Abstract
DYT1 dystonia, the most common inherited form of primary dystonia, is a neurodevelopmental disease caused by a dominant mutation in TOR1A. This mutation (‘ΔE’) removes a single glutamic acid from the encoded protein, torsinA. The effects of this mutation, at the molecular and circuit levels, and the reasons for its neurodevelopmental onset, remain incompletely understood. To uniquely address key questions of disease pathogenesis, we generated a conditional Tor1a knock-in allele that is converted from wild-type to DYT1 mutant (‘induced’ ΔE: Tor1ai-ΔE), following Cre recombination. We used this model to perform a gene dosage study exploring the effects of the ΔE mutation at the molecular, neuropathological and organismal levels. These analyses demonstrated that ΔE-torsinA is a hypomorphic allele and showed no evidence for any gain-of-function toxic properties. The unique capabilities of this model also enabled us to test a circuit-level hypothesis of DYT1 dystonia, which predicts that expression of the DYT1 genotype (Tor1aΔE/+) selectively within hindbrain structures will produce an overtly dystonic animal. In contrast to this prediction, we find no effect of this anatomic-specific expression of the DYT1 genotype, a finding that has important implications for the interpretation of the human and mouse diffusion tensor-imaging studies upon which it is based. These studies advance understanding of the molecular effects of the ΔE mutation, challenge current concepts of the circuit dysfunction that characterize the disease and establish a powerful tool that will be valuable for future studies of disease pathophysiology.
Introduction
Dystonia is a prolonged abnormal involuntary movement that typically causes twisting or abnormal postures. Current treatments are empiric and only partially effective, and our limited knowledge of disease pathogenesis and pathophysiology has slowed the development of targeted therapies. Although dystonia often results from neuronal injury or disease in the context of additional neurological symptoms (‘secondary’ dystonia), several inherited forms cause isolated dystonia without associated neurological findings (‘primary’ dystonia). Studies of genetic forms of primary dystonia have advanced understanding of dystonia biology because of the experimental power of mouse genetics and ability to link molecular defects, cellular and circuit dysfunction and neurological phenotypes with abnormal behaviors.
The most common form of inherited primary dystonia, DYT1, is caused by an in-frame 3 bp deletion within exon 5 of the gene TOR1A (OMIM: 605204) (1). This mutation (‘ΔE’) results in the loss of a single glutamic acid from the encoded protein torsinA. One effect of the ΔE mutation is to impair normal torsinA function. Germline homozygous knock-in mice (Tor1aΔE/ΔE) phenocopy the lethality and nuclear envelope (NE) abnormalities of Tor1a null mice (2). ΔE-torsinA retains some function, however, as isolated central nervous system (CNS) expression of this allele (Nestin Cre+ Tor1aFLX/ΔE) rescues the early post-natal lethality observed when Tor1a is deleted from the CNS (Nestin Cre+ Tor1aFLX/FLX) (3). Consistent with these findings, biochemical studies indicate that the torsinA-interacting proteins LAP1 and LULL1 (4,5) are essential for the torsinA ATPase activity, but only very weakly activate the mutant protein, ΔE-torsinA.
Mutation gain-of-function (GOF) and loss-of-function (LOF) effects are not mutually exclusive (6,7). Several features of DYT1 dystonia and torsinA biology are consistent with ΔE-mediated GOF effects. DYT1 dystonia is dominantly inherited, and the ΔE mutation is the only clearly identified pathogenic mutation—a scenario typical of GOF mutations. Re-localization of torsinA from the endoplasmic reticulum to the nuclear membrane is an effect of the ΔE mutation, which depends on the association with the protein SUN1 (8). ΔE-torsinA is found within perinuclear inclusions invitro, and similar inclusions have been reported in DYT1 subject post-mortem tissue (9). Determining whether these molecular features of ΔE-torsinA contribute to adverse effects in vivo (e.g. for neurological function or histopathology) is critical for conceptualizing novel therapeutics, as GOF and LOF effects would necessitate distinct approaches.
Defining the anatomic circuits disrupted by ΔE-torsinA is also critical for future development of therapies, as it would help focus efforts on key brain regions or cell types. Although several lines of evidence implicate the striatum as the major site of dysfunction in dystonia (reviewed in 10,11), an emerging literature suggests that the cerebellum is also an important region, and aberrant communication between cerebellum and forebrain structures is implicated in disease pathogenesis (12–15). Only 30% of DYT1 mutation carriers develop dystonic symptoms. Diffusion tensor-imaging (DTI) magnetic resonance imaging studies comparing ‘manifesting’ and ‘non-manifesting’ DYT1 mutation carriers show that both subject groups exhibit cerebellothalamic white matter tract abnormalities (16,17). Only non-manifesting carriers have an additional thalamocortical tract defect, and this ‘second hit’ is hypothesized to block the effect of the cerebellothalamic lesion (17). Germline Tor1aΔE/+ mice that mimic the human DYT1 genotype do not develop abnormal twisting movements and exhibit the ‘non-manifesting’ carrier DTI signature (18). These findings predict that ‘selective’ hindbrain expression of the DYT1 genotype at endogenous levels would produce an overtly dystonic model, but the mouse genetic reagents necessary to test this prediction do not exist.
To address these molecular- and circuit-level questions of disease pathogenesis, we generated a novel line of conditional DYT1 knock-in mice that allowed us to pursue a series of in vivo studies not possible with existing models (Table 1). These mice express wild-type torsinA until ‘induction’ by Cre recombination, which swaps out a floxed wild-type Tor1a exon 5 and brings a previously silent downstream ΔE-containing exon 5 into frame. This mutant allowed us to pursue a gene dosage study in vivo to rigorously test for organismal GOF effects of ΔE-torsinA. We hypothesized that if the ΔE mutation conferred meaningful GOF toxic effects, more severe or novel phenotypes would emerge as the number of ΔE-Tor1a knock-in alleles increased. Conversely, increasing levels of a solely hypomorphic (LOF) torsinA molecule might, paradoxically, ameliorate ΔE-torsinA-mediated phenotypes. Analyses of the growth, behavioral and histopathological characteristics of these mice demonstrated that increased dosage of the ΔE-Tor1a allele ‘suppressed’ ΔE-torsinA-mediated phenotypes, suggesting that the ΔE mutation exerts an exclusive LOF effect. These mice also enabled us to test the pathophysiological importance of expressing the DYT1 genotype selectively within hindbrain structures; against predictions of the ‘two hit’ model (30), these mice did not exhibit any overt twisting movements. Considered together, these in vivo studies advance knowledge of the molecular and circuit abnormalities underlying primary dystonia and establish a unique genetic tool valuable for future studies of dystonia pathogenesis and pathophysiology.
Table 1.
Tor1a rodent models
| Rodent model | Viability/growth | Behavior | Histology |
|---|---|---|---|
| Transgenic models overexpressing human ΔE-TOR1A | |||
| Rat neuron-specific enolase promotor (19) | Normal | Incomplete penetrance; limb clasping, hyperkinesia | Ubiquitin, torsinA and lamin-positive brainstem neuron inclusion bodies |
| Human CMV promoter (20) | Not reported | Rotarod learning deficit | Normal |
| Mouse prion protein promoter (21) | Not reported | Rotarod deficit; hyperkinesia | NE structural abnormalities |
| Tyrosine hydroxylase promoter (22) | Normal | Beam walking deficits | Not reported |
| Human Tor1a promoter (23) | Viable; retarded post-natal growth | Progressive hind limb clasping, gait abnormalities | Perinuclear torsinA staining, ultrastructural NE defects |
| Germline manipulation of Tor1a | |||
| Tor1aΔE/+ (24) | Not reported | Beam walking deficits and hyperkinesia in males | Increased brainstem torsinA and ubiquitin staining |
| Tor1aΔE/ΔE and Tor1a−/− (2) | Early post-natal lethality | N/A | Neuronal-specific NE budding |
| Tor1a knockdown (25) | Viable; growth not reported | Beam walking deficits and hyperkinesia in males | Not reported |
| Tor1aΔE/+ (26) | Not reported | Normal | Not reported |
| Conditional deletion of Tor1a | |||
| Emx1 Cre (excitatory cortical neurons) (27) | Viable; growth not reported | Beam walking deficits and hyperkinesia | Normal |
| Pcp2 Cre (purkinje cells) (28) | Not reported | Not reported | Purkinje cell morphological abnormalities |
| Rgs9 Cre (medium spiny neurons) (29) | Not reported | Beam walking deficits | Normal |
| Nestin Cre (neural progenitors) (3) | Post-natal lethality; retarded post-natal growth | Limb clasping, abnormal twisting postures | Selective degeneration of sensorimotor regions, perinuclear ubiquitin inclusions, reactive gliosis |
| Nestin Cre-selective expression of ΔE-Tor1a in CNS (Tor1aΔE/−) (3) | Viable; retarded post-natal growth | Limb clasping, abnormal twisting postures | Selective degeneration of sensorimotor regions, perinuclear ubiquitin inclusions, reactive gliosis |
| Dlx5/6 Cre (GABAergic forebrain neurons) (11) | Normal | Limb clasping, hyperkinesia, grid hang deficits | Selective degeneration of striatal cholinergic interneurons |
Results
A unique line of conditional DYT1 knock-in mice recapitulates molecular and ultrastructural features of ΔE-torsinA
To establish a model enabling the expression of ΔE-torsinA at endogenous levels in an anatomical- and temporal-specific manner, we constructed a conditional Tor1a allele that is converted from a wild-type to a DYT1 mutant allele when acted upon by Cre recombinase. In this ‘Swap allele (Tor1aSwap)’, the final exon is flanked by LoxP sites and is followed by a downstream copy of the same exon harboring the ΔE mutation (an in-frame GAG deletion that removes a single glutamic acid, E; Fig. 1A). Importantly, the endogenous stop codon (TGA) located in the floxed exon 5 ensures that in the absence of Cre, Tor1aSwap/Swap mice (Fig. 1E and F) express normal levels of wild-type torsinA protein (data not shown). To test the predicted Cre inducibility of this model, we crossed mice expressing the ‘gene-targeted construct allele’ (Fig. 1A) with germline-expressing Hprt-Cre mice (31). As expected, progeny from this cross contain the ‘induced’ Tor1ai-ΔE/+ genotype in all tissues. We then intercrossed these Tor1ai-ΔE/+ mice to obtain Tor1ai-ΔE/i-ΔE animals. Similar to originally reported Tor1aΔE/+ knock-in lines (2,24), this intercross yielded all genotypes at the expected Mendelian frequency, a finding confirmed by direct sequencing (Fig. 1C). As expected, Tor1ai-ΔE/+ mice were indistinguishable from littermate controls. Similar to previously reported germline homozygous knock-in animals, Tor1ai-ΔE/i-ΔE pups failed to nurse and perished within 24 h of birth (Table 1) (2,24).
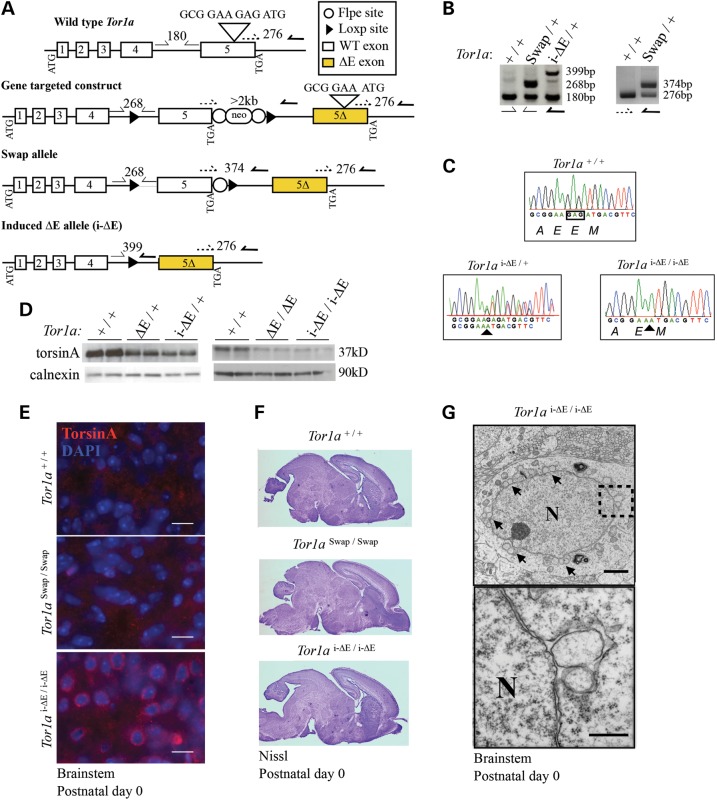
Figure 1.
A novel conditional ΔE-Tor1a knock-in model recapitulates key torsinA loss-of-function phenotypes. (A) Schematic representations of the modified Tor1a gene at different stages of development (wild-type, gene-targeted construct, Swap allele and the induced knock-in ‘i-ΔE’ allele). Half arrows indicate genotyping primer locations. Expected band sizes are shown. Inverted triangle above exon 5 demonstrates the wild-type sequence and deletion of GAG from the gene-targeted construct. (B) Left, to distinguish each Tor1a allele (Tor1a+: 180 bp; Tor1aSwap: 268 bp; Tor1ai-ΔE: 399 bp). Right, the Swap allele was generated from the original gene-targeted construct by using Flpe-expressing mice to remove the neomycin cassette. PCR confirms removal of neomycin cassette from the Swap allele (374 bp). (C) Sanger sequencing of cDNA from P0 brains. Double peaks indicate the presence of the Tor1a+ and Tor1ai-ΔE alleles following Hprt Cre recombinase induction in heterozygous animals (Tor1ai-ΔE/+). Intercrossing Tor1ai-ΔE/+ mice result in offspring with an Hprt Cre-induced homozygous genotype (Tor1ai-ΔE/i-ΔE) with single peaks, indicating deletion of GAG nucleotides and loss of a single glutamic acid, E. (D) Western blot analysis of P0 mouse brain lysate from Tor1a+/+, Hprt Cre-induced mutants (Tor1ai-ΔE/+ and Tor1ai-ΔE/i-ΔE) and previously published germline knock-in Tor1aΔE mice (2). Calnexin is used as loading control (20 µg loaded in technical duplicates). (E) TorsinA immunohistochemistry of P0 brains. Scale bars = 10 µm. This analysis demonstrates the expected perinuclear concentration of ΔE-torsinA in Tor1ai-ΔE/i-ΔE sections. (F) Nissl-stained sagittal sections from P0 animals. (G) Transmission electron microscopic images of NE budding in Tor1ai-ΔE/i-ΔE P0 brainstem neurons demonstrating nuclear membrane buds. Scale bar = 2 µm. N, nucleus. Inset shows higher magnification, scale bar = 500 nm.
We next examined whether our novel germline Tor1ai-ΔE/i-ΔE mice exhibit the molecular and morphological features of established Tor1aΔE/ΔE models (2). Similar to Tor1aΔE/ΔE mice (and DYT1 subjects), Tor1ai-ΔE/i-ΔE mutants show decreased steady-state levels of ΔE-torsinA, which abnormally concentrates in a perinuclear pattern (Fig. 1D and E). Analysis of Nissl-stained brains showed that at birth, brain structure was grossly normal, as reported for Tor1aΔE/ΔE mutants (Fig. 1F). TorsinA null and germline Tor1aΔE/ΔE mice exhibit characteristic neuronal NE abnormalities (NE buds) (2), which were also observed in Tor1ai-ΔE/i-ΔE neurons (Fig. 1G). Considered together, these data confirm that the genetic strategy functions as designed and that Cre-mediated induction of Tor1ai-ΔE recapitulates all major phenotypic features of the previously reported Tor1aΔE allele.
Increased Tor1ai-ΔE gene dosage suppresses growth deficiency
To begin to evaluate ΔE-torsinA for toxic GOF effects in vivo, we utilized the Nestin Cre transgene to generate a cohort of mice that conditionally express one or two DYT1 mutant alleles in the CNS. To generate this cohort, we utilized a cross that employed the Tor1aSwap and previously reported floxed Tor1a allele (Tor1aFLX; Supplementary Material, Fig. S1) (3). Crossing Nestin Cre+ Tor1aSwap/+ and Cre− Tor1aSwap/FLX mice allowed us to collect all experimental and control mice from the same litters (Fig. 2A). For Figures 2–5, mouse genotypes are labeled using the status of Tor1a in the CNS (see Fig. 2A for key). The experimental mice were those expressing either one (Tor1ai-ΔE/−) or two (Tor1ai-ΔE/i-ΔE) DYT1 mutant alleles in the CNS; other genotypes were controls. All genotypes were born at the expected Mendelian frequency (χ2 value = 9.905; df = 7). TorsinA protein levels corresponded to the gene dosage of the Tor1a alleles (Fig. 2B).
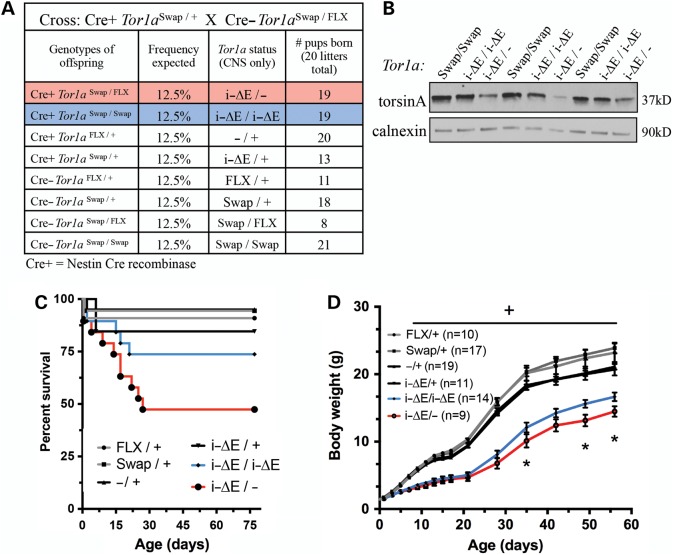
Figure 2.
Increased Tor1ai-ΔE gene dosage suppresses torsinA loss-of-function growth deficiency. (A) The table describing breeding strategy and nomenclature used for gene dosage experiments. All pups were born in the expected Mendelian ratio (χ2 = 9.905; df = 7). (B) Western analysis of P0 brain lysate confirms that torsinA protein levels vary with gene dosage. Three biological replicates are shown. Calnexin is used as loading control (20 µg loaded/sample). (C) Survival curves for offspring of Nestin Cre+ Tor1aSwap/+ × Cre- Tor1aSwap/FLX cross outlined in Figure 1A. Survival analysis indicates significant difference in survival curves, log-rank Mantel–Cox test; P = 0.0017. Post hoc analysis demonstrated that Tor1ai-ΔE/− and Tor1ai-ΔE/i-ΔE mice had decreased survival compared with controls (Fisher's exact test; Tor1ai-ΔE/− versus controls: P < 0.0001 and Tor1ai-ΔE/i-ΔE versus controls: P = 0.0492); however, survival of mutant genotypes did not differ from each other (Fisher's exact test; P = 0.1837). (D) Growth curve demonstrates that increased Tor1ai-ΔE gene dosage suppresses growth deficiency (two-way ANOVA; F5,74 = 50.09, P < 0.0001 and Tukey's multiple comparison test). Tor1ai-ΔE/i-ΔE and Tor1ai-ΔE/− mice were significantly smaller than littermate controls beginning at P7 (denoted by ‘+’ over graph). Tor1ai-ΔE/i-ΔE mice were significantly heavier than Tor1ai-ΔE/− mice at multiple ages (indicated by *). Mice that died prior to P56 were excluded from the growth curve analysis.
Figure 5.
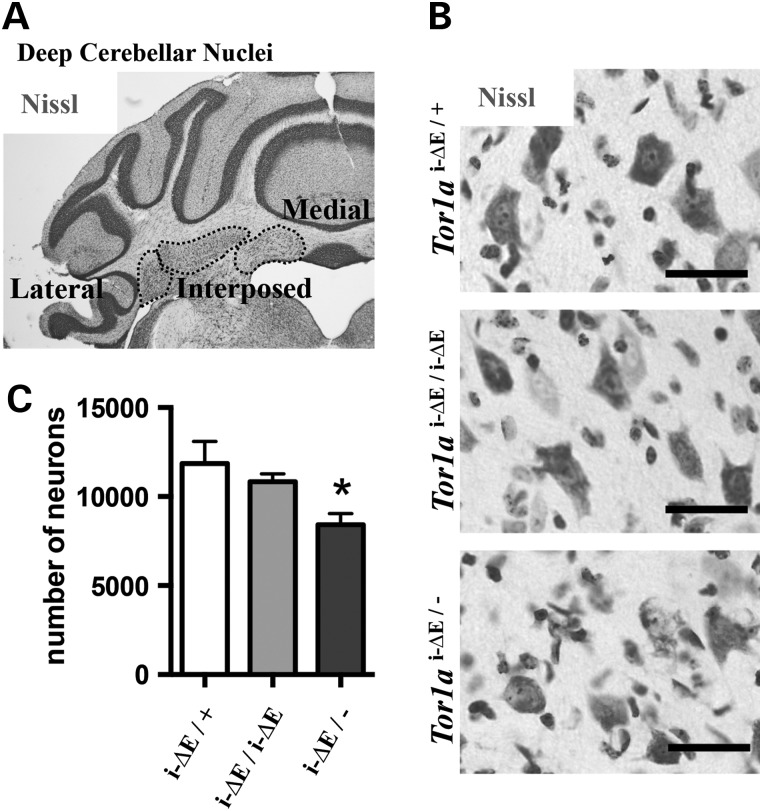
Increasing Tor1ai-ΔE gene dosage rescues neurodegeneration. (A) Representative image of Nissl-stained coronal brain section displaying DCN counted. (B) Examples of Nissl-stained DCN neurons counted using unbiased stereology. Scale bars = 20 µm. (C) Quantification of DCN neurons demonstrates that increasing Tor1ai-ΔE gene dosage rescues neurodegeneration. Means ± SEM for each genotype are displayed (n = 5 per genotype). One-way ANOVA; F2,12 = 4.425, P = 0.0363 and Tukey's multiple comparisons test, *.
For all subsequent analyses, we used four genotypes as controls (Cre+ and Cre− versions of Tor1aFLX/+ and Tor1aSwap/+), discarding Cre-negative Tor1aSwap/Swap and Cre-negative Tor1aSwap/FLX mice. No significant differences in birth weight were observed among the different genotypes. Survival of both experimental groups (Tor1ai-ΔE/− and Tor1ai-ΔE/i-ΔE) was significantly reduced when compared with controls (log-rank Mantel–Cox test; P = 0.0017, post hoc analysis Fisher's exact test; Tor1ai-ΔE/− versus controls: P < 0.0001 and Tor1ai-ΔE/i-ΔE versus controls: P = 0.0492) (Fig. 2C). The survival rate for mice expressing one Tor1ai-ΔE allele (Tor1ai-ΔE/−) was 47.37%, and in mice containing a second Tor1ai-ΔE allele, it was 73.68% (Tor1ai-ΔE/i-ΔE). The difference between survival curves for Tor1ai-ΔE/− and Tor1ai-ΔE/i-ΔE genotypes was not statistically significant (Fisher's exact test; P = 0.1837), demonstrating that increased Tor1ai-ΔE gene dosage does not significantly impair survival. Assessment of growth demonstrated a significant improvement with increased Tor1ai-ΔE gene dosage (Fig. 2D). Both experimental groups were significantly lighter than control animals [two-way analysis of variance (ANOVA); F5,74 = 50.09, P < 0.0001 and Tukey's multiple comparison test] and required an extended weaning period (Supplementary Material, Fig. S2). However, this growth defect was significantly suppressed in mice containing two Tor1ai-ΔE alleles (Tor1ai-ΔE/i-ΔE, Tukey's multiple comparison test). Mice that died prior to the final time point of P56 were excluded from growth analysis. Significant post-natal lethality in experimental groups means that the sickest animals were omitted from the growth curve, suggesting that our results are a conservative representation. These data demonstrate that increasing amounts of ΔE-torsinA—when analyzed in the absence of any wild-type protein—suppress torsinA LOF-mediated growth deficiency.
Increased Tor1ai-ΔE gene dosage does not significantly exacerbate motor abnormalities
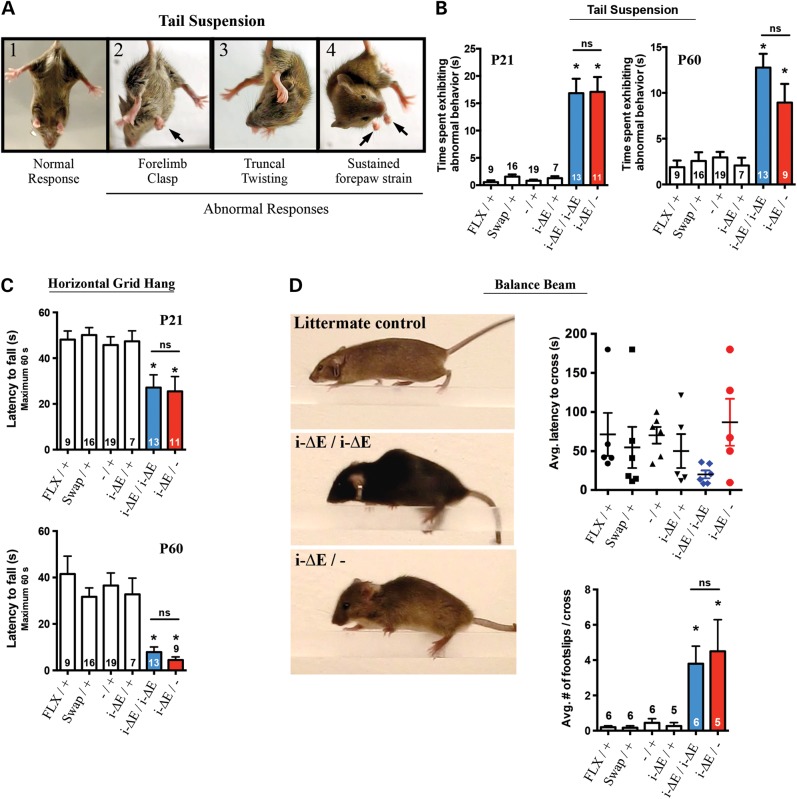
In the early post-natal period, both experimental groups (Tor1ai-ΔE/− and Tor1ai-ΔE/i-ΔE) exhibited delays in neurodevelopmental milestones such as eye opening and were visibly weak and tremulous (Supplementary Material, Video S1). Assessment of motor behavior demonstrated that both mutant groups were significantly impaired compared with controls, but never differed significantly from each other (Fig. 3). Both experimental groups exhibited several abnormal behaviors during tail suspension including forelimb clasping, truncal twisting and sustained forepaw straining (Fig. 3A; Supplementary Material, Video S2). Assessment of videos of this behavior by an observer blinded to genotype demonstrated that both experimental groups differed significantly from controls at P21 (one-way ANOVA; F5,69 = 28.84, P < 0.0001 and Tukey's multiple comparison test, *) and P60 (one-way ANOVA; F5,67 = 15.75, P < 0.0001 and Tukey's multiple comparison test, *), but not from each other (Fig. 3B). A similar pattern was observed in the horizontal grid hang test, which assesses forelimb dexterity, strength and coordination by requiring the mouse to hang upside down from a mesh grid (adapted from 32). Both experimental groups were significantly impaired in their ability to remain suspended from the grid, but did not differ significantly from each other at either age tested (P21: one-way ANOVA; F5,76 = 5.817, P = 0.0001 and Tukey's multiple comparison test, * and P60: one-way ANOVA; F5,73 = 8.402, P < 0.0001 and Tukey's multiple comparison test, *) (Fig. 3C). Falling was often caused by a twisting phenotype as they attempted to traverse the grid (Supplementary Material, Video S3). The ability to traverse a balance beam was also used to assess motor function. All mice had 3 days of training prior to testing. Control animals displayed no difficulties performing this task with their tail and feet remaining above the beam as they crossed (Supplementary Material, Video S4). In contrast, both experimental groups displayed a hunched posture on the beam and typically crossed the beam using their forepaws, while dragging their hindpaws (Fig. 3D). The beam-crossing behavior was also assessed by videos taken on the last day of testing. There was no significant difference in the time to cross the beam, but both experimental genotypes differed from controls by exhibiting abnormal truncal postures during the task and displaying significantly more footslips per cross (one-way ANOVA; F5,25 = 7.766, P = 0.0002 and Tukey's multiple comparison test, *). Considered together, these analyses assessing Tor1ai-ΔE gene dosage do not support a ΔE-torsinA GOF effect on motor behavior.
Figure 3.
Increased Tor1ai-ΔE gene dosage does not significantly exacerbate motor abnormalities. (A) Representative photographs showing normal (1) and abnormal (2–4) responses to tail suspension testing. (B) Quantification of tail suspension behavior at P21 (one-way ANOVA; F5,69 = 28.84, P < 0.0001 and Tukey's multiple comparison test, *) and P60 (one-way ANOVA; F5,67 = 15.75, P < 0.0001 and Tukey's multiple comparison test, *). Experimental mice did not differ from each other at either time point, ns. Means ± SEM for each genotype are displayed, with N above or within bar graph. (C) Quantification of horizontal grid hang behavior at P21 (one-way ANOVA; F5,76 = 5.817, P = 0.0001 and Tukey's multiple comparison test, *) and P60 (one-way ANOVA; F5,73 = 8.402, P < 0.0001 and Tukey's multiple comparison test, *). Means ± SEM for each genotype are displayed, with N above or within bar graph. Experimental mice did not differ from each other at either time point, ns. (D) Quantification of balance beam behavior. Representative images of beam crossing (littermate control, Tor1ai-ΔE/i-ΔE and Tor1ai-ΔE/− mice). Note hunched posture and gripping of beam by hindlimbs in both mutant genotypes. There were no significant differences in average latency to cross the beam, but both experimental groups displayed significantly more footslips per cross compared with controls (one-way ANOVA; F5,25 = 7.766, P = 0.0002 and Tukey's multiple comparison test, *). Means ± SEM for each genotype are displayed, with N above or within bar graph. Experimental mice did not differ from each other, ns.
Increased Tor1ai-ΔE gene dosage rescues neurodegeneration
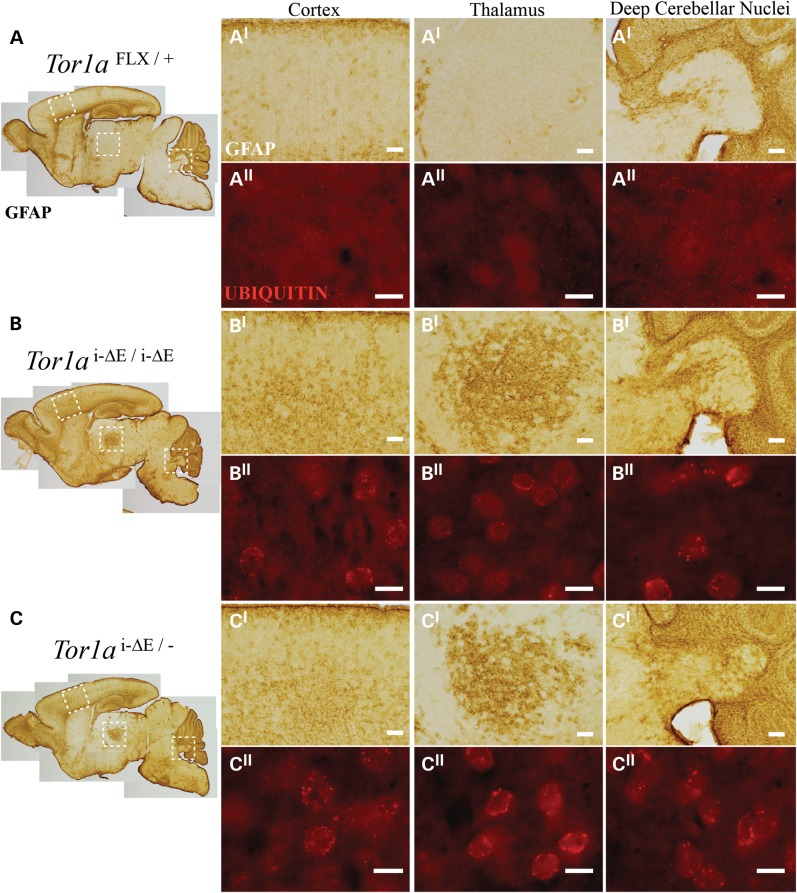
We reported previously that torsinA LOF causes neurodegeneration in distinct sensorimotor structures (3). Affected regions became gliotic and susceptible neurons within those regions exhibited a distinctive perinuclear accumulation of ubiquitin. A single Tor1aΔE allele significantly reduced the magnitude of neuronal loss caused by conditional CNS deletion of torsinA (3). We took advantage of these histopathological features to further examine the effect of ΔE-torsinA on cellular phenotypes linked to abnormal twisting behavior. Consistent with this previous work, both experimental groups (Tor1ai-ΔE/− and Tor1ai-ΔE/i-ΔE) recapitulated the region-specific gliosis and ubiquitin accumulation (Fig. 4). Glial fibrillary acidic protein (GFAP) and ubiquitin immunohistochemistry demonstrated discrete abnormal foci in several sensorimotor structures in both experimental groups, including in deep cerebellar nuclei (DCN), cortex, thalamus, red nucleus and facial motor nucleus (Fig. 4; Supplementary Material, Figs. S3 and S4). The abnormal immunostaining appeared qualitatively similar in both experimental groups. To assess neurodegenerative effects of ΔE-torsinA quantitatively, we used unbiased stereology to determine the extent of cell loss associated with different dosages of Tor1ai-ΔE. We performed this analysis on the DCN (Fig. 5), a structure for which the extent of torsinA dysfunction-related cell loss has been established (3). Analysis of 2-month-old experimental (Tor1ai-ΔE/− and Tor1ai-ΔE/i-ΔE) and control mice confirmed that, as expected, mice expressing a single Tor1ai-ΔE allele show a significant reduction in the number of DCN neurons (one-way ANOVA; F2,12 = 4.425, P = 0.0363 and Tukey's multiple comparisons test) (Fig. 5C). In contrast, mice carrying two Tor1ai-ΔE alleles (Tor1ai-ΔE/i-ΔE) did not show a significant loss of DCN neurons, demonstrating a protective effect of increased Tor1ai-ΔE gene dosage, as observed for weight gain (Fig. 2D). These findings are consistent with the conclusion that the ΔE mutation creates a hypomorphic form of torsinA that does not exert toxic GOF effects.
Figure 4.
Conditional knock-in mice recapitulate DYT1-related neuropathology. (A–C) Sagittal sections of P10 brains immunostained with antibody against GFAP. (AI–CI) Higher magnification images of vulnerable regions (outlined in whole brain sagittal section: cortex, ventral posterior thalamus and DCN) stained for GFAP (brown, scale bars are 100 µm) and (AII–CII) ubiquitin (red, scale bars are 10 µm). Similar appearing abnormalities were observed in Tor1ai-ΔE/i-ΔE and Tor1ai-ΔE/− mice.
Hindbrain-selective induction of the DYT1 genotype does not cause abnormal twisting movements
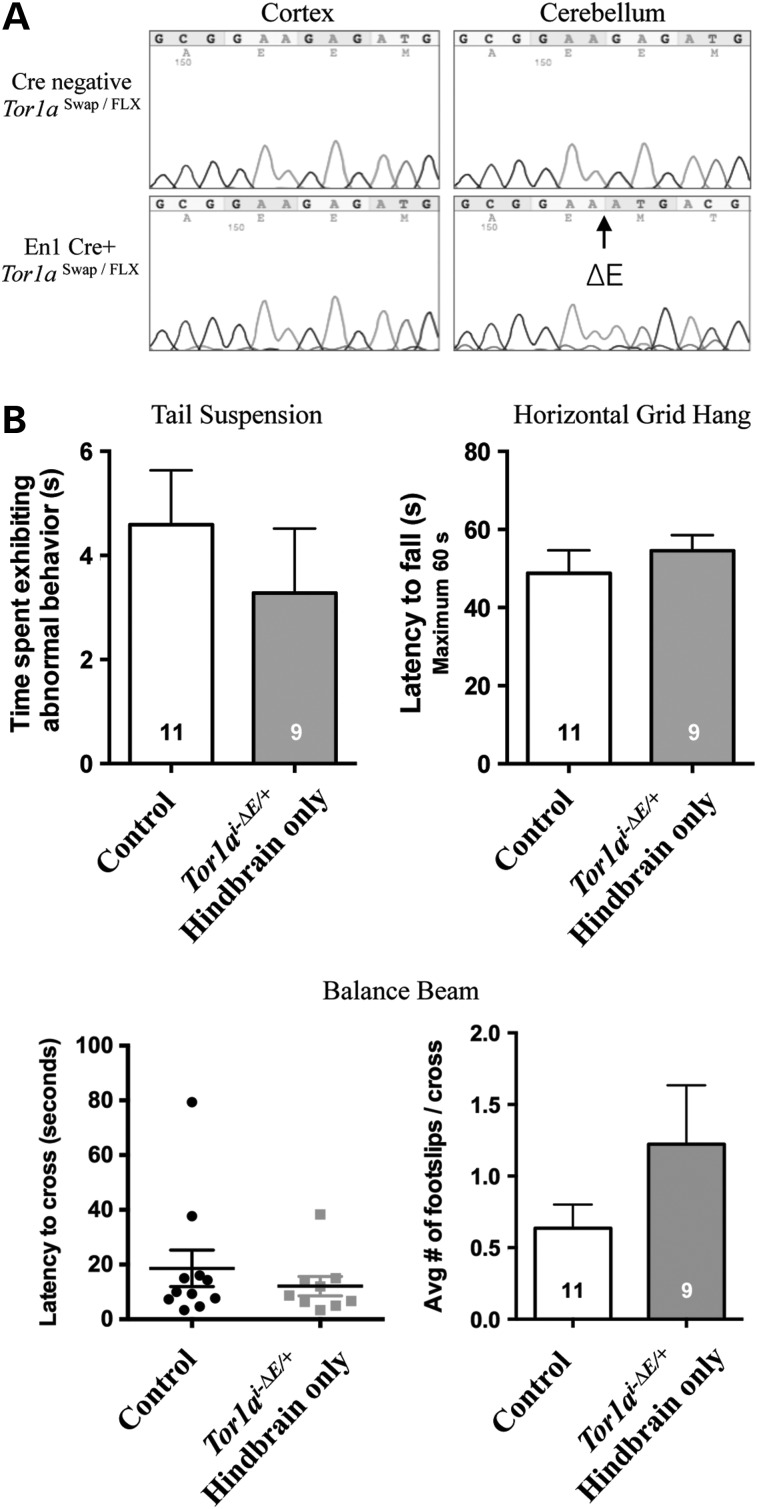
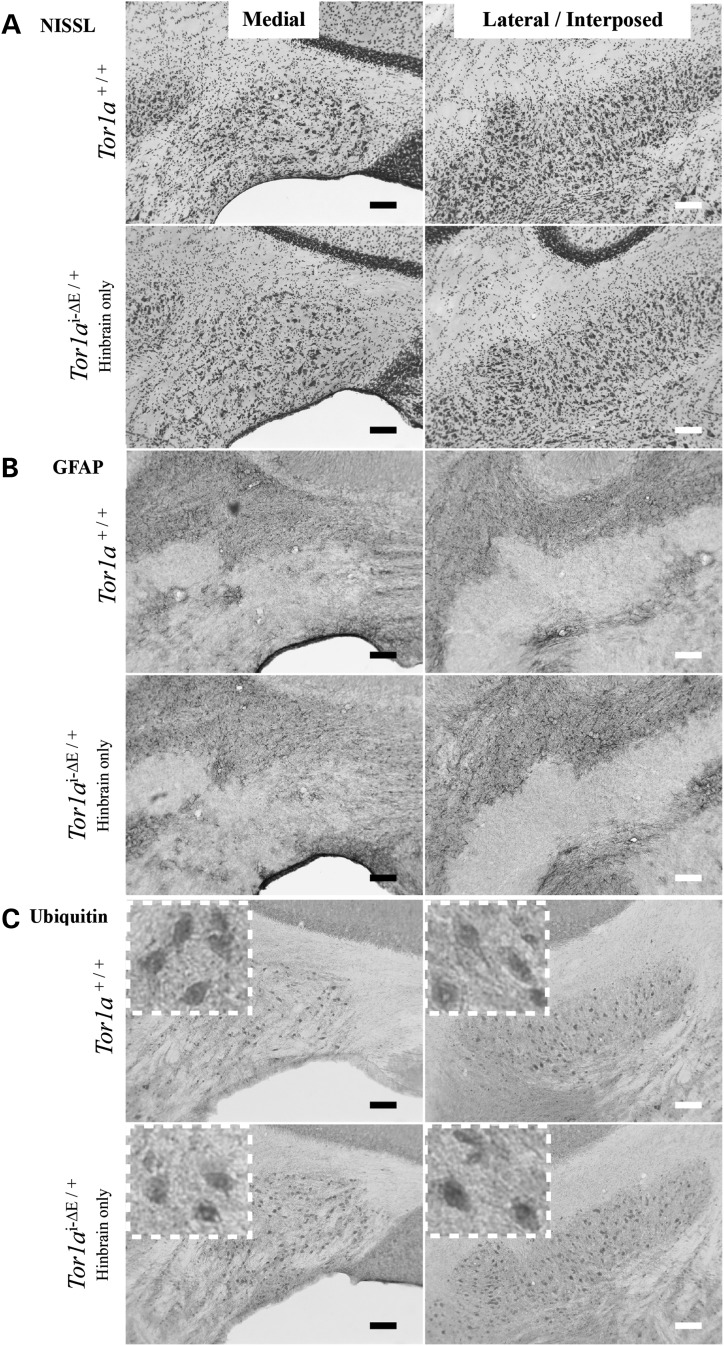
Generation of the Tor1aSwap allele also enabled us to test a model of circuit dysfunction proposed to explain the reduced penetrance characteristic of the DYT1 mutation. This model is based on DTI studies of human DYT1 subjects (16,33) and DYT1 knock-in mice (18). DTI of DYT1 subjects who manifest dystonia exhibits microstructural abnormalities primarily of cerebellothalamic projections, whereas unaffected DYT1 mutation carriers and asymptomatic DYT1 knock-in mice (Tor1aΔE/+) (26) exhibit both cerebellothalamic and thalamocortical defects. These data suggest that dystonia-related signaling could originate in cerebellothalamic projections and that the thalamocortical abnormality prevents such aberrant signaling from reaching forebrain output nuclei. This model predicts that selective induction of the DYT1 genotype in hindbrain structures should cause abnormal twisting movements. We tested this prediction by crossing Tor1aSwap/+ and engrailed1-Cre (En1 Cre) mice that express Cre recombinase in hindbrain structures, including all cerebellar neurons (34). We directly sequenced cerebellar and frontal cortical tissue to confirm that En1 Cre converts Tor1aSwap to a Tor1ai-ΔE allele selectively in the hindbrain and that forebrain tissue remains unaffected (Fig. 6A; see Materials and Methods for details on confirming selectivity of Cre action). En1 Cre+ Tor1aSwap/+(Tor1ai-ΔE/+ selectively in cerebellum/brainstem) mice were born at the expected Mendelian frequency and were indistinguishable from their littermate controls up to 12 months of age. These hindbrain-specific Tor1ai-ΔE/+ mice did not differ from their littermate controls in tail suspension, grid hang or balance beam testing (Fig. 6B). Similarly, these mutants did not differ from their littermate controls in histopathological assessments, including Nissl staining and immunostaining for GFAP and ubiquitin (Fig. 7).
Figure 6.
Midbrain/hindbrain-selective induction of the DYT1 genotype (Tor1ai-ΔE/+) does not cause abnormal twisting movements. (A) Sequencing of cDNA from mouse brain lysates of cortex or cerebellum. Wild-type Tor1a sequence was detected in all samples, except where En1 Cre recombinase acts to induce the ΔE mutation (e.g. cerebellum of En1 Cre+ Tor1aSwap/FLX mice). (B) Behavioral analysis of En1 Cre+ Tor1aSwap/+ mice. Control animals (Cre- Tor1a+/+, Cre- Tor1aSwap/+ and En1 Cre+ Tor1a+/+) did not differ significantly from each other, so their data were combined for clarity. No significant differences were observed between control and En1 Cre+ Tor1aSwap/+ mice in any behavioral tests: tail suspension; horizontal grid hang tests; latency to cross balance beam and footslips per cross on balance beam. Means ± SEM for each genotype are displayed, with N within bar graph.
Figure 7.
Midbrain/hindbrain-selective induction of the DYT1 genotype (Tor1ai-ΔE/+) does not cause overt neuropathology. Cerebellar coronal sections from Tor1a+/+ and En1 Cre+ Tor1aSwap/+ mice: (A) Nissl staining, (B) GFAP and (C) ubiquitin immunostaining. Scale bars = 100 µm.
Discussion
We sought to address fundamental questions regarding the effects of the ΔE mutation at the molecular and circuit levels and to assess the contribution of these effects to motor dysfunction. We pursued these questions by developing a novel genetic strategy that enabled us to convert Tor1a from a wild-type to a DYT1 mutant allele in an anatomic- and temporal-specific manner. Using this powerful approach, we pursued a unique series of studies to assess potential GOF and LOF effects of the ΔE mutation. These studies demonstrate that the ΔE mutation impairs torsinA function, as all phenotypes observed are those tied to torsinA LOF (3). None of the phenotypes we assessed—survival, growth, motor function and histopathology—was exacerbated by increased Tor1ai-ΔE gene dosage (Table 2). Indeed, increased Tor1ai-ΔE gene dosage suppressed the extent of neurodegeneration, which is consistent with a strictly LOF effect of the DYT1 mutation. We also utilized the anatomic-selective capability of our model to explore the prediction that hindbrain-specific expression of the DYT1 genotype (Tor1ai-ΔE/+) at endogenous levels would cause overtly abnormal behavior, which was refuted by this study. These experiments demonstrate the unique power of this conditional knock-in model to address essential questions of DYT1 pathogenesis at the molecular and circuit levels and establish a platform for future studies of dystonia pathophysiology.
Table 2.
Summary of phenotypes in new inducible knock-in mouse models
| Model | Viability/growth | Behavior | Histology |
|---|---|---|---|
| Inducible knock-in ΔE-Tor1a mouse models | |||
| Nestin Cre Tor1aΔE/ΔE knock-in (neural progenitors) | Partial perinatal lethality; retarded post-natal growth | Limb clasping, grid hang deficits, beam walking deficits | Selective pathology of sensorimotor regions, perinuclear ubiquitin inclusions, reactive gliosis |
| Nestin Cre Tor1aΔE/−-isolated knock-in (neural progenitors) | Partial perinatal lethality; retarded post-natal growth | Limb clasping, grid hang deficits, beam walking deficits | Degeneration of deep cerebellar nuclei; selective pathology of sensorimotor nuclei, perinuclear ubiquitin inclusions, reactive gliosis |
| En1 Cre Tor1aΔE/+ knock-in (hindbrain) | Normal | Normal | Normal |
We are aware of only four other conditional knock-in models in the scientific literature (35–38). This strategy avoids the pitfalls that accompany transgenic or viral-based methods that overexpress a non-physiological amount of protein. Several of these previous reports utilized an approach in which a multi-exon wild-type ‘minigene’ is inserted upstream of a knock-in-containing exon (35,37,38). Without cre, the minigene (including stop codon) is transcribed. Following Cre removal of this wild-type minigene, the mutant knock-in exon (and downstream exons) is utilized. Skvorak et al. (36) used an approach similar to ours, but they fused the final two exons. In contrast, the location of the DYT1 mutation within the final Tor1a exon enabled us to avoid the use of a minigene, or fused exons, and leaves the endogenous gene entirely intact. The success of our approach is demonstrated by our use of the Tor1aSwap allele to recapitulate previously reported behavioral and histopathological phenotypes and to conditionally induce the DYT1 mutant torsinA in an anatomically selective manner. Anatomical- and temporal-selective manipulation of endogenous levels of ΔE-torsinA will be valuable for future studies dissecting the cellular anatomy of primary dystonia and assessing the role of neural development in disease pathogenesis, both of which are poorly understood.
DYT1 dystonia is dominantly inherited, but mouse models that mimic the human genotype fail to recapitulate a manifesting state (Table 1) (26). The reasons for the absence of an overt phenotype in heterozygous mice (Tor1aΔE/+) are unclear, but may include differences in the developmental timing or levels of torsinA-pathway molecules such as torsinB, LAP1 or LULL1. The conditional knock-in model enabled us to generate the first homozygous ΔE-Tor1a model that is viable, which displays overtly abnormal motor behavior and dystonia-like movements. The fact that this model recapitulates the behavioral and histological phenotypes identified using independent genetic strategies to manipulate Tor1a (3) further strengthens those findings and the link between developmental neurodegeneration of discrete sensorimotor regions and abnormal twisting movements (Table 1). In contrast to that report, we observe significant lethality prior to weaning using this conditional knock-in strategy. This difference may reflect the fact that in the approach used by Liang et al. (3), the Tor1aΔE allele is expressed constitutively, allowing compensatory mechanisms to develop prior to the post-natal vulnerable period.
To date, few studies have probed the mechanism of action of the ΔE mutation in vivo. Comparisons between different versions of ΔE knock-in and Tor1a knock-out mice demonstrate that the ΔE mutation impairs torsinA function and link torsinA hypofunction to abnormal twisting movements (Tables 1 and 2) (2,3,28). These studies do not exclude the possibility that the ΔE mutation also exerts neomorphic toxic GOF effects (distinct from dominant negative effect, which results in LOF). We devised a novel strategy that enabled us to obtain the first viable homozygous ΔE-Tor1a model and exploited this model to directly investigate toxic GOF effects. We found no evidence for a toxic GOF effect in comparisons of mice harboring one (Nestin Cre+ Tor1aSwap/FLX) or two (Nestin Cre+ Tor1aSwap/Swap) ΔE alleles in the CNS, including assessments of growth and viability, motor behavior and histopathology (Table 2). In fact, increasing Tor1ai-ΔE gene dosage significantly attenuated some phenotypes (growth and neuronal cell number). These results are most consistent with a model in which ΔE exerts LOF effects exclusively, and that higher levels of the hypomorphic protein are beneficial because they boost the overall amount of torsinA activity. This finding has important implications for devising therapeutic approaches for DYT1 dystonia. Our findings do not address whether increasing levels of mutant torsinA would be beneficial in DYT1 subjects, wherein the presence of wild-type protein, dominant negative effects of mutant torsinA would likely prove harmful.
The Tor1aSwap allele also allowed us to test a model of DYT1 circuit dysfunction based on DTI data from human subjects and a mouse model (16,18,33). This model predicts that selective hindbrain expression of the DYT1 genotype (Tor1aΔE/+) would cause overtly abnormal behavior. Our studies are not consistent with this prediction, despite the fact that the Cre driver utilized expresses in all cerebellar cell types, including those vulnerable to more marked degrees of torsinA LOF (3). One potential reason for the difference between the predicted and experimental results concerns the interpretation of DTI changes. Rather than reflecting lesions per se, DTI changes may represent a functional compensation (39) to primary changes that begin elsewhere [e.g. in striatum (11)].
Taken together, our findings provide knowledge of the effects of the ΔE mutation at the molecular and circuit levels that will be important for future studies of disease pathogenesis and therapeutic development. We developed a novel line of Tor1a mutant mice that will enable a range of future studies into the mechanisms underlying the unique vulnerability of discrete cell types to torsinA LOF and the role of neural development in the manifestation of dystonia.
Materials and Methods
Animal husbandry
Mice were housed in the University of Michigan animal care facilities on a 12 h light/dark cycle. Food and water were provided ad libitum. Animal care was in accordance with the University Committee on Use and Care of Animals.
Mice
Novel conditional knock-in mice were generated in collaboration with the Gene Targeting and Transgenic Facility at UCONN Health. A BAC clone RP337K9 (from Children's Hospital Oakland Research Institute) was used with a recombineering-based method to insert the region of interest into plasmid pl253. This modified bluescript plasmid contains a negative selection marker for embryonic stem cell (ES) targeting. Multi-step polymerase chain reaction (PCR) was used to create the mutated exon 5 (3 bp deletion ‘GAG’) + 3′ UTR. After sequencing verification, the correct product was cloned into the pI253-Tor1a vector downstream of the wild-type exon 5 + 3′ UTR. To establish the ‘gene-targeted construct’ (Fig. 1A), a loxp site was inserted upstream of the wild-type exon 5 and an frt-neomycin-frt-loxp cassette was inserted downstream of the wild-type exon 5 + 3′ UTR and before the mutated exon 5 + 3′ UTR. Electroporation of the gene-targeted construct into a hybrid B6/129SVEV ES line followed by screens using long-range PCR allowed for collection of positive clones to generate a chimera. To generate the clean ‘Swap allele’ (Fig. 1A), the chimera were bred to Flpe-expressing mice to remove neomycin through recombination of Frt sites.
Nestin Cre+ (003771), Hprt Cre+ (004302) and En1 Cre+ (007916) mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). Germline DYT1 knock-in mice (Tor1aΔE) were used in this study to verify our new mouse model (2). Mice used to conditionally delete Tor1a (Tor1aFLX) were previously published by our laboratory (3).
Genotyping
Ear (post-natal day 17 and older) and tail samples (post-natal day 0) were digested in 50 mm NaOH for 15 min at 95°C, followed by neutralization with 1 m Tris–HCl (Sigma, St. Louis, MO, USA). Resulting DNA was used for genotyping PCR with the necessary primers and Taq 2× Master Mix (Empirical Bioscience, Grand Rapids, MI, USA; TP-MM-1000). All primers were obtained from Integrated DNA Technologies (Coralville, IA, USA). Genotyping primers to distinguish Tor1a+ and germline Tor1ai-ΔE alleles are loxpF 5′-TCCTCCCCCAAGTACATCAG-3′; LoxpR 5′-CGTCCAGTCCTGGAAACACT-3′ and FrtR 5′-TGGAACTGACGACCACTCAG-3′. PCR program is as follows: 4°C, 3 min; (94°C, 30 s; 65°C, 30 s; 72°C, 30 s) × 33 cycles; 72°C, 5 min; 4°C, 2 min; band sizes: 180 bp+; 268 bp gene-targeted construct and 399 bp germline i-ΔE (Fig. 1B). Genotyping primers to detect deletion of neomycin cassette are frtF 5′-GCCTCTGTGCTCTTCTCTGG-3′ and FrtR 5′-TGGAACTGACGACCACTCAG-3′. PCR program is as follows: 94°C, 3 min; (94°C, 30 s; 69°C, 30 s; 72°C, 30 s) × 33 cycles; 72°C, 5 min; 4°C, 2 min; band sizes: 276 bp internal positive control; 374 bp (Swap allele) neomycin cassette removed and >2 kb (gene-targeted construct) neomycin cassette included. Other than validation experiments described in Figure 1, all animals used for this study were derived from the Swap allele after neomycin cassette deletion.
Genotyping primers to distinguish Tor1a+, Tor1aSwap and Tor1aFLX alleles are loxgtF 5′-CCTGCCTCAGCCTAACTACG-3′; LoxgtR 5′-TGTGTGCATTTACCCAGAGC-3′; LoxpF2 5′-GGACACATTGGGCCACCTTG-3′ and LoxpR 5′-CGTCCAGTCCTGGAAACACT-3′. PCR program is 94°C, 3 min; (94°C, 30 s; 65°C, 30 s; 72°C, 30 s) × 33 cycles; 72°C, 5 min; 4°C, 2 min. Band sizes are 213 and 519 bp Tor1a+/+; 213, 307 and 519 bp Tor1aFLX/+; 213, 519 and 607 bp Tor1aSwap/+; 307 and 519 bp Tor1aFLX/FLX; 213, 307, 519 and 607 bp Tor1aSwap/FLX and 213 and 607 bp Tor1aSwap/Swap (Supplementary Material, Fig. S1). Genotyping methods to detect a Cre-recombinase-positive genotype and constitutive germline knock-in mice (Tor1aΔE) were as previously reported (2,3).
DNA sequencing
RNA extraction was performed on whole brain lysates using RNeasy Plus Mini Kit (Qiagen, Venlo, The Netherlands; 74134). RNA was converted to cDNA using SMART™ MMLV Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA; PT4045-2). To isolate cDNA encompassing the DYT1 mutation, PCR was performed using forward primer 5′-GCCGTGTCGGTCTTCAATAA-3′ and reverse primer 5′-ACAGTCTTGCAGCCCTTGTC-3′. Bands of 262 and 259 bp (3 bp deletion; appear as identical bands after electrophoresis) were isolated using Wizard SV Gel and PCR Clean-up system (Promega, Madison, WI, USA; A9281). These samples were sent to the University of Michigan's DNA Sequencing Core with sequencing primer 5′-GCCGTGTCGGTCTTCAATAA-3′. The sequencing primer is located within exon 4 of Tor1a, which ensures that sequencing is of cDNA derived from mRNA and not from genomic DNA. Resulting sequences were analyzed using 4Peaks software (A. Griekspoor and Tom Groothuis, mekentosj.com). For confirmation of hindbrain-specific induction by En1 Cre, we chose to analyze cDNA from En1 Cre+ Tor1aSwap/FLX mice opposed to En1 Cre+ Tor1aSwap/+ mice (our experimental genotype) in order to visualize single peaks.
Western blot
Protein lysates were made by homogenizing whole brain tissue in 500 µl of lysis buffer (0.1% sodium dodecyl sulfate, 2.5 mm Tris–HCl, 1 mm ethylenediaminetetraacetic acid) and Complete Mini Protease Inhibitor Cocktail (Roche, Basel, Switzerland, 11836170001). Protein concentrations were determined using BCA Protein Assay Kit (Thermo Scientific, Wilmington, MA, USA, 23227) and analyzed by NanoDrop™ (Thermo Scientific, Wilmington, DE, USA). Lysates were diluted in Laemmli sample buffer, and 20 µg of protein was separated on a 10% Mini-PROTEAN® TGX™ polyacrylamide gel (Bio-Rad, Hercules, CA, USA, 456–1033). Western blot transfer onto Immun-Blot PVDF membrane (Bio-Rad, Hercules, CA, USA, 1620177) was followed by blocking with 5% non-fat dry milk dissolved in phosphate-buffered saline (PBS)-T (0.01 m PBS, Sigma, St. Louis, MO, USA, P-3813; 0.1% Tween® 20, Sigma, St. Louis, MO, USA, P9416) for 1 h. The membrane was incubated at 4°C overnight with primary antibodies (torsinA, Abcam, Cambridge, United Kingdom, ab34540 1:10 000; calnexin, Enzo Life Sciences, Ann Arbor, MI, USA, SPA-860 1:20 000) diluted in PBS-T followed by a 1 h incubation with anti-rabbit IgG-horseradish peroxidase-conjugated secondary antibody (Cell Signaling, Danvers, MA, USA, 7074S 1:10 000) diluted in PBS-T. Developing was performed using SuperSignal® West Femto (Thermo Scientific, Wilmington, MA, USA, 34095) for torsinA blots and SuperSignal® West Dura (Thermo Scientific, Wilmington, MA, USA, 34075) for calnexin blots. Blots were exposed to GE™ Healthcare Amersham™ Hyperfilm ECL (Thermo Fisher Scientific, Wilmington, MA, USA, 45-001-508).
Immunohistochemistry
Animals were transcardially perfused with PB (0.1 m PB, Sigma, St. Louis, MO, USA, P7994), followed by 4% paraformaldehyde (Sigma, St. Louis, MO, USA, P6148) in PB. Brains were post-fixed overnight in this fixative and then cryoprotected in 20% sucrose dissolved in PB. Sectioning was performed on a Leica CM3050 S (Leica Biosystems, Buffalo Grove, IL, USA) cryostat to collect sections of 50 µm (P0 and P10 brains) and 40 µm (adult ages). Sections were permeabilized by washing in PBS-Tx (0.01 m PBS; 0.01% Triton™ X-100, Fisher Scientific, Wilmington, MA, USA, BP151-500). Sections to be stained using peroxidase/diaminobenzidine (DAB) methods were incubated with 0.3% hydrogen peroxide (Sigma, St. Louis, MO, USA, H1009) diluted in PBS followed by PBS washes. For both immunofluorescence and peroxidase staining, sections were blocked with 5% normal donkey serum (NDS, Jackson ImmunoResearch, West Grove, PA, USA, 017-000-121) in PBS-Tx for 1 h. Overnight incubation with primary antibodies diluted in PBS-Tx/1.5% NDS (torsinA, Abcam, Cambridge, United Kingdom, ab34540 1:100; ubiquitin, Dako, Carpinteria, CA, USA, Z0458 1:500; GFAP, Dako, Carpinteria, CA, USA, Z0443 1:2000) was followed by washes in PBS-Tx and a 1 h incubation in secondary antibody (Alexa Fluor® 555 Donkey Anti-Rabbit IgG, Life Technologies, Carlsbad, CA, USA, A-31572 1:500; Biotinylated Donkey Anti-Rabbit, Jackson ImmunoResearch, West Grove, PA, USA, 711-065-152). For immunofluorescent tissue, ProLong® Gold Antifade Reagent with DAPI (Life Technologies, Carlsbad, CA, USA, P36935) was used for mounting on Fisherbrand® Superfrost® Plus microscope slides (Fisher Scientific, Wilmington, MA, USA, 12-550-15). Sections to be stained for peroxidase activity were incubated with VECTASTAIN Elite ABC kit (Vector Laboratories, Burlingame, CA, USA, PK-6100), followed by DAB (SIGMAFAST™ 3,3′-Diaminobenzidine tablets, Sigma, St. Louis, MO, USA, D4418). DAB-stained sections were then mounted on slides and dried overnight before dehydrating in ethanol and clearing with Fisherbrand™ Citrosolv™ Clearing Agent (Fisher Scientific, Wilmington, MA, USA, 22-143-975) and cover slipping with Fisher Scientific™ Permount™ Mounting Medium (Fisher Scientific, Wilmington, MA, USA, SP15-500). Nissl staining with Cresyl Violet Acetate (Sigma, St. Louis, MO, USA, C5042) was performed on unstained sections that were mounted on slides followed by cover slipping as described. Imaging was performed on an Axioskop2 microscope (Carl Zeiss Microscopy, Jena, Germany) and an Olympus digital camera (model DP70, Olympus, Tokyo, Japan).
Stereology
Stereological investigations were completed on both mutant genotypes (Nestin Cre+ Tor1aSwap/Swap and Nestin Cre+ Tor1aSwap/FLX) and one littermate control (Nestin Cre+ Tor1aSwap/+). Five animals were used per genotype. Animals were transcardially perfused as described earlier at 2 months of age. Prior to cryosectioning, the primary researcher was re-blinded to the animal genotype and remained blinded until all stereological counts were made. Brains were sectioned coronally at 30 µm and Nissl-stained. Stereo Investigator (MBF Bioscience, Williston, VT, USA) software was used to perform stereological analysis of neurons within the lateral, interposed and medial regions of the DCN. Every third section was counted using a counting frame of 80 µm2 and a sampling grid of 125 µm2. These parameters allowed for a coefficient of Gundersen less than 0.1. Counts from individual regions were summed and averaged per genotype to allow for comparison of total DCN.
Ultrastructural analysis
Post-natal day 0 pups were transcardially perfused with 4% paraformaldehyde/2.5% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA, USA, 15710 and 16220, respectively) in PB. Brains were collected and post-fixed for 2 weeks in the same solution. Preparation of brains for electron microscopy was performed in collaboration with the Robert P. Apkarian Integrated Electron Microscopy Core at Emory University, as described previously (40). Imaging was performed at University of Michigan's Microscopy and Image Analysis Laboratory using a Philips CM-100 transmission electron microscope (Philips, Amsterdam, The Netherlands).
Animal viability and growth
To assess perinatal lethality in our Hprt Cre-recombinase-induced knock-in Tor1ai-ΔE mouse model, we generated homozygous mice (Tor1ai-ΔE/i-ΔE) by intercrossing two mice heterozygous for the germline mutation. Litters were genotyped at post-natal day 0 and monitored for lethality.
To compare Nestin Cre+ Tor1aSwap/Swap and Nestin Cre+ Tor1aSwap/FLX born in the same litters, we employed the breeding strategy of Nestin Cre+ Tor1aSwap/+ mice mated with Cre- Tor1aSwap/FLX mice. This resulted in eight possible genotypes as offspring (Fig. 2A). The following genotypes were kept for this study: Nestin Cre+ Tor1aSwap/Swap, Nestin Cre+ Tor1aSwap/FLX, Nestin Cre+ Tor1aSwap/+, Nestin Cre+ Tor1aFLX/+, Cre- Tor1aSwap/+ and Cre- Tor1aFLX/+. Two genotypes were omitted from this study and sacrificed at post-natal day 0: cre- Tor1aSwap/Swap and Cre- Tor1aSwap/FLX. Breeding cages housing two females and one male were used to generate this cohort, and 1 week prior to giving birth, each female was separated into her own cage. This arrangement allowed for accurate data collection, prevented overcrowding and reduced competition for nutrients. To measure survival and growth, each pup was weighed and genotyped at post-natal day 0 (birth weight). Date of birth was determined to be the day in which pups were observed between 6:00 a.m. and 12:00 p.m. Pups included in this study were weighed every other day until 21 days, and then mice were weighed weekly until post-natal day 56. Litters were observed each day in order to collect dead animals for survival analysis.
Motor behavior
Mixed housing was used for all mice in this study to eliminate environmental bias. Male and female mice were used for the analysis of survival, growth, tail suspension and grid hang behavior tests. Only males were used for balance beam behavior tests.
Tail suspension
Individual mice were held by the tail 20 cm above a table top, and 15 s videos were recorded. Abnormal behaviors included forelimb clasping, truncal twisting and sustained straining of forepaws. Videos were analyzed by a researcher blinded to the genotype. Time spent clasping, twisting and forepaw straining (in seconds) were calculated separately and combined to generate an overall severity score.
Horizontal grid hang
The horizontal grid apparatus was modeled after a previously published grid hang test (32). The grid's measurements are: 21 cm height, 21.5 cm width and 21.5 cm length. Testing involved placing a mouse in the middle of the grid, followed by careful turning of the apparatus and placing it on the table top. This required the mouse to hang upside down from the grid mesh. Latency to fall from the grid was measured using video recording. A maximum of 1 min was filmed for this test.
Balance beam
Mice received 3 consecutive days of training and a fourth testing day. On each day, the mouse was placed on an open platform (20 cm2) and allowed to cross a plexiglass beam of 44 cm in length at a height of 53 cm to a dark box (20 cm3) three times. On the first day, a 2 cm wide balance beam was used for the first two crossings followed by a crossing on a 1 cm wide beam. The second, third and fourth (test) days used a bar of 1 cm width. For testing, each mouse was filmed as it performed the beam crossing. The videos were then analyzed for abnormal postures and crossing behaviors by investigators blinded to the genotype. The number of footslips per cross was calculated and averaged per animal. Latency to cross was measured for each crossing and averaged per animal.
Data analysis
GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA, USA) was used to analyze all data. Means are displayed with error bars representing SEM. All one-way ANOVA tests were followed by post hoc Tukey's multiple comparison tests. All Kruskal–Wallis non-parametric tests were followed by post hoc Dunn's multiple comparison tests. Significance was determined to be P < 0.05.
Supplementary Material
Funding
This work was supported by the National Institutes of Health (T32-GM007315 to C.E.W.), the National Institute of Neurological Disorders and Stroke (RO1NS077730 to W.T.D.) and a Dr Edward V. Staab Memorial Grant from Tyler's Hope for a Dystonia Cure Foundation to W.T.D.
Supplementary Material
Acknowledgements
The authors thank the staff of University of Michigan's Core Facilities (DNA Sequencing Core, Microscopy and Image Analysis Laboratory and Unit of Laboratory Animal Medicine), the laboratory of Stanley Watson at the University of Michigan for use of their stereology equipment, the Robert P. Apkarian Integrated Electron Microscopy Core at Emory University and the Gene Targeting and Transgenic Facility at UCONN Health. The authors also thank past and present co-workers in the Dauer laboratory, as this work would not be possible without their guidance and dedication.
Conflict of Interest statement. None declared.
References
- 1.Ozelius L.J., Hewett J.W., Page C.E., Bressman S.B., Kramer P.L., Shalish C., de Leon D., Brin M.F., Raymond D., Corey D.P. et al. (1997) The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet., 17, 40–48. [DOI] [PubMed] [Google Scholar]
- 2.Goodchild R.E., Kim C.E., Dauer W.T. (2005) Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron, 48, 923–932. [DOI] [PubMed] [Google Scholar]
- 3.Liang C.C., Tanabe L.M., Jou S., Chi F., Dauer W.T. (2014) TorsinA hypofunction causes abnormal twisting movements and sensorimotor circuit neurodegeneration. J. Clin. Invest., 124, 3080–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao C., Brown R.S., Chase A.R., Eisele M.R., Schlieker C. (2013) Regulation of Torsin ATPases by LAP1 and LULL1. Proc. Natl Acad. Sci. USA, 110, 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sosa B.A., Demircioglu F.E., Chen J.Z., Ingram J., Ploegh H.L., Schwartz T.U. (2014) How lamina-associated polypeptide 1 (LAP1) activates Torsin. Elife, 3, e03239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun S., Ling S., Qiu J., Albuquerque C.P., Zhou Y., Tokunaga S., Li H., Qiu H., Bui A., Yeo G.W. et al. (2015) ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat. Commun., 6, 6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fakhouri W.D., Rahimov F., Attanasio C., Kouwenhoven E.N., Ferreira De Lima R.L., Felix T.M., Nitschke L., Huver D., Barrons J., Kousa Y.A. et al. (2014) An etiologic regulatory mutation in IRF6 with loss- and gain-of-function effects. Hum. Mol. Genet., 23, 2711–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jungwirth M.T., Kumar D., Jeong D.Y., Goodchild R.E. (2011) The nuclear envelope localization of DYT1 dystonia torsinA-DeltaE requires the SUN1 LINC complex component. BMC Cell Biol., 12, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNaught K.S., Kapustin A., Jackson T., Jengelley T., JnoBaptiste R., Shashidharan P., Perl D.P., Pasik P., Olanow C.W. (2004) Brainstem pathology in DYT1 primary torsion dystonia. Ann. Neurol., 56, 540–547. [DOI] [PubMed] [Google Scholar]
- 10.Pappas S.S., Leventhal D.K., Albin R.L., Dauer W.T. (2014) Mouse models of neurodevelopmental disease of the basal ganglia and associated circuits. Curr. Top. Dev. Biol., 109, 97–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pappas S.S., Darr K., Holley S.M., Cepeda C., Mabrouk O.S., Wong J.M., LeWitt T.M., Paudel R., Houlden H., Kennedy R.T. et al. (2015) Forebrain deletion of the dystonia protein torsinA causes dystonic-like movements and loss of striatal cholinergic neurons. Elife, 4, e08352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C.H., Fremont R., Arteaga-Bracho E.E., Khodakhah K. (2014) Short latency cerebellar modulation of the basal ganglia. Nat. Neurosci., 17, 1767–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prudente C.N., Hess E.J., Jinnah H.A. (2014) Dystonia as a network disorder: what is the role of the cerebellum? Neuroscience, 260, 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raike R.S., Pizoli C.E., Weisz C., van den Maagdenberg A.M., Jinnah H.A., Hess E.J. (2013) Limited regional cerebellar dysfunction induces focal dystonia in mice. Neurobiol. Dis., 49, 200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raike R.S., Hess E.J., Jinnah H.A. (2015) Dystonia and cerebellar degeneration in the leaner mouse mutant. Brain Res., 1611, 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carbon M., Kingsley P.B., Su S., Smith G.S., Spetsieris P., Bressman S., Eidelberg D. (2004) Microstructural white matter changes in carriers of the DYT1 gene mutation. Ann. Neurol., 56, 283–286. [DOI] [PubMed] [Google Scholar]
- 17.Argyelan M., Carbon M., Niethammer M., Ulug A.M., Voss H.U., Bressman S.B., Dhawan V., Eidelberg D. (2009) Cerebellothalamocortical connectivity regulates penetrance in dystonia. J. Neurosci., 29, 9740–9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulug A.M., Vo A., Argyelan M., Tanabe L., Schiffer W.K., Dewey S., Dauer W.T., Eidelberg D. (2011) Cerebellothalamocortical pathway abnormalities in torsinA DYT1 knock-in mice. Proc. Natl Acad. Sci. USA, 108, 6638–6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shashidharan P., Sandu D., Potla U., Armata I.A., Walker R.H., McNaught K.S., Weisz D., Sreenath T., Brin M.F., Olanow C.W. (2005) Transgenic mouse model of early-onset DYT1 dystonia. Hum. Mol. Genet., 14, 125–133. [DOI] [PubMed] [Google Scholar]
- 20.Sharma N., Baxter M.G., Petravicz J., Bragg D.C., Schienda A., Standaert D.G., Breakefield X.O. (2005) Impaired motor learning in mice expressing torsinA with the DYT1 dystonia mutation. J. Neurosci., 25, 5351–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grundmann K., Reischmann B., Vanhoutte G., Hubener J., Teismann P., Hauser T.-K., Bonin M., Wilbertz J., Horn S., Nguyen H.P. et al. (2007) Overexpression of human wildtype torsinA and human DeltaGAG torsinA in a transgenic mouse model causes phenotypic abnormalities. Neurobiol. Dis., 27, 190–206. [DOI] [PubMed] [Google Scholar]
- 22.Page M.E., Bao L., Andre P., Pelta-Heller J., Sluzas E., Gonzalez-Alegre P., Bogush A., Khan L.E., Lacovitti L., Rice M.E., Ehrlich M.E. (2010) Cell-autonomous alteration of dopaminergic transmission by wild type and mutant (DeltaE) TorsinA in transgenic mice. Neurobiol. Dis., 39, 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grundmann K., Glockle N., Martella G., Sciamanna G., Hauser T.-K., Yu L., Castaneda S., Pichler B., Fehrenbacher B., Schaller M. et al. (2012) Generation of a novel rodent model for DYT1 dystonia. Neurobiol. Dis., 47, 61–74. [DOI] [PubMed] [Google Scholar]
- 24.Dang M.T., Yokoi F., McNaught K.S., Jengelley T.A., Jackson T., Li J., Li Y. (2005) Generation and characterization of Dyt1 DeltaGAG knock-in mouse as a model for early-onset dystonia. Exp. Neurol., 196, 452–463. [DOI] [PubMed] [Google Scholar]
- 25.Dang M.T., Yokoi F., Pence M.A., Li Y. (2006) Motor deficits and hyperactivity in Dyt1 knockdown mice. Neurosci. Res., 56, 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanabe L.M., Martin C., Dauer W.T. (2012) Genetic background modulates the phenotype of a mouse model of DYT1 dystonia. PLoS ONE, 7, e32245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoi F., Dang M.T., Mitsui S., Li J., Li Y. (2008) Motor deficits and hyperactivity in cerebral cortex-specific Dyt1 conditional knockout mice. J. Biochem., 143, 39–47. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L., Yokoi F., Jin Y.H., DeAndrade M.P., Hashimoto K., Standaert D.G., Li Y. (2011) Altered dendritic morphology of Purkinje cells in Dyt1 DeltaGAG knock-in and purkinje cell-specific Dyt1 conditional knockout mice. PLoS ONE, 6, e18357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokoi F., Dang M.T., Li J., Standaert D.G., Li Y. (2011) Motor deficits and decreased striatal dopamine receptor 2 binding activity in the striatum-specific Dyt1 conditional knockout mice. PLoS ONE, 6, e24539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niethammer M., Carbon M., Argyelan M., Eidelberg D. (2011) Hereditary dystonia as a neurodevelopmental circuit disorder: evidence from neuroimaging. Neurobiol. Dis., 42, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang S.H., Silva F.J., Tsark W.M., Mann J.R. (2002) A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis, 32, 199–202. [DOI] [PubMed] [Google Scholar]
- 32.Tillerson J.L., Miller G.W. (2003) Grid performance test to measure behavioral impairment in the MPTP-treated-mouse model of parkinsonism. J. Neurosci. Methods, 123, 189–200. [DOI] [PubMed] [Google Scholar]
- 33.Trost M., Carbon M., Edwards C., Ma Y., Raymond D., Mentis M.J., Moeller J.R., Bressman S.B., Eidelberg D. (2002) Primary dystonia: is abnormal functional brain architecture linked to genotype? Ann. Neurol., 52, 853–856. [DOI] [PubMed] [Google Scholar]
- 34.Kimmel R.A., Turnbull D.H., Blanquet V., Wurst W., Loomis C.A., Joyner A.L. (2000) Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev., 14, 1377–1389. [PMC free article] [PubMed] [Google Scholar]
- 35.Wingate A.D., Martin K.J., Hunter C., Carr J.M., Clacher C., Arthur J.S. (2009) Generation of a conditional CREB Ser133Ala knockin mouse. Genesis, 47, 688–696. [DOI] [PubMed] [Google Scholar]
- 36.Skvorak K., Vissel B., Homanics G.E. (2006) Production of conditional point mutant knockin mice. Genesis, 44, 345–353. [DOI] [PubMed] [Google Scholar]
- 37.Bayascas J.R., Sakamoto K., Armit L., Arthur J.S., Alessi D.R. (2006) Evaluation of approaches to generation of tissue-specific knock-in mice. J. Biol. Chem., 281, 28772–28781. [DOI] [PubMed] [Google Scholar]
- 38.Parra S., Huang X., Charbeneau R.A., Wade S.M., Kaur K., Rorabaugh B.R., Neubig R.R. (2014) Conditional disruption of interactions between Galphai2 and regulator of G protein signaling (RGS) proteins protects the heart from ischemic injury. BMC Pharmacol. Toxicol., 15, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackey A.P., Whitaker K.J., Bunge S.A. (2012) Experience-dependent plasticity in white matter microstructure: reasoning training alters structural connectivity. Front. Neuroanat., 6, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodchild R.E., Dauer W.T. (2005) The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J. Cell Biol., 168, 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.