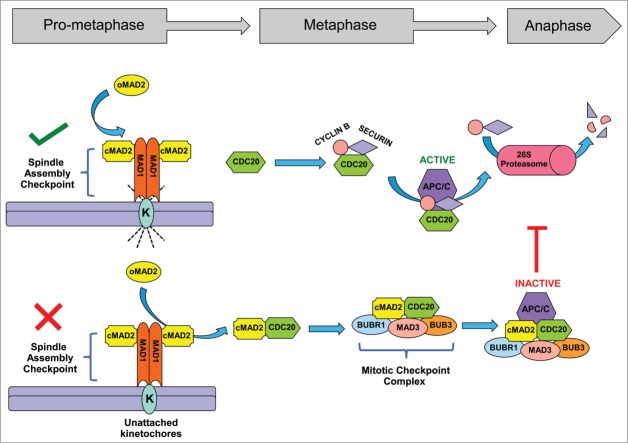
Cell division is a tightly controlled process. A multitude of cellular factors, including signaling proteins and enzymatic modifications work in concert to accomplish the error-proof segregation of chromosomes during mitosis. One of the most critical failsafe mechanisms that operates at the onset of metaphase is the spindle assembly checkpoint (SAC). The SAC complex senses erroneously attached or unattached spindle microtubules at the kinetochores and initiates a chain of events that halts chromosome separation until all the kinetochores are properly bound.19 Of all the essential proteins that directly or indirectly participate in SAC signaling, the contribution of MAD2 and MAD1 proteins are best understood and play key roles in this process. A conformational change in MAD2 from an “open” form (inactive) to a “closed” form (active) acts as a trigger for the initiation of a cascade of events that guides the cell through the process of cell division. At the kinetochore the MAD2-MAD1 complex stimulates the rate of MAD2 conversion from open to closed form, which provides the driving force to generate a diffusible anaphase wait signal, the mitotic checkpoint complex (MCC).20-25 The core components of the MCC machinery comprise MAD2, BUBR1, CDC20, MAD3 and BUB3, which disrupts CDC20-mediated activation of anaphase promoting complex (APC/C) and thus prevents the degradation of SECURIN and CYCLIN B1 that is required for metaphase exit19,21,26 (Fig. 1).
Figure 1.
Spindle assembly and mitotic checkpoints control anaphase entry. The spindle assembly checkpoint (SAC) proteins MAD1 and MAD2 monitor the spindle microtubule-kinetochore attachments. A proper attachment satisfies the spindle checkpoint (green tick), CDC20 can then activate APC/C and promote entry into anaphase. If the SAC detects any unattached or improperly attached kinetochore (K), the spindle checkpoint is ‘unsatisfied’ (red cross). Inactive, Open-MAD2 (O-MAD2) undergoes a conformational change to active, Closed-MAD2 (C-MAD2). The C-MAD2 then binds to CDC20 and along with other proteins MAD3, BUB3, BUBR1 form the mitotic checkpoint complex (MCC). The MCC binds and inhibits the APC/C function and results in metaphase arrest until all the kinetochores are properly attached to the spindle microtubules.
The expression and function of several accessory proteins can influence SAC/MCC signaling. One well-characterized example is the p31 (comet) protein, which competes with MAD1 and CDC20 to bind with MAD2 and results in abrogation of SAC activation. Overexpression of p31 (comet) leads to a failure of MAD2-dependent checkpoint function, early anaphase entry, and subsequent chromosome-segregation defects.20,26-29
We have identified WT1 as a novel player in the mitotic checkpoint-signaling pathway.13 This is mediated by a direct interaction between WT1 and MAD2. WT1 preferentially binds to the active C-MAD2 conformer and leads to stabilization of the interaction between MAD2 and the checkpoint components. The MAD2-binding region is present in all the 4 major isoforms of WT1. Expression of WT1 prolongs the MCC-mediated inhibition of APC/C and further delays mitotic exit. Although WT1 can also interact with inactive O-MAD2 it does not compete with either MAD1 or CDC20 for the same binding site in MAD2, and it is not yet clear if WT1 binding influences the conformational stability of MAD2. It is also not clear whether WT1 stays bound to MAD2 in both the SAC and the MCC complexes and if/when it dissociates from the complex (Fig. 2). It will also be interesting to determine the role of other MAD2-binding proteins such as p31 (comet) in modulating WT1-mediated checkpoint function.
Figure 2.
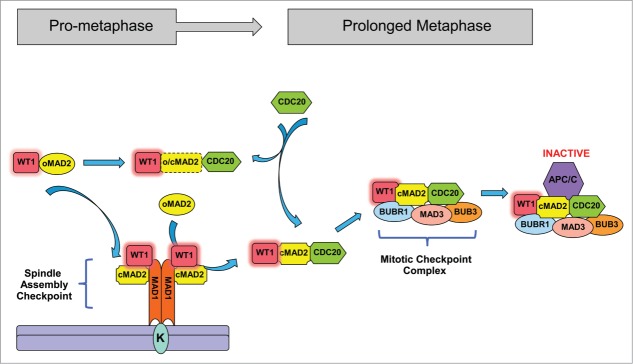
WT1 interacts with MAD2 and delays anaphase entry. WT1 interacts with MAD2 during mitosis. WT1 amplifies SAC signaling by promoting MAD2 and MAD1 association. Presence of WT1 stabilizes the MCC-mediated inhibition of APC/C and thereby prolongs the metaphase stage of the cell cycle.
Deregulation of the checkpoint pathway by either mis-expression of crucial factors or by hyper/hypo-stimulation of SAC signaling leads to chromosome-segregation defects and eventually chromosomal instability (CIN).14,30-33 How WT1 regulates the mitotic checkpoint function during cancer is not known. The relative expression and functional regulation of critical checkpoint proteins can significantly influence how aggressively tumor cells proliferate and accumulate chromosomal defects with each division. The role of WT1 in regulating the mitotic checkpoint will be important to consider when targeting WT1 in cancer therapies, in addition to the functions of WT1 in embryonic development.
WT1- p53- MAD2 Axis
Tumor suppressor proteins such as p53 and retinoblastoma (Rb) guard genomic integrity by stringently regulating cell cycle progression. They are active at different cell cycle checkpoints and control the expression of several genes with products involved in the cell cycle (Table 1). p53 or Rb loss-of-function mutations result in checkpoint failure and uncontrolled cell division that leads to tumorous growth. Most tumor suppressors including p53, Rb, ARF regulate the cell cycle by controlling the expression of proteins crucial for mitotic checkpoint function.15,34-39 The tumor suppressor, breast cancer gene-1 (BRCA1) directly or indirectly regulates several key steps of the cell cycle. Loss-of-function mutation of BRCA1 causes abnormalities in S-phase, G2/M and the spindle assembly checkpoints that promote the development of breast and ovarian cancers.40 Interestingly, multiple tumor suppressors such as BRCA1, p53 and WT1 modulate IGF1R expression during cell growth, postnatal development and oncogenesis. Deregulated IGF1R results in defective cell division and chromosomal instability.40,41 Studies have shown that MAD2 expression is negatively regulated by tumor suppressors p53 and Rb and overexpression of MAD2 is observed in cells lacking functional Rb and p53. Inactivation of p53/Rb tumor suppressor pathways in combination with MAD2 overexpression promotes chromosomal instability and tumorigenesis.31,37,42,43 WT1 also regulates genes engaged in cell cycle progression such as CYCLIN E. WT1 interacts with the tumor suppressors p53 and p73 through its zinc finger regions. Interaction of WT1 and p53 mutually influences the transcriptional function of each other. Expression of the GADD45a gene is regulated by WT1 in both p53-dependent and -independent pathways in response to genotoxic agents.1,44-46 Recently it was found that certain gain-of-function mutations in the WT1 gene could activate several cell cycle genes and promote cell proliferation.47 Additionally, we have shown that WT1 can also directly bind to MAD2 and regulate the fidelity of cell division via a transcription-independent pathway.13 It is not known whether other tumor suppressor pathway(s) negatively or positively affect this function of WT1. Regulation of MAD2 in Wilms’ tumor and other WT1-dependent adult tumors in presence or absence of functional p53 will shed light on the factors driving chromosomal instability and cancer progression.
Table 1.
Tumor suppressors and cell cycle regulation
| Tumor Suppressor | Gene Target | Protein-protein interaction | Function |
|---|---|---|---|
| P53 | p21, GADD45 | G1/S checkpoint | |
| Rb | Cyclin A, Cyclin E, Cdk2 | E2F | G1/S checkpoint |
| BRCA1 | P21 | Cyclin D1, Cdk2, Cdk4 | S-phase checkpoint, G2/M checkpoint, spindle checkpoint |
| WT1 | Cyclin E | MAD2 | Spindle checkpoint |
| ARF | Aurora B | Mitotic checkpoint |
Role of WT1-interacting Partners in Mitotic Checkpoint Regulation
The transcription regulatory function of WT1 is fine-tuned by its interacting partners that include general transcription factors, activators, repressors, and chromatin modifying enzymes.14,15,45 Brain acid soluble protein 1 (BASP1), is a significant binding partner of WT1. BASP1 associates with WT1 at the promoters of WT1-target genes and represses their transcription.48,49 Different subcellular localization pattern and relative abundance of WT1 cofactors determine the binding partner preference. Although WT1 and BASP1 predominantly interact at the gene promoters and regulate transcriptional output, it is also possible that BASP1 influences WT1-mediated mitotic checkpoint function and affects the fidelity of chromosome segregation during cell cycle. In several cell lines BASP1 is known to have significant cytoplasmic localization. Interestingly, immunofluorescence analysis of BASP1 during different stages of mitosis in WiT49 cells showed its predominant association with condensed metaphase chromosomes (Fig. 3). Such localization pattern of BASP1 is reminiscent of mitotic checkpoint proteins and suggests a possible role for BASP1 in checkpoint regulation. It thus remains to be tested whether BASP1 affects the direct binding of WT1 with MAD2 and regulates the checkpoint signaling in normal and in malignant conditions.
Figure 3.
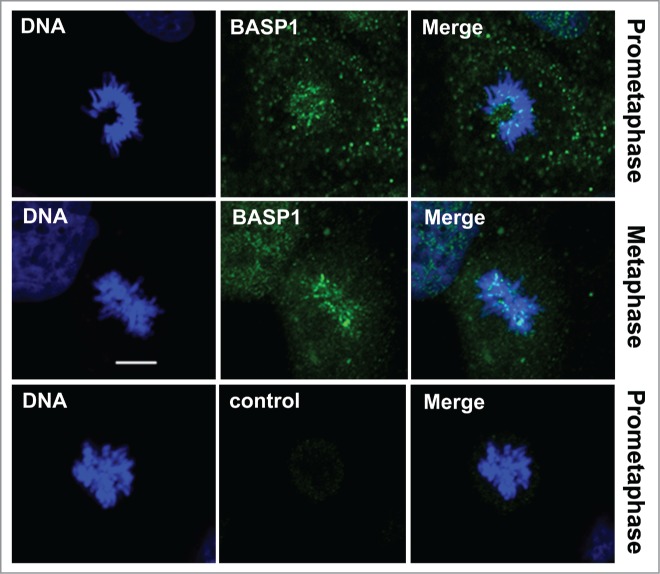
BASP1 colocalizes with metaphase chromosomes. Immunofluorescence analysis was carried out in WiT49 cells with anti-BASP1 antibodies at the prometaphase and metaphase stages of mitosis. Anti-GST antibody was used as a negative control. DNA was stained with Hoechst. Scale bar is 10 microns.
Earlier work from our lab showed that WT1 expression contributes to protection of cells from apoptosis.50 Treatment of cells with cytotoxic drugs stimulates the proteolytic processing of WT1 by HtrA2 and promotes apoptosis.50,51 We analyzed the efficiency of WT1 processing and apoptosis that is induced by several cytotoxic agents including Doxorubicin, Etoposide, Topotecan and Vincristine (Fig. 4). Generally, the reduction in the level of WT1 following treatment with the cytotoxic agents is congruent with the level of cell death observed (as indicated by viable cell count and PARP cleavage). Strikingly, however, we observed that Vincristine did not stimulate WT1 processing, but still significantly induced apoptosis. Vincristine elicits its effects through a distinct mechanism when compared to the other drugs used in this analysis. It inhibits tubulin polymerisation, while the other agents either directly, or indirectly, induce DNA damage. Vincristine is the choice first-line agent in Wilms’ tumor and is generally an effective treatment.52 However, more aggressive tumors such as anaplastic Wilms’ tumor are treated with a combination of Vincristine and Doxorubicin. It is clear from the data in Figure 4 that Doxorubicin is a highly effective inducer of WT1 degradation and also of apoptosis. Anaplastic Wilms’ tumors express high levels of wild-type WT1 that likely acts as an oncogene.52 Our data suggest that Vincristine may be a less effective agent against anaplastic Wilms’ tumor because it does not stimulate the degradation of WT1. Thus, not all cytotoxic agents stimulate WT1 processing. As stated above, WT1 can act as both a tumor suppressor and an oncogene and so effective therapies will need to enhance WT1 activity in the former and ablate it in the latter.
Figure 4.
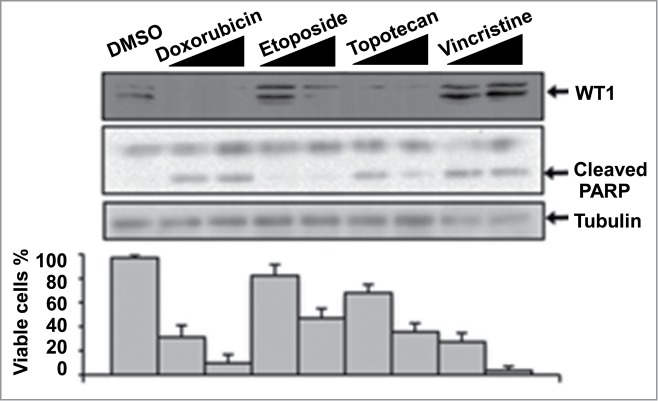
The effect of cytotoxic drugs on WT1 and apoptosis in MCF7 cells. Cells were treated with 1 μg/ml or 5 μg/ml of each drug for 20 hours and whole cell extracts prepared, then immunoblotted with the antibodies indicated. To measure cell viability, cells were treated as above, stained with trypan blue and the stained/excluded cells counted and expressed graphically as percentage viable.
It is interesting that Vincristine, which exerts its effects on the mitotic spindle, does not induce proteolysis of WT1 as part of its mechanism of action. Indeed, we have found that other proapoptotic drugs that function at the mitotic spindle (Nocodazole and Paclitaxel) also do not stimulate WT1 processing (data not shown). It is interesting to speculate that mitosis-specific functions of WT1 might protect WT1 from degradation. Indeed, the level of MAD2 profoundly influences the response of cancer cells to therapy and prognosis.52-55 Reduced expression of MAD2 protein is known to abrogate mitotic checkpoint function and promote chromosomal instability in certain types of human cancers. Ectopic overexpression of MAD2 increased the sensitivity of a checkpoint-defective nasopharyngeal carcinoma cell line to Vincristine treatment and resulted in G2/M cell cycle arrest.56 Analysis of the efficacy of these therapeutic agents in the presence or absence of WT1 in a MAD2 overexpression background will be useful to reveal its potential as a prognostic indicator and also in drug resistance.
Post-translational Modification of WT1 and Mitotic Checkpoint Function
WT1 is regulated by post-translational modification, such as phosphorylation and sumoylation.57,58 Several studies have indicated the potential implications of such modifications in the cellular localization and functions of WT1. Post-translational modifications, in combination with the binding partner preference, could significantly influence the tumor suppressor and oncogenic properties of WT1 during embryonic development and cellular homeostasis.45,57-59 Compared to the sumoylation of WT1, phosphorylation is relatively better understood. WT1 is targeted by Protein kinases A and C (PKA and PKC). Phosphorylation of WT1 at Ser-365 and Ser-393 within the zinc finger domain negatively influences the DNA-binding and transcriptional activation functions of WT1. Additionally phosphorylated WT1 showed cytoplasmic retention.58 The exact molecular function of the cytoplasmic pool of phosphorylated WT1 is not known. It will therefore be important to determine if the phosphorylation status of WT1 influences the mitotic checkpoint signaling.
Phosphorylation-dephosphorylation switching is one of the crucial regulators of the key steps of SAC/MCC signaling. Dynamic activities of different mitotic kinases and phosphatases control the binding of checkpoint proteins onto the kinetochores and monitor the microtubule attachment errors. Kinases such as Aurora B and Mps1 phosphorylate multiple microtubule binding proteins and ensure an error-free cell cycle progression.60
Protein kinase C is activated by tumor-promoting phorbol esters. Hyper-activated PKC can aberrantly phosphorylate transcription factors leading to overexpression of oncogenes and hence aid cancer progression.61-63 It would be of interest to study a potential role for PKC in cancers where WT1 is overexpressed and to determine if phosphorylation of WT1 affects the mitotic checkpoint function and promotes CIN. PKC-mediated phosphorylation of WT1 could potentially affect its association with MAD2. This regulation could be an important factor to determine the net effect of WT1-mediated transcription regulation and cell division control during development and carcinogenesis. Hence the phosphorylation status of WT1 at a given time could be an indicator of cell proliferation and survival.
Conclusions
WT1 plays crucial roles in several cellular processes including transcription regulation, apoptosis and cell survival. WT1 can function either as a tumor suppressor during embryogenesis or an oncogene driving tumor growth in adults. Identification of WT1 as direct regulator of mitotic checkpoint further highlights its importance in fundamental processes of cellular homeostasis. Future studies aimed at understanding the role of WT1 in checkpoint failure and chromosomal instability during cancer would be helpful in designing better anti-cancer therapies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Hohenstein P, Hastie ND. The many facets of the Wilms' tumour gene, WT1. Hum Mol Genet 2006; 15 Spec No 2:R196-201; PMID:16987884; http://dx.doi.org/ 10.1093/hmg/ddl196 [DOI] [PubMed] [Google Scholar]
- 2. Discenza MT, Pelletier J. Insights into the physiological role of WT1 from studies of genetically modified mice. Physiol Genomics 2004; 16:287-300; PMID:14966251; http://dx.doi.org/ 10.1152/physiolgenomics.00164.2003 [DOI] [PubMed] [Google Scholar]
- 3. Kreidberg JA. WT1 and kidney progenitor cells. Organogenesis 2010; 6:61-70; PMID:20885852; http://dx.doi.org/ 10.4161/org.6.2.11928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roberts SG. Transcriptional regulation by WT1 in development. Curr Opin Genet Dev 2005; 15:542-7; PMID:16099645; http://dx.doi.org/ 10.1016/j.gde.2005.08.004 [DOI] [PubMed] [Google Scholar]
- 5. Rudat C, Kispert A. Wt1 and epicardial fate mapping. Circ Res 2012; 111:165-9; PMID:22693350; http://dx.doi.org/ 10.1161/CIRCRESAHA.112.273946 [DOI] [PubMed] [Google Scholar]
- 6. Ozdemir DD, Hohenstein P. Wt1 in the kidney–a tale in mouse models. Pediatr Nephrol 2014; 29:687-93; PMID:24240471; http://dx.doi.org/ 10.1007/s00467-013-2673-7 [DOI] [PubMed] [Google Scholar]
- 7. Scholz H, Kirschner KM. A role for the Wilms' tumor protein WT1 in organ development. Physiology 2005; 20:54-9; PMID:15653840; http://dx.doi.org/ 10.1152/physiol.00048.2004 [DOI] [PubMed] [Google Scholar]
- 8. Gao Y, Toska E, Denmon D, Roberts SG, Medler KF. WT1 regulates the development of the posterior taste field. Development 2014; 141:2271-8; PMID:24803588; http://dx.doi.org/ 10.1242/dev.105676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang XN, Li ZS, Ren Y, Jiang T, Wang YQ, Chen M, Zhang J, Hao JX, Wang YB, Sha RN, et al. The Wilms tumor gene, Wt1, is critical for mouse spermatogenesis via regulation of sertoli cell polarity and is associated with non-obstructive azoospermia in humans. PLoS Genet 2013; 9:e1003645; PMID:23935527; http://dx.doi.org/ 10.1371/journal.pgen.1003645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wagner N, Wagner KD, Hammes A, Kirschner KM, Vidal VP, Schedl A, Scholz H. A splice variant of the Wilms' tumour suppressor Wt1 is required for normal development of the olfactory system. Development 2005; 132:1327-36; PMID:15716344; http://dx.doi.org/ 10.1242/dev.01682 [DOI] [PubMed] [Google Scholar]
- 11. Morrison AA, Viney RL, Ladomery MR. The post-transcriptional roles of WT1, a multifunctional zinc-finger protein. Biochim Biophy Acta 2008; 1785:55-62; PMID:17980713 [DOI] [PubMed] [Google Scholar]
- 12. Hartkamp J, Roberts SG. The role of the Wilms' tumour-suppressor protein WT1 in apoptosis. Biochem Soc Trans 2008; 36:629-31; PMID:18631130; http://dx.doi.org/ 10.1042/BST0360629 [DOI] [PubMed] [Google Scholar]
- 13. Shandilya J, Toska E, Richard DJ, Medler KF, Roberts SG. WT1 interacts with MAD2 and regulates mitotic checkpoint function. Nat Commun 2014; 5:4903; PMID:25232865; http://dx.doi.org/ 10.1038/ncomms5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chau YY, Hastie ND. The role of Wt1 in regulating mesenchyme in cancer, development, and tissue homeostasis. Trends Genet 2012; 28:515-24; PMID:22658804; http://dx.doi.org/ 10.1016/j.tig.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 15. Huff V. Wilms' tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer 2011; 11:111-21; PMID:21248786; http://dx.doi.org/ 10.1038/nrc3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang L, Han Y, Suarez Saiz F, Minden MD. A tumor suppressor and oncogene: the WT1 story. Leukemia 2007; 21:868-76; PMID:17361230 [DOI] [PubMed] [Google Scholar]
- 17. Owen C, Fitzgibbon J, Paschka P. The clinical relevance of Wilms Tumour 1 (WT1) gene mutations in acute leukaemia. Hematol Oncol 2010; 28:13-9; PMID:20013787 [DOI] [PubMed] [Google Scholar]
- 18. Sugiyama H. WT1 (Wilms' tumor gene 1): biology and cancer immunotherapy. Japan J Clin Oncol 2010; 40:377-87; PMID:20395243; http://dx.doi.org/ 10.1093/jjco/hyp194 [DOI] [PubMed] [Google Scholar]
- 19. Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol 2012; 22:R966-80; PMID:23174302; http://dx.doi.org/ 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 20. Fava LL, Kaulich M, Nigg EA, Santamaria A. Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J 2011; 30:3322-36; PMID:21772247; http://dx.doi.org/ 10.1038/emboj.2011.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han JS, Holland AJ, Fachinetti D, Kulukian A, Cetin B, Cleveland DW. Catalytic assembly of the mitotic checkpoint inhibitor BubR1-Cdc20 by a Mad2-induced functional switch in Cdc20. Mol Cell 2013; 51:92-104; PMID:23791783; http://dx.doi.org/ 10.1016/j.molcel.2013.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luo X, Yu H. Protein metamorphosis: the two-state behavior of Mad2. Structure 2008; 16:1616-25; PMID:19000814; http://dx.doi.org/ 10.1016/j.str.2008.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell 2007; 131:730-43; PMID:18022367; http://dx.doi.org/ 10.1016/j.cell.2007.08.049 [DOI] [PubMed] [Google Scholar]
- 24. Schuyler SC, Wu YF, Kuan VJ. The Mad1-Mad2 balancing act–a damaged spindle checkpoint in chromosome instability and cancer. J Cell Sci 2012; 125:4197-206; PMID:23093575; http://dx.doi.org/ 10.1242/jcs.107037 [DOI] [PubMed] [Google Scholar]
- 25. Skinner JJ, Wood S, Shorter J, Englander SW, Black BE. The Mad2 partial unfolding model: regulating mitosis through Mad2 conformational switching. J Cell Biol 2008; 183:761-8; PMID:19029339; http://dx.doi.org/ 10.1083/jcb.200808122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol Cell 2011; 44:710-20; PMID:22152475; http://dx.doi.org/ 10.1016/j.molcel.2011.11.014 [DOI] [PubMed] [Google Scholar]
- 27. Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci 2011; 124:3905-16; PMID:22100920; http://dx.doi.org/ 10.1242/jcs.093286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang M, Li B, Tomchick DR, Machius M, Rizo J, Yu H, Luo X. p31comet blocks Mad2 activation through structural mimicry. Cell 2007; 131:744-55; PMID:18022368; http://dx.doi.org/ 10.1016/j.cell.2007.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yun M, Han YH, Yoon SH, Kim HY, Kim BY, Ju YJ, Kang CM, Jang SH, Chung HY, Lee SJ, et al. p31comet Induces cellular senescence through p21 accumulation and Mad2 disruption. Mol Cancer Res 2009; 7:371-82; PMID:19276188; http://dx.doi.org/ 10.1158/1541-7786.MCR-08-0056 [DOI] [PubMed] [Google Scholar]
- 30. Bakhoum SF, Compton DA. Chromosomal instability and cancer: a complex relationship with therapeutic potential. J Clin Invest 2012; 122:1138-43; PMID:22466654; http://dx.doi.org/ 10.1172/JCI59954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc Natl Acad Sci U S A 2005; 102:11296-301; PMID:16055552; http://dx.doi.org/ 10.1073/pnas.0505053102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep 2012; 13:501-14; PMID:22565320; http://dx.doi.org/ 10.1038/embor.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rao CV, Yamada HY, Yao Y, Dai W. Enhanced genomic instabilities caused by deregulated microtubule dynamics and chromosome segregation: a perspective from genetic studies in mice. Carcinogenesis 2009; 30:1469-74; PMID:19372138; http://dx.doi.org/ 10.1093/carcin/bgp081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hong B, van den Heuvel AP, Prabhu VV, Zhang S, El-Deiry WS. Targeting tumor suppressor p53 for cancer therapy: strategies, challenges and opportunities. Curr Drug Targets 2014; 15:80-9; PMID:24387333; http://dx.doi.org/ 10.2174/1389450114666140106101412 [DOI] [PubMed] [Google Scholar]
- 35. Britigan EM, Wan J, Zasadil LM, Ryan SD, Weaver BA. The ARF tumor suppressor prevents chromosomal instability and ensures mitotic checkpoint fidelity through regulation of Aurora B. Mol Biol Cell 2014; 25:2761-73; PMID:25057018; http://dx.doi.org/ 10.1091/mbc.E14-05-0966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dick FA, Rubin SM. Molecular mechanisms underlying RB protein function. Nat Rev Mol Cell Biol 2013; 14:297-306; PMID:23594950; http://dx.doi.org/ 10.1038/nrm3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Malumbres M. Oncogene-induced mitotic stress: p53 and pRb get mad too. Cancer Cell 2011; 19:691-2; PMID:21665141; http://dx.doi.org/ 10.1016/j.ccr.2011.05.023 [DOI] [PubMed] [Google Scholar]
- 38. van Deursen JM. Rb loss causes cancer by driving mitosis mad. Cancer Cell 2007; 11:1-3; PMID:17222786; http://dx.doi.org/ 10.1016/j.ccr.2006.12.006 [DOI] [PubMed] [Google Scholar]
- 39. Giacinti C, Giordano A. RB and cell cycle progression. Oncogene 2006; 25:5220-7; PMID:16936740; http://dx.doi.org/ 10.1038/sj.onc.1209615 [DOI] [PubMed] [Google Scholar]
- 40. Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucl Acids Res 2006; 34:1416-26; PMID:16522651; http://dx.doi.org/ 10.1093/nar/gkl010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Werner H, Sarfstein R. Transcriptional and epigenetic control of IGF1R gene expression: implications in metabolism and cancer. Growth Horm IGF Res 2014; 24:112-8; http://dx.doi.org/ 10.1016/j.ghir.2014.03.006 [DOI] [PubMed] [Google Scholar]
- 42. Chi YH, Ward JM, Cheng LI, Yasunaga J, Jeang KT. Spindle assembly checkpoint and p53 deficiencies cooperate for tumorigenesis in mice. Int J Cancer 2009; 124:1483-9; PMID:19065665; http://dx.doi.org/ 10.1002/ijc.24094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell 2011; 19:701-14; PMID:21665145; http://dx.doi.org/ 10.1016/j.ccr.2011.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Al-Hussain T, Ali A, Akhtar M. Wilms tumor: an update. Adv Anatomic Pathol 2014; 21:166-73; PMID:24713986; http://dx.doi.org/ 10.1097/PAP.0000000000000017 [DOI] [PubMed] [Google Scholar]
- 45. Toska E, Roberts SG. Mechanisms of transcriptional regulation by WT1 (Wilms' tumour 1). Biochem J 2014; 461:15-32; PMID:24927120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Johnson D, Hastwell PW, Walmsley RM. The involvement of WT1 in the regulation of GADD45a in response to genotoxic stress. Mutagenesis 2013; 28:393-9; PMID:23476008; http://dx.doi.org/ 10.1093/mutage/get015 [DOI] [PubMed] [Google Scholar]
- 47. Busch M, Schwindt H, Brandt A, Beier M, Gorldt N, Romaniuk P, Toska E, Roberts S, Royer HD, Royer-Pokora B. Classification of a frameshift/extended and a stop mutation in WT1 as gain-of-function mutations that activate cell cycle genes and promote Wilms tumour cell proliferation. Hum Mol Genet 2014; 23:3958-74; PMID:24619359; http://dx.doi.org/ 10.1093/hmg/ddu111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Toska E, Campbell HA, Shandilya J, Goodfellow SJ, Shore P, Medler KF, Roberts SG. Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell Rep 2012; 2:462-9; PMID:22939983; http://dx.doi.org/ 10.1016/j.celrep.2012.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Toska E, Shandilya J, Goodfellow SJ, Medler KF, Roberts SG. Prohibitin is required for transcriptional repression by the WT1-BASP1 complex. Oncogene 2014; 33:5100-8; PMID:24166496; http://dx.doi.org/ 10.1038/onc.2013.447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hartkamp J, Carpenter B, Roberts SG. The Wilms' tumor suppressor protein WT1 is processed by the serine protease HtrA2/Omi. Mol Cell 2010; 37:159-71; PMID:20122399; http://dx.doi.org/ 10.1016/j.molcel.2009.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hartkamp J, Roberts SG. HtrA2, taming the oncogenic activities of WT1. Cell Cycle 2010; 9:2508-14; PMID:20543571; http://dx.doi.org/ 10.4161/cc.9.13.12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kondo S, Kaji M. Overexpression of MAD2 predicts clinical outcome in primary lung cancer patients. Lung Cancer 2011; 74:124-31; PMID:21376419; http://dx.doi.org/ 10.1016/j.lungcan.2011.01.025 [DOI] [PubMed] [Google Scholar]
- 53. McGrogan BT, Gilmartin B, Carney DN, McCann A. Taxanes, microtubules and chemoresistant breast cancer. Biochim Biophys Acta 2008; 1785:96-132; PMID:18068131 [DOI] [PubMed] [Google Scholar]
- 54. Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 2010; 464:436-40; PMID:20173739; http://dx.doi.org/ 10.1038/nature08803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu L, Liu S, Guo W, Zhang B, Liang Y, Feng Q. Upregulation of Mad2 facilitates in vivo and in vitro osteosarcoma progression. Oncol Rep 2012; 28:2170-6; PMID:22992948 [DOI] [PubMed] [Google Scholar]
- 56. Wang X, Jin DY, Wong HL, Feng H, Wong YC, Tsao SW. MAD2-induced sensitization to vincristine is associated with mitotic arrest and Raf/Bcl-2 phosphorylation in nasopharyngeal carcinoma cells. Oncogene 2003; 22:109-16; PMID:12527913; http://dx.doi.org/ 10.1038/sj.onc.1206069 [DOI] [PubMed] [Google Scholar]
- 57. Smolen GA, Vassileva MT, Wells J, Matunis MJ, Haber DA. SUMO-1 modification of the Wilms' tumor suppressor WT1. Cancer Res 2004; 64:7846-51; PMID:15520190; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-1502 [DOI] [PubMed] [Google Scholar]
- 58. Ye Y, Raychaudhuri B, Gurney A, Campbell CE, Williams BR. Regulation of WT1 by phosphorylation: inhibition of DNA binding, alteration of transcriptional activity and cellular translocation. EMBO J 1996; 15:5606-15; PMID:8896454 [PMC free article] [PubMed] [Google Scholar]
- 59. Green LM, Wagner KJ, Campbell HA, Addison K, Roberts SG. Dynamic interaction between WT1 and BASP1 in transcriptional regulation during differentiation. Nucleic Acids Res 2009; 37:431-40; PMID:19050011; http://dx.doi.org/ 10.1093/nar/gkn955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Funabiki H, Wynne DJ. Making an effective switch at the kinetochore by phosphorylation and dephosphorylation. Chromosoma 2013; 122:135-58; PMID:23512483; http://dx.doi.org/ 10.1007/s00412-013-0401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yamasaki T, Takahashi A, Pan J, Yamaguchi N, Yokoyama KK. Phosphorylation of Activation Transcription Factor-2 at Serine 121 by Protein Kinase C Controls c-Jun-mediated Activation of Transcription. J Biol Chem 2009; 284:8567-81; PMID:19176525; http://dx.doi.org/ 10.1074/jbc.M808719200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mochly-Rosen D, Das K, Grimes KV. Protein kinase C, an elusive therapeutic target? Nat Rev Drug Discov 2012; 11:937-57; PMID:23197040; http://dx.doi.org/ 10.1038/nrd3871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Toton E, Ignatowicz E, Skrzeczkowska K, Rybczynska M. Protein kinase Cepsilon as a cancer marker and target for anticancer therapy. Pharmacol Rep 2011; 63:19-29; PMID:21441608; http://dx.doi.org/ 10.1016/S1734-1140(11)70395-4 [DOI] [PubMed] [Google Scholar]