Abstract
There is extensive evidence from model systems that disrupting associations between co-adapted mitochondrial and nuclear genotypes can lead to deleterious and even lethal consequences. While it is tempting to extrapolate from these observations and make inferences about the human-health effects of altering mitonuclear associations, the importance of such associations may vary greatly among species, depending on population genetics, demographic history and other factors. Remarkably, despite the extensive study of human population genetics, the statistical associations between nuclear and mitochondrial alleles remain largely uninvestigated. We analysed published population genomic data to test for signatures of historical selection to maintain mitonuclear associations, particularly those involving nuclear genes that encode mitochondrial-localized proteins (N-mt genes). We found that significant mitonuclear linkage disequilibrium (LD) exists throughout the human genome, but these associations were generally weak, which is consistent with the paucity of population genetic structure in humans. Although mitonuclear LD varied among genomic regions (with especially high levels on the X chromosome), N-mt genes were statistically indistinguishable from background levels, suggesting that selection on mitonuclear epistasis has not preferentially maintained associations involving this set of loci at a species-wide level. We discuss these findings in the context of the ongoing debate over mitochondrial replacement therapy.
Keywords: assisted reproductive technologies, coevolution, cytonuclear interactions, mitochondrial genome, mtDNA, sex chromosomes
1. Introduction
The biology of eukaryotic cells depends upon highly coevolved interactions between nuclear and mitochondrial genomes [1,2]. There is a growing body of evidence that disrupting these interactions has harmful consequences for organismal fitness [3–5]. In fact, incompatibilities between interacting nuclear and mitochondrial genes from diverging populations may represent one of the primary mechanisms responsible for reproductive isolation and the origin of species [6,7]. With increasing awareness about the biomedical importance of mitochondrial DNA [8–10], there is also greater interest in how epistatic interactions between mitochondrial variants and the nucleus may affect human health [11,12]. There are numerous examples in which the severity of mitochondrial genetic diseases is dependent upon the nuclear background or vice versa [13–24], and the use of human cell lines to generate cytoplasmic hybrids (cybrids) has provided additional evidence for mitonuclear interactions [25–29]. To our knowledge, however, there are no documented cases within humans of true mitonuclear mismatch, in which mitochondrial haplotypes have been shown to be superior on their own ‘native’ nuclear backgrounds and compromised when found on foreign nuclear backgrounds (i.e. sign epistasis; figure 1).
Figure 1.
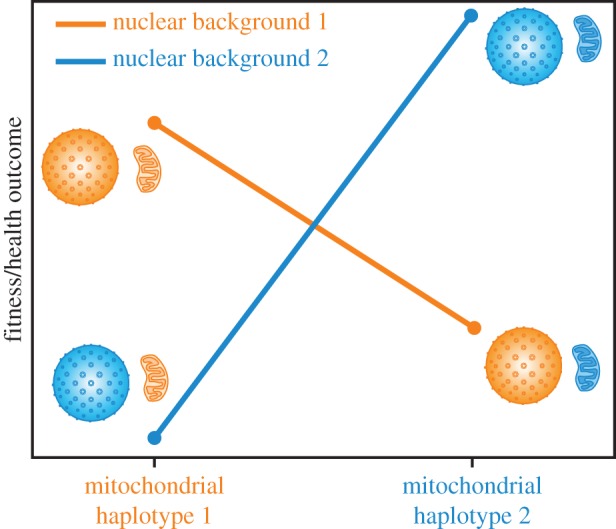
Illustration of sign epistasis in mitonuclear interactions. The coloured organelles reflect the concept of matched and mismatched pairings between mitochondria and nuclei.
Interest in the effects of mitonuclear interactions on human health has become particularly acute in recent years because of the controversy surrounding the approval of mitochondrial replacement (MR) therapy as an assisted reproductive technology in the UK [30]. MR therapy aims to prevent the transmission of diseases resulting from mitochondrial mutations by transferring the nuclear genetic material from an affected egg, polar body or fertilized zygote into a healthy donor's cytoplasm [31–35]. Much of the controversy about these techniques has centred on ethical concerns and the potential side effects associated with physical manipulation of reproductive cells, but an additional concern has emerged from the field of evolutionary biology—specifically that MR could produce genetic incompatibilities associated with novel pairings of nuclear and mitochondrial genotypes [36–38].
The idea that MR causes increased risk of genetic incompatibilities is supported by genetic crossing and cybrid studies in model systems, including yeast, fruit flies, copepods and rodents [39–47]. In these and other eukaryotic lineages, mitochondrial-targeted proteins such as OXPHOS complex subunits [48–50], ribosomal subunits [51,52], tRNA synthetases [44] and RNA polymerases [53] have been found to exhibit incompatibilities with certain mitochondrial backgrounds and/or accelerated evolution indicative of positive selection and compensatory mitonuclear coevolution. One of the more comprehensive bodies of work has established the copepod Tigriopus californicus as a model for studying mitonuclear incompatibilities [54] and left little doubt that performing MR as an assisted reproductive technique in T. californicus could cause more health problems for the copepods than it would fix. Of course, humans are not copepods and, unfortunately, questions about the health effects of disrupting mitonuclear interactions in humans are less clear-cut. There is difficulty in generalizing the results from model systems because the extent of mitonuclear co-adaptation is likely to be highly species-specific and depend on population genetic factors such as mitochondrial and nuclear mutation rates, effective population size, mating system, generation time and population subdivision [55–59]. There is some reason to think that mitonuclear co-adaptation within human populations may be less extensive because of recent demographic expansion and a relatively small amount of genetic structure [60–62], but the controversy over MR has thus far received little scrutiny through the lens of human population genetics.
A history of selection favouring specific combinations of mitochondrial and nuclear alleles should leave detectable signatures in the genetic diversity of a species [63–65]. For example, we would expect N-mt genes to be subject to more intense selection for efficient mitonuclear interactions than other nuclear genes and, therefore, to exhibit stronger statistical associations with the mitochondrial genome (mitonuclear linkage disequilibrium, LD). Such population genetic information could offer a valuable complement to phenotypic and biomedical data. It has been proposed that human mitonuclear incompatibilities are often cryptic, with poorly matched mitonuclear combinations being eliminated during oogenesis [38]. While such a mechanism would preclude directly observing the phenotypic consequences of mitonuclear mismatch, the elimination of specific mitonuclear genotypes from the population by natural selection would still alter the statistical associations between mitochondrial and nuclear loci and potentially leave detectable footprints in patterns of mitonuclear LD.
The recent flood of human population genomic data has been accompanied by detailed analysis of LD throughout the nuclear genome [60,66–70] as well as phylogeographic and functional analysis of variation in the mitochondrial genome [71–74]. Remarkably, however, genome-wide patterns of mitonuclear LD remain almost entirely unexplored. Here, we take advantage of existing human population genomic data to analyse patterns of mitonuclear LD and to test hypotheses about the history of selection on mitonuclear interactions—a pertinent topic in light of the current controversy surrounding MR.
2. Material and methods
(a). Human population genomic data
Genotypic data were obtained from the Human Genome Diversity Project (HGDP) [60]. This dataset comprises 660 918 single nucleotide polymorphism (SNP) genotypes for each of 1043 individuals, including 163 SNPs from the mitochondrial genome. The sampled individuals are broadly representative of human genetic and geographical diversity, encompassing 51 different populations [75]. We restricted our analysis to 585 300 nuclear SNPs with a minor allele frequency of at least 5% to avoid biases associated with measures of LD between loci with rare alleles (electronic supplementary material, figure S1) [76].
The chromosome and nucleotide position for each SNP were extracted from the SNP142 database obtained from the UCSC human genome downloads site [77]. SNPs found in coding sequences, introns, untranslated regions, or 5′ and 3′ flanking regions were associated with corresponding Entrez Gene IDs using SNPnexus [78] with the GRCh37/h19 human genome assembly. N-mt genes were identified based on the human MitoCarta classification [79]. In addition, we manually selected a smaller set of N-mt genes which code for proteins that form multi-subunit complexes with mitochondrial-encoded gene products or directly interact with mitochondrial DNA or RNAs. This subset included genes that encode OXPHOS subunits, ribosomal proteins, tRNA synthetases, and DNA and RNA polymerases (electronic supplementary material, table S1). In total, 237 156 SNPs were assigned to genes in the MitoCarta dataset. Of those, 7677 were assigned to the entire set of MitoCarta N-mt genes, and 754 were found in the more targeted subset.
(b). Determination of mitochondrial haplogroups
Illumina SNP genotyping probe sequences were obtained from the NCBI Probe Database and used to identify nucleotide positions for each mitochondrial SNP, which were then converted to the position numbering for the revised Cambridge Reference Sequence (rCRS) [80]. The resulting SNP data were analysed with HaploGrep v. 2.0 Beta [81] with build 16 of PhyloTree [82] to assign a mitochondrial haplogroup for each individual (electronic supplementary material, table S2). Haplogroups with fewer than 20 individuals were combined with closely related haplogroup(s) following the mitochondrial genealogy described by van Oven & Kayser [82] (electronic supplementary material, table S3). The L5 haplogroup was excluded from subsequent analyses, because it contained only two individuals in the HGDP dataset, and its position within the human mitochondrial genealogy precludes merging it with any other haplogroups.
(c). Principal component analysis
To provide a visual clustering of nuclear genetic diversity to compare with mitochondrial haplotype variation, a principal component analysis of all nuclear SNPs was performed using SmartPCA, which is part of the Eigensoft v. 6.0.1 package [83]. We employed default settings for the number of eigenvectors calculated and for the removal of outlier individuals. Formatting for the HGDP data for input into SmartPCA was performed with a custom bash script and Plink v. 1.07 [84].
(d). Analysis of mitonuclear linkage disequilibrium
A custom Perl script was used to calculate LD between each nuclear SNP locus and the mitochondrial genome (electronic supplementary material, file S1). Because there are more than two mitochondrial alleles (haplogroups), we calculated multi-allelic LD using the following measures as defined by Zhao et al. [76]: D′, r2, D*, χ2df and χ2′. We accounted for differences in ploidy between autosomes, sex chromosomes and the mitochondrial genome by randomly sampling one allele per individual for each diploid nuclear locus. D′ was only moderately correlated with each of the other measures of LD (r < 0.5), reflecting known differences in how low-frequency genotype combinations differentially affect these measures [76]. The other four LD measures were highly correlated with each other (r > 0.9). Therefore, for subsequent analyses, we considered D′ and only one of the four other measures (r2). D′ is the standard LD coefficient (D) scaled to be between 0 and 1 based on the maximum possible absolute value of D for the given allele frequencies [85]. By contrast, r2 is the square of the correlation coefficient between a pair of loci and represents the amount of predictive information that one locus can provide about another [67,76,86].
For each nuclear SNP, the significance of the observed association between mitochondrial haplogroups and nuclear alleles was assessed with a Fisher's exact test, which was implemented in R v. 3.1.2 [87], using the fisher.test function with p-values calculated by Monte Carlo simulations with 105 replicates. The false discovery rate (FDR) for identifying non-random mitonuclear associations was calculated with a Benjamini–Hochberg step-up procedure [88]. Mitonuclear LD values were calculated for each gene by averaging D′ and r2 estimates across all SNPs associated with that gene. ANOVAs were implemented in R to test whether chromosomes maintain significantly different levels of mitonuclear LD based on mean values from all genes containing a minimum of five SNPs. These values were log-transformed to meet normality assumptions. We also performed Fisher's exact tests to determine whether chromosomes differed significantly in the proportion of genes that were found in the upper 5% tail of mitonuclear LD values across the entire genome. Because of the small number of Y-linked SNPs in the HGDP dataset, the Y chromosome was excluded from these analyses.
(e). Functional enrichment analysis
To test the hypothesis that selection on co-adapted mitonuclear interactions has preferentially maintained LD between N-mt genes and the mitochondrial genome, we used t-tests implemented in R to compare log-transformed levels of mitonuclear LD (D′ and r2) between N-mt genes and all other genes. We also performed Fisher's exact tests to determine whether N-mt genes were overrepresented in the 5% tail of the distribution of mitonuclear LD values. These tests were performed with the entire set of N-mt genes and with the targeted subset of N-mt genes (electronic supplementary material, table S1) that were chosen based on their more intimate molecular interactions with mitochondrial-encoded gene products or the mitochondrial genome itself. To identify any functional characteristics that are over-represented in genes with high levels of mitonuclear LD, we performed a gene ontology (GO) enrichment analysis with the David 6.7 web server [89]. We used the list of genes with the top 5% of r2-values as the test set for this analysis. We restricted each of the above analyses to genes with a minimum of five SNPs.
3. Results
SNP genotypes from 1041 geographically and genetically diverse individuals revealed that mitonuclear LD is prevalent throughout the human genome. The vast majority (89%) of nuclear SNPs exhibited a significantly non-random association with mitochondrial haplogroups at an FDR threshold of 0.05, and genome-wide measures of genetic diversity tended to cluster with mitochondrial haplogroups (figure 2). Although significant, the observed levels of mitonuclear LD were generally low. The median and mean values of D′ were 0.2360 and 0.2541, respectively. Because the D′ statistic is known to yield inflated values when certain genotypic combinations are rare or unobserved [76], r2 is probably the more informative measure of LD in this case. The median and mean values of r2 were only 0.0033 and 0.0041, respectively, with a maximum observed value of 0.0304 (SNP rs8179271).
Figure 2.
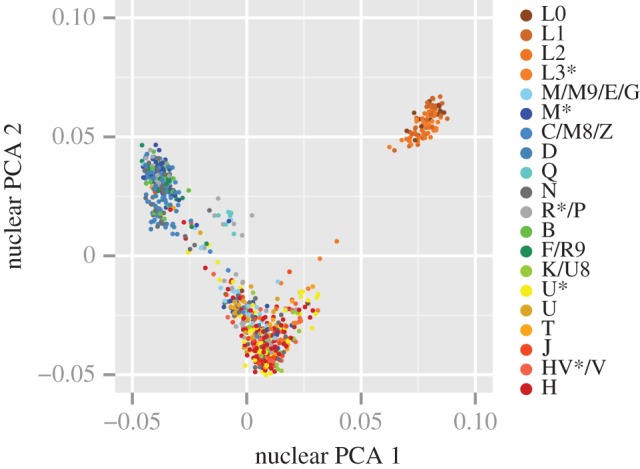
Principal component analysis (PCA) of SNPs in N-mt genes. Individuals are colour-coded based on their mitochondrial haplogroups. The N group consists of haplogroups N, N*, I, W, Y, A, S, X and O.
The magnitude of mitonuclear LD varied across different regions of the genome (figure 3). There were significant differences among chromosomes for gene-averaged values of both D′ and r2 (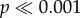 for both measures; ANOVA). Most notably, levels of mitonuclear LD were substantially higher in X-linked genes (mean D′ = 0.31; mean r2 = 0.0062) than autosomal genes (figures 3 and 4) (mean D′ = 0.26; mean r2 = 0.0041). There were also significant (albeit smaller) differences among autosomes for both D′ and r2 (
for both measures; ANOVA). Most notably, levels of mitonuclear LD were substantially higher in X-linked genes (mean D′ = 0.31; mean r2 = 0.0062) than autosomal genes (figures 3 and 4) (mean D′ = 0.26; mean r2 = 0.0041). There were also significant (albeit smaller) differences among autosomes for both D′ and r2 (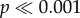 for both measures; ANOVA) (electronic supplementary material, table S4). On average, genes on chromosome 15 exhibited the strongest mitonuclear LD (mean D′ = 0.272; mean r2 = 0.0048), while genes on chromosome 19 (mean D′ = 0.250; mean r2 = 0.0040) and chromosome 14 (mean D′ = 0.252; mean r2 = 0.0039) had the weakest associations in terms of D′ and r2, respectively.
for both measures; ANOVA) (electronic supplementary material, table S4). On average, genes on chromosome 15 exhibited the strongest mitonuclear LD (mean D′ = 0.272; mean r2 = 0.0048), while genes on chromosome 19 (mean D′ = 0.250; mean r2 = 0.0040) and chromosome 14 (mean D′ = 0.252; mean r2 = 0.0039) had the weakest associations in terms of D′ and r2, respectively.
Figure 3.
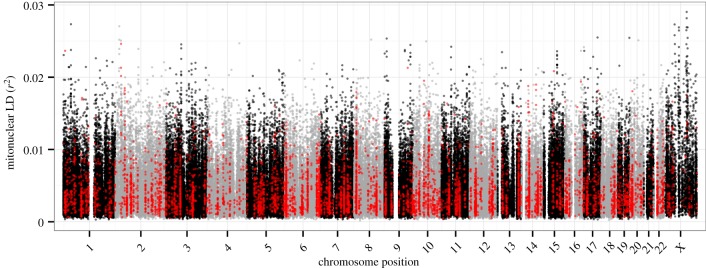
Variation in mitonuclear LD across the nuclear genome. Only SNPs associated with annotated genes are plotted, with those in N-mt genes shown in red.
Figure 4.
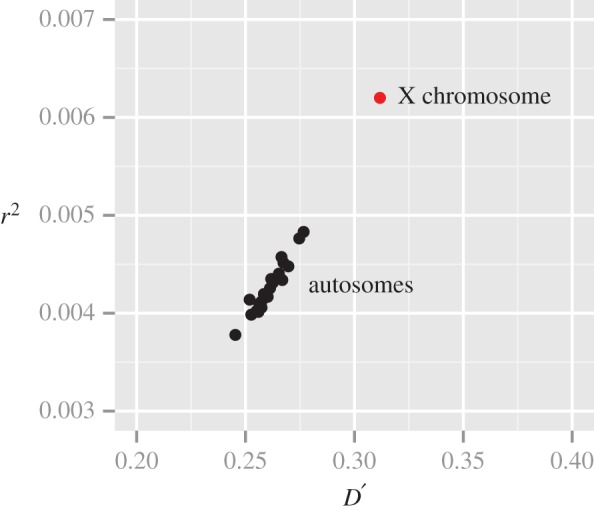
Mitonuclear LD averaged by chromosome. Each point represents a chromosome, and LD statistics (D′ and r2) are calculated based on the mean of all genes with at least five SNPs.
We did not find evidence that levels of mitonuclear LD were significantly different for N-mt genes than for the rest of the genome. Epistatic selection on mitonuclear interactions is expected to preferentially involve nuclear-encoded proteins that are targeted to mitochondria, yielding the prediction that N-mt genes would exhibit elevated levels of mitonuclear LD. However, this was not observed when LD was measured across the global sample of individuals that make up the HGDP dataset. Gene-averaged levels of mitonuclear LD were nearly identical between N-mt genes (mean D′ = 0.260; mean r2 = 0.00419) and the rest of the genes in the genome (mean D′ = 0.258; mean r2 = 0.00419), resulting in non-significant differences for both D′ and r2 (p = 0.83 and 0.54, respectively; ANOVA). N-mt genes were slightly overrepresented in the set of ‘outlier’ genes with the highest levels of mitonuclear LD, with 5.8 and 5.3% of N-mt genes being found in the 5% upper tail of the mitonuclear LD distribution based on D′ or r2, respectively, but these values did not deviate significantly from the random expectation (p = 0.48 and 0.72 for D′ and r2, respectively). Overall, N-mt genes were statistically indistinguishable from the rest of the genome with respect to their mitonuclear LD distributions (figures 3 and 5). Restricting these analyses to N-mt genes that are expected to be involved in the most direct interactions with the mitochondrial genome and its gene products (electronic supplementary material, table S1) also yielded non-significant results (electronic supplementary material, table S5). There is a potential for bias in these analyses because mitonuclear LD levels differ substantially between the X chromosome and the autosomes (figures 3 and 4), and it has been shown that N-mt genes are under-represented on the mammalian X chromosome [90]. To account for this bias, we repeated the analysis of N-mt genes after excluding the X chromosome and found no qualitative change in the results.
Figure 5.
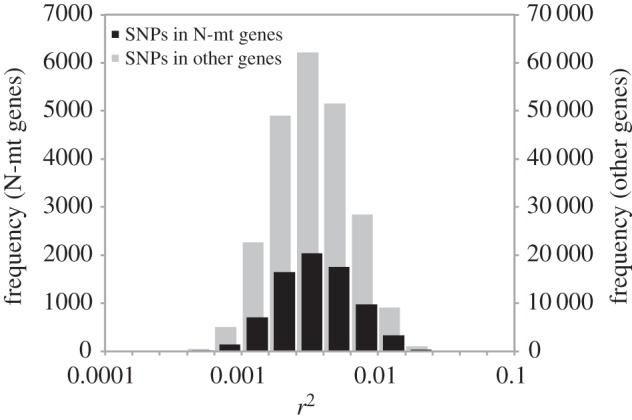
Frequency distribution of mitonuclear LD (r2) for SNPs in N-mt genes and other genes that are not targeted to the mitochondria.
In the absence of a detectable effect associated with N-mt genes, we considered the possibility that other functional characteristics could be responsible for variation in levels of mitonuclear LD among genes. However, the subset of genes with the top 5% highest levels of mitonuclear LD was not significantly enriched for terms in any of the three GO categories (biological processes, cellular components or molecular functions).
4. Discussion
(a). Human mitonuclear linkage disequilibrium
For mitochondrial and nuclear loci (or any loci that are not physically linked), the maintenance of LD requires either epistatic selection for certain combinations of alleles or non-random mating. In the absence of these forces, existing LD would decay rapidly (50% loss each generation) [91]. By itself, selection for specific combinations of alleles would have to be very strong to counteract this decay and maintain significant LD. Although it is conceivable that loci across the entire nuclear genome are all involved in epistatic interactions with the mitochondrial genome that produce such large fitness effects, we consider this very unlikely. Instead, the prevalence of significant mitonuclear LD throughout the entire human genome almost certainly reflects non-random mating resulting from the history of isolation between human populations [60,61].
There is obvious genetic differentiation among human populations, but levels of population structure are generally low [60,61]. In an analysis of the same HGDP dataset, Li et al. [60] found that the among-population component represents only 9% of total human genetic diversity. Population structure is more pronounced for genetic variation in the mitochondrial genome. Because of its asexual and effectively haploid mode of inheritance, the mitochondrial genome has a lower effective population size (Ne) and, therefore, should experience higher rates of genetic divergence between populations as a result of drift [92]. Accordingly, large geographical regions such as Europe and East Asia are highly enriched for a small fraction of mitochondrial haplotype diversity, reflecting the history of repeated bottlenecks as human populations migrated out of Africa [10,82,93]. The geographically structured distribution of mitochondrial genetic variation may also have been shaped by local adaptation to environmental conditions such as climate [72,74,94]. Therefore, the significant but generally weak statistical associations between nuclear and mitochondrial polymorphisms that are prevalent throughout the human genome are consistent with our understanding of human population genetics and demographic history.
The elevated level of mitonuclear LD that we observed for X-linked genes is probably caused by the fact that the X chromosome has a lower Ne than autosomes, resulting in faster rates of population divergence by drift [60,95]. Estimates of the among-population component of total diversity in the HGDP sample are significantly higher for the X chromosome (13%) than autosomes (9%) [60]. As both mitochondrial and X-linked loci accumulate differences between populations at a high rate, they will more rapidly build up associations with each other. Another relevant issue is that two-thirds of X chromosomes in a population are found in females and are, therefore, co-transmitted with mtDNA, whereas autosomes are shared equally by males and females [96]. As a result, in populations with a recent history of admixture, there may be a slower rate of decay of mitonuclear LD that involves X-linked genes. It has also been proposed that higher rates of co-transmission should facilitate co-adaptations between X-linked genes and the mitochondrial genome [90,96]. Therefore, it is also conceivable that selection against disrupting co-adapted mitonuclear genotypes may be partially responsible for the higher levels of mitonuclear LD on the X chromosome. Teasing apart the relative contributions of Ne, inheritance, and selection is an important motivation for additional empirical and theoretical work on the generation and maintenance of mitonuclear LD on autosomes and sex chromosomes.
(b). Mitonuclear interactions and the risks of mitochondrial replacement therapy
The development and approval of MR as a clinical treatment has reaffirmed the importance of understanding mitonuclear interactions in humans [30–35]. One source of controversy surrounding this technology is its potential to create maladapted pairings of mitochondrial and nuclear alleles, which is viewed as a serious concern by some researchers [36–38] but has been dismissed by others based on the limited evidence for such incompatibilities in humans and the fact that the genetic shuffling resulting from MR is similar (though not identical [38]) to the normal results of biparental sexual reproduction [62,97,98]. We propose that taking a species' population genetic history into account is important when assessing the potential for mitonuclear interactions and incompatibilities. Our analysis revealed significant non-random associations between mitochondrial haplotypes and nuclear alleles throughout the human genome. In principle, these findings lend credibility to the argument that MR has the potential to disrupt co-adapted mitonuclear interactions. However, as noted above, the statistical associations were generally weak as would be expected given the limited amounts of population genetic structure found in humans [60,61] and the fact that mitochondrial haplotypes are only partially informative in predicting continental ancestry [93] or controlling for population stratification in association studies [99,100].
To further assess the importance of mitonuclear coevolution in human populations, we hypothesized that the effects of epistatic selection (specifically mitonuclear LD) would vary across the genome and be most pronounced in N-mt genes. The logic behind this prediction is that selection for efficient mitonuclear interactions would: (i) disproportionately affect genes which encode proteins that are targeted to the mitochondria and involved in direct mitonuclear interactions, (ii) lead to faster sorting of ancestral genetic diversity among isolated populations at these loci, and (iii) preferentially restrict introgression at these loci during periods of genetic admixture. This line of logic is consistent with the observation that N-mt genes can exhibit steep clines in hybrid zones that mirror the distribution of mitochondrial genetic variation [101,102] and that mitonuclear interactions play a disproportionate role in generating reproductive isolation between species [6,7].
We did not detect elevated levels of mitonuclear LD for N-mt genes, indicating that selection has not systematically maintained stronger mitonuclear associations involving these loci when assessed at the global scale of human genetic diversity. However, there are reasons that this test may be conservative, which point to important avenues for future investigation. First, our separation of nuclear genes into two categories (N-mt and ‘other’) is surely an oversimplification that may mask more subtle variation among genes. Given the extent to which mitochondria are integrated into the broader metabolic, signalling and regulatory pathways within the eukaryotic cell, it is possible that gene products that are not physically targeted to the mitochondria could still create epistatic fitness effects that depend on the mitochondrial background, thereby reducing any signal that differentiates N-mt genes from the rest of the genome. Second, major mitonuclear incompatibilities can result from individual substitutions in a single N-mt gene. For example, Meiklejohn et al. [44] identified mitonuclear incompatibility with major fitness consequences in Drosophila melanogaster that is based on a SNP in a mitochondrial-encoded tRNA gene and a single amino acid polymorphism in the corresponding N-mt tRNA synthetase gene. Such effects that are localized to a single gene may get lost in the noise when pooled with hundreds of other N-mt genes in the genome. Therefore, even though we did not detect greater mitonuclear LD in N-mt genes, the genes exhibiting the highest levels of mitonuclear LD represent candidates for involvement in mitonuclear interactions and incompatibilities (electronic supplementary material, table S6). Third, because the published data used in our analyses were derived from SNP genotyping arrays, they are biased towards high-frequency polymorphisms (which we exacerbated by filtering based on minor allele frequency). This bias is relevant because alleles that are involved in epistatic interaction with highly deleterious effects are less likely to rise to high frequencies. Therefore, datasets based on whole genome or exome re-sequencing could provide a valuable complement to this analysis.
Future analysis of mitonuclear LD in humans may benefit from a targeted approach that focuses on specific N-mt genes and specific populations with mitochondrial haplotypes that have been implicated in mitonuclear epistasis [13–24]. Analysis of human cybrid cell lines is also a promising method for manipulating mitonuclear genotypes and identifying loci responsible for deleterious mismatched mitonuclear interactions [26–28]. To date, however, cybrid studies in humans have each been performed on a single nuclear background, which does not allow for distinguishing any mitonuclear interaction effects from the additive effects of mitochondrial genotype. Generating a full matrix of pairwise combinations of nuclear and mitochondrial genotypes as has been done in yeast [45] could be used to test for mitonuclear sign epistasis (figure 1) in human cell lines and eventually to identify the specific loci responsible for any epistatic effects.
Another important approach for expanding our understanding of mitonuclear interactions will be to measure LD within relatively panmictic populations. Incorporating data from human populations with a recent history of admixture would be particularly valuable. One of the expected effects of selection on mitonuclear interactions is to slow the decay of LD at loci responsible for mitonuclear incompatibilities when populations interbreed. This would be difficult to observe in more isolated populations because alleles associated with strong mitonuclear incompatibilities are likely to have already been purged. Examining recently admixed populations will be especially valuable for further testing the hypothesis that mitonuclear incompatibilities commonly occur without ever being observed, because they are selectively eliminated during oogenesis [38].
Overall, our analysis did not find compelling evidence for a history of strong selection to conserve co-adapted mitonuclear interactions in humans, which is in line with the arguments that mitonuclear incompatibilities may be less prevalent in humans than many other eukaryotic lineages because of human demographic history [62]. Perhaps our most striking observation, however, is how little we currently know about mitonuclear LD in humans. It is clear that this gap in our understanding of the population genetics of our own species needs to be addressed. As researchers and clinicians evaluate MR techniques, this will help assess the risks associated with mitonuclear incompatibilities alongside other concerns about the ethics of genetic engineering, the technical risks associated with manipulating reproductive cells and the efficient allocation of biomedical resources. We suggest that taking advantage of the ever-increasing availability of human population genomic data to analyse patterns of mitonuclear LD will offer a novel approach to identify loci that may have been historical targets of selection for efficient mitonuclear interactions and to determine the epistatic component of human phenotypes and diseases with a complex genetic basis.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank John Brookfield, Adam Eyre-Walker and an anonymous reviewer for insightful comments that helped us improve an earlier version of the manuscript.
Data accessibility
The SNP genotype data from the HGDP were previously published [60], and the data are available via the HGDP website: http://www.hagsc.org/hgdp. The code used to analyse these data and the resulting data tables are available as electronic supplementary material accompanying this paper.
Authors' contributions
D.B.S. conceived of and designed the study, performed analyses and drafted the manuscript. P.D.F. and J.C.H. designed the study and performed analyses. All authors critically revised the manuscript and gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
Our research on the evolution of mitonuclear interactions is supported by the National Science Foundation (MCB-1412260) and Colorado State University. J.C.H. is supported by a National Institutes of Health Postdoctoral Fellowship (F32GM116361).
References
- 1.Rand DM, Haney RA, Fry AJ. 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19, 645–653. ( 10.1016/j.tree.2004.10.003) [DOI] [PubMed] [Google Scholar]
- 2.Levin L, Blumberg A, Barshad G, Mishmar D. 2014. Mito-nuclear co-evolution: the positive and negative sides of functional ancient mutations. Front. Genet. 5, 448 ( 10.3389/fgene.2014.00448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burton RS, Pereira RJ, Barreto FS. 2013. Cytonuclear genomic interactions and hybrid breakdown. Annu. Rev. Ecol. Evol. Syst. 44, 281–302. ( 10.1146/annurev-ecolsys-110512-135758) [DOI] [Google Scholar]
- 4.Wolff JN, Ladoukakis ED, Enríquez JA, Dowling DK. 2014. Mitonuclear interactions: evolutionary consequences over multiple biological scales. Phil. Trans. R. Soc. B 369, 20130443 ( 10.1098/rstb.2013.0443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill GE. 2015. Mitonuclear ecology. Mol. Biol. Evol. 32, 1917–1927. ( 10.1093/molbev/msv104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gershoni M, Templeton AR, Mishmar D. 2009. Mitochondrial bioenergetics as a major motive force of speciation. Bioessays 31, 642–650. ( 10.1002/bies.200800139) [DOI] [PubMed] [Google Scholar]
- 7.Burton RS, Barreto FS. 2012. A disproportionate role for mtDNA in Dobzhansky-Muller incompatibilities? Mol. Ecol. 21, 4942–4957. ( 10.1111/mec.12006) [DOI] [PubMed] [Google Scholar]
- 8.Larsson NG. 2010. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 79, 683–706. ( 10.1146/annurev-biochem-060408-093701) [DOI] [PubMed] [Google Scholar]
- 9.Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. 2010. Mitochondrial DNA mutations and human disease. Biochim. Biophys. Acta 1797, 113–128. ( 10.1016/j.bbabio.2009.09.005) [DOI] [PubMed] [Google Scholar]
- 10.Wallace DC, Chalkia D. 2013. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb. Perspect. Med. 3, a021220 ( 10.1101/cshperspect.a021220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dowling DK. 2014. Evolutionary perspectives on the links between mitochondrial genotype and disease phenotype. Biochim. Biophys. Acta 1840, 1393–1403. ( 10.1016/j.bbagen.2013.11.013) [DOI] [PubMed] [Google Scholar]
- 12.Horan MP, Cooper DN. 2014. The emergence of the mitochondrial genome as a partial regulator of nuclear function is providing new insights into the genetic mechanisms underlying age-related complex disease. Hum. Genet. 133, 435–458. ( 10.1007/s00439-013-1402-4) [DOI] [PubMed] [Google Scholar]
- 13.Hudson G, et al. 2005. Identification of an X-chromosomal locus and haplotype modulating the phenotype of a mitochondrial DNA disorder. Am. J. Hum. Genet. 77, 1086–1091. ( 10.1086/498176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rai E, Sharma S, Koul A, Bhat AK, Bhanwer AJ, Bamezai RN. 2007. Interaction between the UCP2–866G/A, mtDNA 10398G/A and PGC1α p.Thr394Thr and p.Gly482Ser polymorphisms in type 2 diabetes susceptibility in North Indian population. Hum. Genet. 122, 535–540. ( 10.1007/s00439-007-0421-4) [DOI] [PubMed] [Google Scholar]
- 15.Feder J, Ovadia O, Blech I, Cohen J, Wainstein J, Harman-Boehm I, Glaser B, Mishmar D. 2009. Parental diabetes status reveals association of mitochondrial DNA haplogroup J1 with type 2 diabetes. BMC Med. Genet. 10, 60 ( 10.1186/1471-2350-10-60) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potluri P, et al. 2009. A novel NDUFA1 mutation leads to a progressive mitochondrial complex I-specific neurodegenerative disease. Mol. Genet. Metab. 96, 189–195. ( 10.1016/j.ymgme.2008.12.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonaïti B, Olsson M, Hellman U, Suhr O, Bonaïti-Pellié C, Planté-Bordeneuve V. 2010. TTR familial amyloid polyneuropathy: does a mitochondrial polymorphism entirely explain the parent-of-origin difference in penetrance? Eur. J. Hum. Genet. 18, 948–952. ( 10.1038/ejhg.2010.36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaweda-Walerych K, et al. 2010. Mitochondrial transcription factor A variants and the risk of Parkinson's disease. Neurosci. Lett. 469, 24–29. ( 10.1016/j.neulet.2009.11.037) [DOI] [PubMed] [Google Scholar]
- 19.Taherzadeh-Fard E, Saft C, Akkad DA, Wieczorek S, Haghikia A, Chan A, Epplen JT, Arning L. 2011. PGC-1alpha downstream transcription factors NRF-1 and TFAM are genetic modifiers of Huntington disease. Mol. Neurodegener. 6, 32 ( 10.1186/1750-1326-6-32) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez-Duran A, Pacheu-Grau D, Martinez-Romero I, Lopez-Gallardo E, Lopez-Perez MJ, Montoya J, Ruiz-Pesini E. 2012. Oxidative phosphorylation differences between mitochondrial DNA haplogroups modify the risk of Leber's hereditary optic neuropathy. Biochim. Biophys. Acta 1822, 1216–1222. ( 10.1016/j.bbadis.2012.04.014) [DOI] [PubMed] [Google Scholar]
- 21.Neeve VC, et al. 2012. What is influencing the phenotype of the common homozygous polymerase-gamma mutation p.Ala467Thr? Brain 135, 3614–3626. ( 10.1093/brain/aws298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo LF, Hou CC, Yang WX. 2013. Nuclear factors: roles related to mitochondrial deafness. Gene 520, 79–89. ( 10.1016/j.gene.2013.03.041) [DOI] [PubMed] [Google Scholar]
- 23.Strauss KA, et al. 2013. Severity of cardiomyopathy associated with adenine nucleotide translocator-1 deficiency correlates with mtDNA haplogroup. Proc. Natl Acad. Sci. USA 110, 3453–3458. ( 10.1073/pnas.1300690110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gershoni M, et al. 2014. Disrupting mitochondrial-nuclear coevolution affects OXPHOS complex I integrity and impacts human health. Genome Biol. Evol. 6, 2665–2680. ( 10.1093/gbe/evu208) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenyon L, Moraes CT. 1997. Expanding the functional human mitochondrial DNA database by the establishment of primate xenomitochondrial cybrids. Proc. Natl Acad. Sci. USA 94, 9131–9135. ( 10.1073/pnas.94.17.9131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin TK, Lin HY, Chen SD, Chuang YC, Chuang JH, Wang PW, Huang ST, Tiao MM, Chen JB, Liou CW. 2012. The creation of cybrids harboring mitochondrial haplogroups in the Taiwanese population of ethnic Chinese background: an extensive in vitro tool for the study of mitochondrial genomic variations. Oxid. Med. Cell. Longev. 2012, 824275 ( 10.1155/2012/824275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenney MC, et al. 2014. Inherited mitochondrial DNA variants can affect complement, inflammation and apoptosis pathways: insights into mitochondrial-nuclear interactions. Hum. Mol. Genet. 23, 3537–3551. ( 10.1093/hmg/ddu065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenney MC, et al. 2014. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: implications for population susceptibility to diseases. Biochim. Biophys. Acta 1842, 208–219. ( 10.1016/j.bbadis.2013.10.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malik D. 2014. Human retinal transmitochondrial cybrids with J or H mtDNA haplogroups respond differently to ultraviolet radiation: implications for retinal diseases. PLoS ONE 9, e99003 ( 10.1371/journal.pone.0099003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couzin-Frankel J. 2015. Eggs’ power plants energize new IVF debate. Science 348, 14–15. ( 10.1126/science.348.6230.14) [DOI] [PubMed] [Google Scholar]
- 31.Craven L, et al. 2010. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature 465, 82–85. ( 10.1038/nature08958) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paull D, et al. 2013. Nuclear genome transfer in human oocytes eliminates mitochondrial DNA variants. Nature 493, 632–637. ( 10.1038/nature11800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tachibana M, et al. 2013. Towards germline gene therapy of inherited mitochondrial diseases. Nature 493, 627–631. ( 10.1038/nature11647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T, Sha H, Ji D, Zhang HL, Chen D, Cao Y, Zhu J. 2014. Polar body genome transfer for preventing the transmission of inherited mitochondrial diseases. Cell 157, 1591–1604. ( 10.1016/j.cell.2014.04.042) [DOI] [PubMed] [Google Scholar]
- 35.Wolf DP, Mitalipov N, Mitalipov S. 2015. Mitochondrial replacement therapy in reproductive medicine. Trends Mol. Med. 21, 68–76. ( 10.1016/j.molmed.2014.12.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reinhardt K, Dowling DK, Morrow EH. 2013. Mitochondrial replacement, evolution, and the clinic. Science 341, 1345–1346. ( 10.1126/science.1237146) [DOI] [PubMed] [Google Scholar]
- 37.Gemmell N, Wolff JN. 2015. Mitochondrial replacement therapy: cautiously replace the master manipulator. BioEssays 37, 584–585. ( 10.1002/bies.201500008) [DOI] [PubMed] [Google Scholar]
- 38.Morrow EH, Reinhardt K, Wolff JN, Dowling DK. 2015. Risks inherent to mitochondrial replacement. EMBO Rep. 16, 541–544. ( 10.15252/embr.201439110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamaoka M, Isobe K, Shitara H, Yonekawa H, Miyabayashi S, Hayashi JI. 2000. Complete repopulation of mouse mitochondrial DNA-less cells with rat mitochondrial DNA restores mitochondrial translation but not mitochondrial respiratory function. Genetics 155, 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rand DM, Fry A, Sheldahl L. 2006. Nuclear-mitochondrial epistasis and Drosophila aging: introgression of Drosophila simulans mtDNA modifies longevity in D. melanogaster nuclear backgrounds. Genetics 172, 329–341. ( 10.1534/genetics.105.046698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dowling DK, Friberg U, Hailer F, Arnqvist G. 2007. Intergenomic epistasis for fitness: within-population interactions between cytoplasmic and nuclear genes in Drosophila melanogaster. Genetics 175, 235–244. ( 10.1534/genetics.105.052050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ellison CK, Burton RS. 2008. Interpopulation hybrid breakdown maps to the mitochondrial genome. Evolution 62, 631–638. ( 10.1111/j.1558-5646.2007.00305.x) [DOI] [PubMed] [Google Scholar]
- 43.Lee HY, Chou JY, Cheong L, Chang NH, Yang SY, Leu JY. 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135, 1065–1073. ( 10.1016/j.cell.2008.10.047) [DOI] [PubMed] [Google Scholar]
- 44.Meiklejohn CD, Holmbeck MA, Siddiq MA, Abt DN, Rand DM, Montooth KL. 2013. An Incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet. 9, e1003238 ( 10.1371/journal.pgen.1003238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paliwal S, Fiumera AC, Fiumera HL. 2014. Mitochondrial-nuclear epistasis contributes to phenotypic variation and coadaptation in natural isolates of Saccharomyces cerevisiae. Genetics 198, 1251–1265. ( 10.1534/genetics.114.168575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deuse T. 2015. SCNT-derived ESCs with mismatched mitochondria trigger an immune response in allogeneic hosts. Cell Stem Cell 16, 33–38. ( 10.1016/j.stem.2014.11.003) [DOI] [PubMed] [Google Scholar]
- 47.Spirek M, Polakova S, Jatzova K, Sulo P. 2015. Post-zygotic sterility and cytonuclear compatibility limits in S. cerevisiae xenomitochondrial cybrids. Front. Genet. 5, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willett CS, Burton RS. 2004. Evolution of interacting proteins in the mitochondrial electron transport system in a marine copepod. Mol. Biol. Evol. 21, 443–453. ( 10.1093/molbev/msh031) [DOI] [PubMed] [Google Scholar]
- 49.Mishmar D, Ruiz-Pesini E, Mondragon-Palomino M, Procaccio V, Gaut B, Wallace DC. 2006. Adaptive selection of mitochondrial complex I subunits during primate radiation. Gene 378, 11–18. ( 10.1016/j.gene.2006.03.015) [DOI] [PubMed] [Google Scholar]
- 50.Osada N, Akashi H. 2012. Mitochondrial-nuclear interactions and accelerated compensatory evolution: evidence from the primate cytochrome c oxidase complex. Mol. Biol. Evol. 29, 337–346. ( 10.1093/molbev/msr211) [DOI] [PubMed] [Google Scholar]
- 51.Barreto FS, Burton RS. 2013. Evidence for compensatory evolution of ribosomal proteins in response to rapid divergence of mitochondrial rRNA. Mol. Biol. Evol. 30, 310–314. ( 10.1093/molbev/mss228) [DOI] [PubMed] [Google Scholar]
- 52.Sloan DB, Triant DA, Wu M, Taylor DR. 2014. Cytonuclear interactions and relaxed selection accelerate sequence evolution in organelle ribosomes. Mol. Biol. Evol. 31, 673–682. ( 10.1093/molbev/mst259) [DOI] [PubMed] [Google Scholar]
- 53.Ellison CK, Burton RS. 2010. Cytonuclear conflict in interpopulation hybrids: the role of RNA polymerase in mtDNA transcription and replication. J. Evol. Biol. 23, 528–538. ( 10.1111/j.1420-9101.2009.01917.x) [DOI] [PubMed] [Google Scholar]
- 54.Burton RS, Ellison CK, Harrison JS. 2006. The sorry state of F2 hybrids: consequences of rapid mitochondrial DNA evolution in allopatric populations. Am. Nat. 168, S14–S24. ( 10.1086/509046) [DOI] [PubMed] [Google Scholar]
- 55.Asmussen MA, Arnold J. 1991. The effects of admixture and population subdivision on cytonuclear disequilibria. Theor. Popul. Biol. 39, 273–300. ( 10.1016/0040-5809(91)90024-A) [DOI] [PubMed] [Google Scholar]
- 56.Wade MJ, Goodnight CJ. 2006. Cyto-nuclear epistasis: two-locus random genetic drift in hermaphroditic and dioecious species. Evolution 60, 643–659. ( 10.1111/j.0014-3820.2006.tb01146.x) [DOI] [PubMed] [Google Scholar]
- 57.Brandvain Y, Wade MJ. 2009. The functional transfer of genes from the mitochondria to the nucleus: the effects of selection, mutation, population size and rate of self-fertilization. Genetics 182, 1129–1139. ( 10.1534/genetics.108.100024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montooth KL, Meiklejohn CD, Abt DN, Rand DM. 2010. Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila. Evolution 64, 3364–3379. ( 10.1111/j.1558-5646.2010.01077.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fields PD, McCauley DE, McAssey EV, Taylor DR. 2014. Patterns of cyto-nuclear linkage disequilibrium in Silene latifolia: genomic heterogeneity and temporal stability. Heredity 112, 99–104. ( 10.1038/hdy.2013.79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li JZ, et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104. ( 10.1126/science.1153717) [DOI] [PubMed] [Google Scholar]
- 61.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. 2002. Genetic structure of human populations. Science 298, 2381–2385. ( 10.1126/science.1078311) [DOI] [PubMed] [Google Scholar]
- 62.Chinnery PF, Craven L, Mitalipov S, Stewart JB, Herbert M, Turnbull DM. 2014. The challenges of mitochondrial replacement. PLoS Genet. 10, e1004315 ( 10.1371/journal.pgen.1004315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohta T. 1982. Linkage disequilibrium due to random genetic drift in finite subdivided populations. Proc. Natl Acad. Sci. USA 79, 1940–1944. ( 10.1073/pnas.79.6.1940) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen R. 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39, 197–218. ( 10.1146/annurev.genet.39.073003.112420) [DOI] [PubMed] [Google Scholar]
- 65.Wade MJ. 2007. The co-evolutionary genetics of ecological communities. Nat. Rev. Genet. 8, 185–195. ( 10.1038/nrg2031) [DOI] [PubMed] [Google Scholar]
- 66.Reich DE, et al. 2001. Linkage disequilibrium in the human genome. Nature 411, 199–204. ( 10.1038/35075590) [DOI] [PubMed] [Google Scholar]
- 67.Ardlie KG, Kruglyak L, Seielstad M. 2002. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 3, 299–309. ( 10.1038/nrg777) [DOI] [PubMed] [Google Scholar]
- 68.International HapMap Consortium. 2003. The International HapMap Project. Nature 426, 789–796. ( 10.1038/nature02168) [DOI] [PubMed] [Google Scholar]
- 69.Conrad DF, Jakobsson M, Coop G, Wen X, Wall JD, Rosenberg NA, Pritchard JK. 2006. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nat. Genet. 38, 1251–1260. ( 10.1038/ng1911) [DOI] [PubMed] [Google Scholar]
- 70.1000 Genomes Project Consortium. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65. ( 10.1038/nature11632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wallace DC, Brown MD, Lott MT. 1999. Mitochondrial DNA variation in human evolution and disease. Gene 238, 211–230. ( 10.1016/S0378-1119(99)00295-4) [DOI] [PubMed] [Google Scholar]
- 72.Mishmar D, et al. 2003. Natural selection shaped regional mtDNA variation in humans. Proc. Natl Acad. Sci. USA 100, 171–176. ( 10.1073/pnas.0136972100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruiz-Pesini E, et al. 2007. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 35, D823–D828. ( 10.1093/nar/gkl927) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Balloux F, Handley LJ, Jombart T, Liu H, Manica A. 2009. Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation. Proc. R. Soc. B 276, 3447–3455. ( 10.1098/rspb.2009.0752) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cann HM, et al. 2002. A human genome diversity cell line panel. Science 296, 261–262. ( 10.1126/science.296.5566.261b) [DOI] [PubMed] [Google Scholar]
- 76.Zhao H, Nettleton D, Soller M, Dekkers JC. 2005. Evaluation of linkage disequilibrium measures between multi-allelic markers as predictors of linkage disequilibrium between markers and QTL. Genet. Res. 86, 77–87. ( 10.1017/S001667230500769X) [DOI] [PubMed] [Google Scholar]
- 77.Rosenbloom KR, et al. 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43, D670–D681. ( 10.1093/nar/gku1177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chelala C, Khan A, Lemoine NR. 2009. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics 25, 655–661. ( 10.1093/bioinformatics/btn653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pagliarini DJ, et al. 2008. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123. ( 10.1016/j.cell.2008.06.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 23, 147 ( 10.1038/13779) [DOI] [PubMed] [Google Scholar]
- 81.Kloss-Brandstätter A, Pacher D, Schonherr S, Weissensteiner H, Binna R, Specht G, Kronenberg F. 2011. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum. Mutat. 32, 25–32. ( 10.1002/humu.21382) [DOI] [PubMed] [Google Scholar]
- 82.van Oven M, Kayser M. 2009. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum. Mutat. 30, E386–E394. ( 10.1002/humu.20921) [DOI] [PubMed] [Google Scholar]
- 83.Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet. 2, e190 ( 10.1371/journal.pgen.0020190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. ( 10.1086/519795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lewontin RC. 1964. The interaction of selection and linkage. I. General considerations; heterotic models. Genetics 49, 49–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hill WG, Robertson A. 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38, 226–231. ( 10.1007/BF01245622) [DOI] [PubMed] [Google Scholar]
- 87.R Core Team. 2014. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 88.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300. [Google Scholar]
- 89.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. ( 10.1038/nprot.2008.211) [DOI] [PubMed] [Google Scholar]
- 90.Drown DM, Preuss KM, Wade MJ. 2012. Evidence of a paucity of genes that interact with the mitochondrion on the X in mammals. Genome Biol. Evol. 4, 763–768. ( 10.1093/gbe/evs064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Slatkin M. 2008. Linkage disequilibrium: understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 9, 477–485. ( 10.1038/nrg2361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Neiman M, Taylor DR. 2009. The causes of mutation accumulation in mitochondrial genomes. Proc. R. Soc. B 276, 1201–1209. ( 10.1098/rspb.2008.1758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Emery LS, Magnaye KM, Bigham AW, Akey JM, Bamshad MJ. 2015. Estimates of continental ancestry vary widely among individuals with the same mtDNA haplogroup. Am. J. Hum. Genet. 96, 183–193. ( 10.1016/j.ajhg.2014.12.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. 2004. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303, 223–226. ( 10.1126/science.1088434) [DOI] [PubMed] [Google Scholar]
- 95.Ramachandran S, Rosenberg NA, Zhivotovsky LA, Feldman MW. 2004. Robustness of the inference of human population structure: a comparison of X-chromosomal and autosomal microsatellites. Hum. Genomics 1, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rand DM, Clark AG, Kann LM. 2001. Sexually antagonistic cytonuclear fitness interactions in Drosophila melanogaster. Genetics 159, 173–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tanaka AJ, Sauer MV, Egli D, Kort DH. 2013. Harnessing the stem cell potential: the path to prevent mitochondrial disease. Nat. Med. 19, 1578–1579. ( 10.1038/nm.3422) [DOI] [PubMed] [Google Scholar]
- 98.Human Fertilisation and Embryology Authority. 2013 HFEA statement regarding the Klaus Reinhardt et al. Science paper ‘Mitochondrial replacement, evolution, and the clinic’. See http://www.hfea.gov.uk/8178.html.
- 99.Biffi A, et al. 2010. Principal-component analysis for assessment of population stratification in mitochondrial medical genetics. Am. J. Hum. Genet. 86, 904–917. ( 10.1016/j.ajhg.2010.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Makowsky R, Yan Q, Wiener HW, Sandel M, Aissani B, Tiwari HK, Shrestha S. 2012. The utility of mitochondrial and Y chromosome phylogenetic data to improve correction for population stratification. Front. Genet. 3, 301 ( 10.3389/fgene.2012.00301) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trier CN, Hermansen JS, Saetre GP, Bailey RI. 2014. Evidence for mito-nuclear and sex-linked reproductive barriers between the hybrid Italian sparrow and its parent species. PLoS Genet. 10, e1004075 ( 10.1371/journal.pgen.1004075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Beck EA, Thompson AC, Sharbrough J, Brud E, Llopart A. 2015. Gene flow between Drosophila yakuba and Drosophila santomea in subunit V of cytochrome c oxidase: a potential case of cytonuclear co-introgression. Evolution 69, 1973–1986. ( 10.1111/evo.12718) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SNP genotype data from the HGDP were previously published [60], and the data are available via the HGDP website: http://www.hagsc.org/hgdp. The code used to analyse these data and the resulting data tables are available as electronic supplementary material accompanying this paper.


