Abstract
Nucleolin is a pleiotropic protein involved in a variety of cellular processes. Although multipolar spindle formation has been observed after nucleolin depletion, the roles of nucleolin in centrosome regulation and functions have not been addressed. Here we report using immunofluorescence and biochemically purified centrosomes that nucleolin co-localized only with one of the centrioles during interphase which was further identified as the mature centriole. Upon nucleolin depletion, cells exhibited an amplification of immature centriole markers surrounded by irregular pericentrin staining; these structures were exempt from maturation markers and unable to nucleate microtubules. Furthermore, the microtubule network was disorganized in these cells, exhibiting frequent non-centrosomal microtubules. At the mature centriole a reduced kinetics in the centrosomal microtubule nucleation phase was observed in live silenced cells, as well as a perturbation of microtubule anchoring. Immunoprecipitation experiments showed that nucleolin belongs to protein complexes containing 2 key centrosomal proteins, γ-tubulin and ninein, involved in microtubule nucleation and anchoring steps. Altogether, our study uncovered a new role for nucleolin in restricting microtubule nucleation and anchoring at centrosomes in interphase cells.
Keywords: centrosome, interphase, mature centriole, microtubules, nucleolin
Introduction
Centrosomes are the main microtubule organizing centers in animal cells. They are formed by 2 barrel shaped microtubule based structures named centrioles, surrounded by a dense protein matrix: the pericentriolar material (PCM). In interphase cells, by organizing the microtubule cytoskeleton, the centrosome controls intracellular transport, organelle positioning, cell shape and polarity, as well as cell motility.1 During mitosis, the duplicated centrosomes are important for bipolar spindle establishment, which in turn is crucial for the correct chromosome segregation in anaphase.
Centrosome duplication is precisely controlled and coordinated with the cell cycle to allow a single duplication per cell cycle.2-4 Centriole duplication occurs in S phase and is initiated in coordination with DNA replication by the cdk2-cyclinE kinase. During centriole duplication a procentriole is assembled orthogonally to the proximal end of each pre-existing centriole. The procentrioles elongate until G2 phase so that the cell possesses 2 functional centrosomes in mitosis. During centrosome maturation, phosphorylation-driven accumulation of γ-tubulin ring complexes and other PCM proteins, like pericentrin, increases their ability to nucleate microtubules.5
Centrosome cycle deregulation can lead to the presence of supernumerary centrioles in the cell. In mitosis, the presence of multiple centrosomes can cause the establishment of a multipolar spindle responsible for chromosome missegregation, which in turn triggers aneuploid cell formation.6 This deregulation is a common characteristic of several cancers.7,8 Therefore, it is particularly critical to go forward in understanding molecular mechanisms regulating the number, shape, and position of centrioles in the cell, for preventing cell transformation.9,10
Within the centrosome, the 2 centrioles are different. The older one (called mother or mature centriole in contrast with the younger one called daughter or immature centriole) possesses 2 kinds of appendages.11 The distal appendages are important during ciliogenesis to anchor the mother centriole to the plasma membrane12,13 whereas the subdistal appendages are involved in microtubule anchoring at the centrosome.13,14 Even though, proteomic analyzes have identified several proteins associated with the centrosomes,15,16 their localization within the centrosome structure has only been characterized for a subset of proteins. Development of super-resolution approaches in optics, have recently contributed to decipher internal centrosomal organization.17,18 Indeed, several reports have lately revealed the presence of functional domains within the PCM, defined as concentric layers enclosing the mature centriole in a tube- or toroid-like arrangement.19-21 The outer PCM layer contains proteins involved in microtubule nucleation such as γ-tubulin.20 Pericentrin adopts an extended conformation and is organized radially with its C terminus associated to the centriole wall and its N terminus extending toward the PCM periphery.20
Microtubule nucleation is initiated by protein complexes containing γ-tubulin and called γTuRC (γ-Tubulin Ring Complexes).22,23 The γTuRC is composed of γTuSC subunits (γ-Tubulin Small Complex) formed by γ-tubulin, GCP2 and GCP3 and assembled by additional proteins such as GCP4, GCP5 and GCP6.24 Several additional proteins interacting with the γTuRC have been identified and implicated in the recruitment of this complex at microtubule nucleation sites like the centrosome.25 When nucleated in the PCM, short microtubules are either released in the cytoplasm or anchored at the mature centriole subdistal appendages.26 Therefore, centrosome-dependent organization of microtubules involves distinct processes such as nucleation, anchoring and release of microtubules. Even though, it is admitted distinct contributions of γ-TuRC and ninein to microtubule nucleation and anchoring processes, it has been proposed that ninein constitutes a molecular link between microtubule-nucleation and -anchoring activities at the centrosome.14
Nucleolin (NCL) is an abundant non-ribosomal protein of the nucleolus where it is involved in ribosome biogenesis. Nevertheless, nucleolin functions are not restricted to nucleoli as it has been identified in different compartments such as in the nucleoplasm, cytoplasm and at the cell membrane (for review see 27). The presence of nucleolin in several cellular compartments reveals its multifunctional nature. The depletion of nucleolin by RNA interference leads to reduced proliferation and increased apoptosis.28,29 In interphase cells, the absence of nucleolin results in an increased nuclear size as well as numerous nuclear alterations.28,29 Interestingly, when nucleolin-silenced cells reach mitosis, abnormal centrosome amplification is observed.28,29 However, the molecular mechanism linking nucleolin to centrosome duplication regulation has not been elucidated to date. By proteomic analysis, nucleolin has been identified in human centrosomes15 and mitotic spindles.30 By immunofluorescence, a phosphorylated form of nucleolin has also been detected in mitosis at the spindle poles28 but its function in these structures has never been addressed.
In the present study, we have explored the functions of nucleolin in key regulatory processes governed by centrosomes. Specifically, we focused on nucleolin's role on interphase centrosome. Our data clearly show that nucleolin is localized at the centrosome and more specifically at the mature centriole throughout interphase. By immunoprecipitation experiments, we also found that nucleolin belongs to protein complexes containing 2 key centrosomal proteins, γ-tubulin and ninein, involved in microtubule nucleation and anchorage steps. To assess centrosomal functions of nucleolin we then investigated the consequences of nucleolin silencing by RNA interference on centrosome structure and function. Nucleolin silencing induces an amplification of immature centriole markers in interphase cells surrounded by pericentrin staining, but found to be devoided of maturation markers and therefore unable to nucleate microtubules. We showed that although the number of microtubules forming the aster around the remaining mature centrosome is unchanged, a dense microtubule network, not connected to the centrosomal aster, is observed around the nucleus of nucleolin-silenced cells. In addition, we highlighted that the microtubule nucleation step in siNCL cells is slowed down compared to control cells. Finally, we collected evidences that the microtubule anchoring step in siNCL cells is perturbed since non-centrosomal microtubules are clearly visible after only 5 minutes of microtubule repolymerization. Altogether, our results point to a role for nucleolin in restricting microtubule nucleation and anchoring to centrosomes.
Results
In interphase cells, nucleolin labels one of the 2 centrioles
In order to determine whether nucleolin is present at the centrosome during interphase, immunofluorescence experiments were performed in osteosarcoma U2OS cells with a monoclonal antibody directed against the full-length human nucleolin. As shown on Fig. 1, nucleolin was not only detected within DAPI depleted regions corresponding to nucleoli (Fig. 1A, merge) but also as a single dot co-distributing with one of the 2 centrioles, labeled with centrin-1-GFP (Fig. 1A, merged zoom, z-stack gallery and the corresponding intensity profile on Fig. 1D). This observation was facilitated when cells were fixed at −20°C in pure methanol (Fig. S1A, C). The presence of nucleolin at interphase centrosome was confirmed using a second antibody directed against the N-terminus domain (first one hundred amino acids) of human nucleolin (Fig. S1E).
Figure 1 (See previous page).
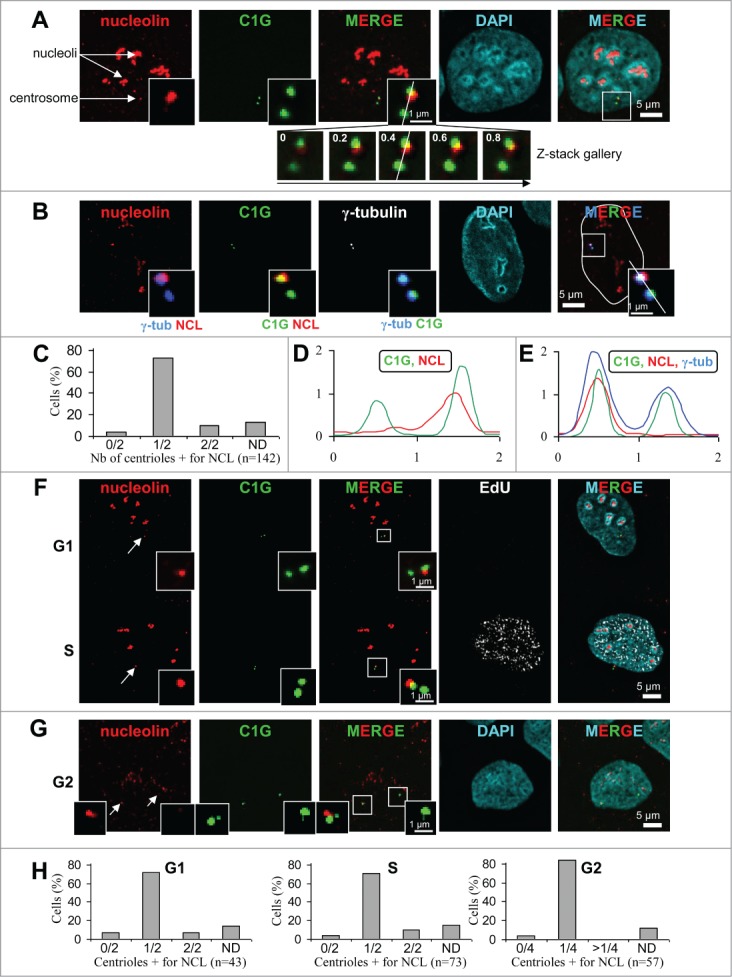
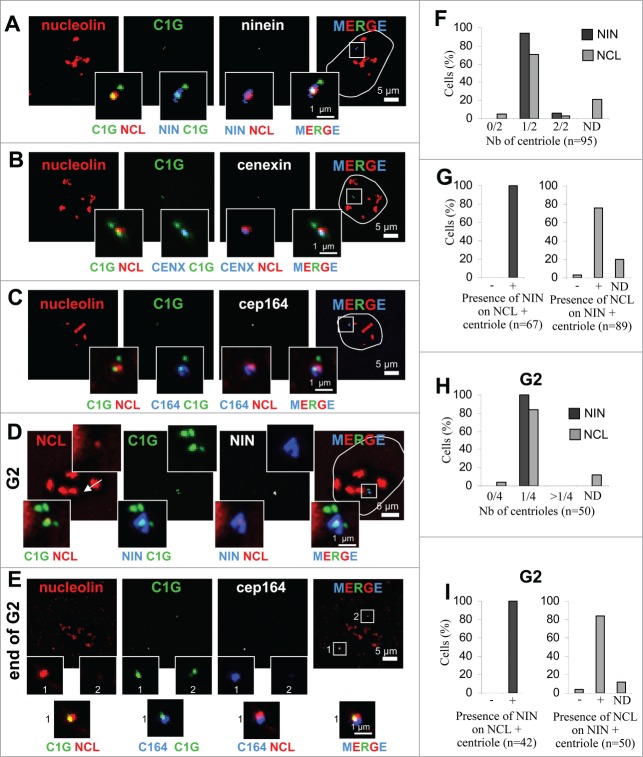
Nucleolin decorates one of the 2 centrioles throughout interphase in U2OS-centrin-1-GFP cells. (A) Co-visualization of nucleolin, centrin-1-GFP (C1G) and nuclear counterstain (DAPI) shown as individual projections and as a 2- and 3-color merged projections. Nucleolin was detected with a monoclonal antibody and revealed with an Alexa555 secondary antibody [red] and centrin-1-GFP detection was enhanced with a GFP booster [green], while nuclei were counterstained with DAPI [cyan]. On the top left image, arrows show nucleoli and the centrosome area. Enlarged images of the centrosome area (boxed on the 3 color image) are shown as insets. Full size images and insets on the first row correspond to the projection of 5 consecutive optical sections centered on the focal plane, that have been previously submitted to a 3D constrained iterative deconvolution process. Insets on second row correspond to the individual optical sections of the centrosome area, separated by 0.2 μm steps, used to obtain the projection on the first row. Scale bars represent 5 μm on full size images and 1 μm on enlarged insets. (B) Co-visualization of nucleolin, γ-tubulin, centrin-1-GFP (C1G) and nuclear counterstain (DAPI) shown as individual projection and as a 3-color merged projection with nuclear outline. Nucleolin, centrin-1-GFP and DAPI were detected similarly as in A, while γ-tubulin was detected with a primary antibody directly coupled to TRITC [in white on the unmerged image and in blue on merged images]. Enlarged projections of the centrosome area (boxed on the 3 color image) are shown as 2-color insets. Scale bars represent 5 μm on full size images and 1 μm on enlarged insets. (C) Quantification of the number of centrioles (centrin-1-GFP label) positive for nucleolin per cell, for A and B experiments. Cells exhibiting 0/2, 1/2 and 2/2 centrioles labeled are reported as percentages of the number n of cells analyzed. ND stands for “not determinable” and mainly corresponds to cases where the centrosomal signal is superimposed to the strong nucleolar signal obtained with nucleolin. (D and E) Fluorescent intensity profiles, along a line (displayed on the 0.4 μm section of the z-stack gallery in A for D and on the 3-color inset in B for E) of an individual section of the centrosome area, obtained for nucleolin [NCL, red], centrin-1-GFP [C1G, green] and for E, γ-tubulin [blue]. The x axes represent the distance along the line in μm and the y axes correspond to fluorescence intensity in arbitrary units. (F and G) Co-visualization of nucleolin and centrin-1-GFP (C1G) shown as individual projections and as a 2-color merged projection in G1 and S cell cycle phases (F) and in G2 phase (G). Asynchronously growing cells were selected under the microscope thanks to the 4 centrin dots specific of G2 cells (see 2 arrows pointing to the 2 separated centrosomes on left image). Asynchronously growing cells were incubated with the nucleotide analog EdU for 30 min before fixation, detected with Alexa647 [white] and visualized as an individual projection and on the 4-color merged projection in F. Nucleolin, centrin-1-GFP and DAPI were detected similarly as in A. Enlarged projections of the centrosome area (boxed on the 2 color merged image) are presented in the insets. Scale bars represent 5 μm on full size images and 1 μm on enlarged insets. (H) Quantification of the number of centrioles (centrin-1-GFP label) positive for nucleolin per cell, for F and G experiments, in G1 (left), S (middle) or G2 (right) cell cycle phases. G1 and S cells exhibiting 0/2, 1/2 and 2/2 centrioles labeled are reported as percentages of the number n of cells analyzed. G2 cells exhibiting 0/4, 1/4 and more than 4 (>1/4) centrioles labeled are reported as percentages of the number n of cells analyzed. As in C, ND stands for “not determinable."
Nucleolin was detected at centrosomes in 95% of interphase cells (Fig. 1C, left column). The majority of the cells show only one of the 2 centrioles positive for nucleolin, while a minority (less than 10%) show both centrioles positive for nucleolin (Fig. 1C). This minority was also found when analysis was performed on G1 or S phase cells, but not on G2 cells (see below). Centrosomal localization of nucleolin was also confirmed with another marker specific to the pericentriolar matrix, γ-tubulin (Fig. 1B). Co-detection of nucleolin with centrin-1-GFP and γ-tubulin highlighted the difference of labeling: centrin-1-GFP and γ-tubulin delineated 2 distinct centrosomal structures, while nucleolin labeled only one of them (Fig. 1B, zoom and intensity profile on Fig. 1E). One can notice that nucleolin only partially colocalized with centrin-1-GFP or with the γ-tubulin signals (Figs. 1A, B, D and E, zooms). Therefore, the centrosomal localization of nucleolin was likely distinct from the centriolar lumen (labeled with centrin-1-GFP) or the pericentriolar matrix (labeled with γ-tubulin). The centrosomal labeling with nucleolin was also observed in various murine and human cell lines, as illustrated for hTERT-RPE1 cells (Fig. S2). To test the specificity of nucleolin's centrosomal labeling (Figs. 1A–E), we quantified centrosomal fluorescence intensity in nucleolin depleted cells (Fig. S3). The nucleolin signal was reduced at background level in silenced cells (Fig. S3H, black curve) compared to control cells (Fig. S3G, black curve). Therefore, our results provide evidence that nucleolin is a specific centrosomal marker of interphase cells, preferentially decorating a single centriole.
In order to address whether the number of centrioles decorated by nucleolin was cell cycle dependent, immunofluorescence experiments were repeated with cell cycle markers (Figs. 1F–H). Cells were grown in the presence of the uridine analog (EdU) prior cell fixation, to label replicating cells (Fig. 1F lower). G1 and G2 cells were further discriminated according to the number of centrioles: 2 in the case of G1 cells (Fig. 1F upper) and 4 in the case of G2 cells (Fig. 1G). In S phase, the percentage of cells exhibiting one out of 2 centrioles positive for nucleolin was almost identical to that of G1 cells (Fig. 1F and 1H, compare G1 and S histograms), suggesting that nucleolin was not recruited during centriole duplication. However, the fact that NCL labeling could be found on both centrioles in G1 and S phase cells (7 and 12 % respectively, see Fig. 1H), suggests that tra-nsient recruitment of NCL occur on the second centriole during those cell cycle phases. In G2 cells, labeling of more than 2 centrioles was not observed, thereby excluding transient recruitment to immature centrioles during G2 (Fig. 1H). Similarly to G1 and S cells, the majority of G2 cells (> 80%) exhibited one out of 4 centrioles positive for nucleolin (Figs. 1G, and H, histogram G2). Therefore, our results indicate that the number of centrioles positive for nucleolin was not cell cycle dependent and implies that nucleolin loading at centrosome might occur after the end of G2 phase, i.e. in mitosis or in early G1 cells.
Nucleolin is a core constituent of centrosomes, independently of microtubules
To decipher whether nucleolin is really part of the centrosome structure, KE37 cells were used to purify centrosomes according to classical protocols.31 Purified centrosomes were analyzed by Western blot using the nucleolin antibody (Fig. 2A). This analysis confirmed the presence of nucleolin in the purified centrosome fraction (Fig. 2A). Because nucleolin is an abundant protein that has several cellular localizations, only a small amount of nucleolin is expected to be associated with the centrosomal structure. Therefore, no enrichment of nucleolin in the centrosomal fraction compared to the cell lysate was expected. As anticipated for a centrosomal preparation, the γ-tubulin band intensity increased in the purified centrosome lane compared to the cell lysate lane (Fig. 2B, upper). B23, another nucleolar protein, also localized at centrosomes 32, was also present in the purified centrosome fraction (Fig. 2B, lower panel).
Figure 2 (See previous page).
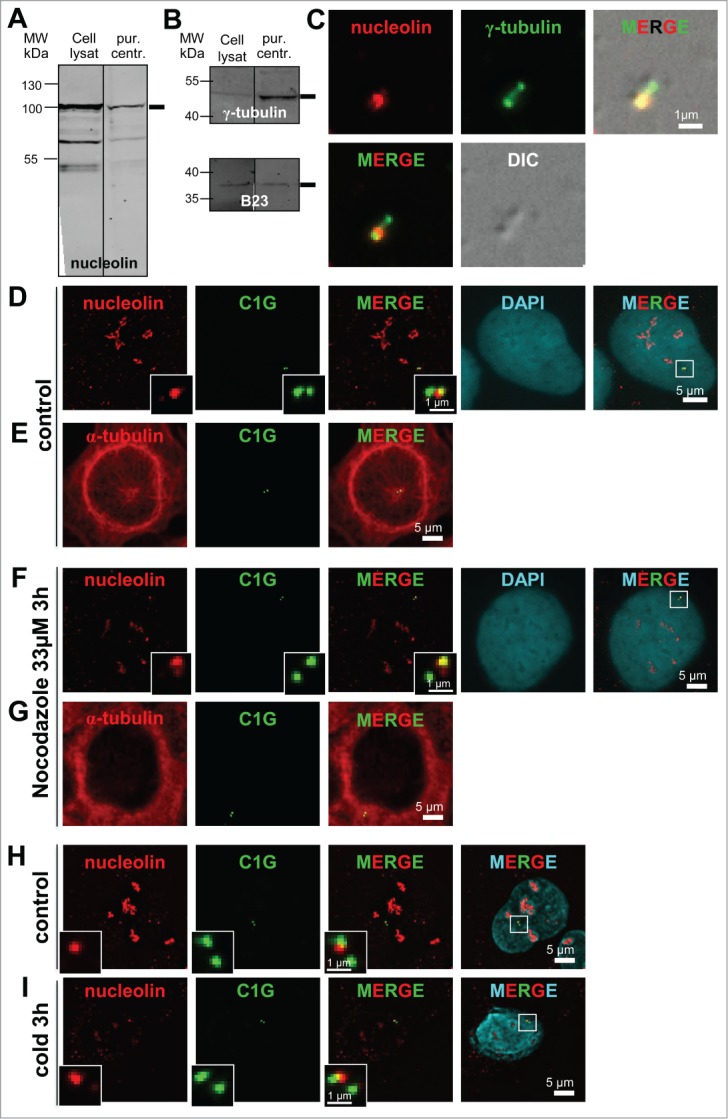
Nucleolin is stably bound to centrosomes independently of microtubules. (A–C) Nucleolin is present in pure centrosome preparations. (A and B) Fluorescent Western blots of cell lysates and biochemically purified centrosomes (pur. centr.) probed with a nucleolin monoclonal antibody (A), an anti γ-tubulin polyclonal antibody (B, upper), and an anti B23 monoclonal antibody (B, lower). Monoclonal antibodies were detected with a secondary antibody coupled to Alexa680, while the polyclonal antibody was detected with a secondary antibody coupled to IRdye800. C/ Co-visualization of nucleolin and γ-tubulin on purified centrosomes, derived from same experiment as in A and B. Purified centrosomes were spun down on coverslips, fixed in cold methanol and submitted to immunofluorescence using a nucleolin monoclonal antibody (detected with a secondary antibody coupled to Alexa488) [red] and an anti γ-tubulin antibody directly coupled to TRITC [green]. The corresponding DIC (Differential Interference Contrast) image is shown in gray. Scale bar represents 1 μm. (D–I) Nucleolin remains associated with centrosomes after nocodazole- (F and G) or cold- (I) induced microtubule depolymerization in U2OS-centrin-1-GFP cells. Co-visualization of nucleolin and centrin-1-GFP (D, F, H and I) or α-tubulin and centrin-1-GFP (E and G). Asynchronously growing cells were treated with nocodazole (F and G) or incubated at 4°C (I) for 3 hours before fixation. Control untreated cells are presented in (D, E and H). Centrin-1-GFP detection was enhanced with a GFP booster [green], nucleolin (D, F, H and I) and α-tubulin (E and G) were detected with a monoclonal antibody (detected with a secondary antibody coupled to Alexa555) [red], while in (D, F, H and I) nuclei were counterstained with DAPI [cyan]. In (D, F, H and I), enlarged images of the centrosome area (displayed on the 3 color merged images) are presented in the insets. In D and F, the DAPI images were not submitted to a 3D constrained iterative deconvolution process. Scale bars represent 5 μm on full size images and 1 μm on enlarged insets.
The purity of centrosome preparation was assessed by microscopic observation. No contaminants like membranes could be detected in the centrosome fraction. Immuno-detection of nucleolin and γ-tubulin on centrosome preparations spread onto coverslips revealed that the nucleolin signal was co-distributed with the γ-tubulin signals (Fig. 2C) resembling the pattern previously observed in whole cells (Fig. 1B). Altogether, our observations clearly show that nucleolin is bound to centrosomes.
To determine whether molecular motors drive nucleolin localization at centrosome, its localization was assessed after microtubule depolymerization (Fig. 2D–I). As expected after microtubule depolymerization, cells displayed a blurry α-tubulin staining, highlighting the disassembled α-β tubulin dimers (Fig. 2G compared to Fig. 2E). Nocodazole treatment did not abolish the original centrosomal localization of nucleolin (Fig. 2D and F). Similarly, no difference in nucleolin localization at the centrosome was observed after a 3h cold treatment (Figs. 2H–I), even though a quite complete disruption of microtubule network was observed (Fig. 7A6). Thus, microtubule network disruption by nocodazole or cold treatments does not affect the centrosomal localization of nucleolin.
As nucleolin is often associated with another nucleolar protein, B23, we investigated whether nucleolin was co-recruited with B23 at the centrosome. The experiments carried out in cells depleted for B23 (Fig. S3A–B), revealed that nucleolin expression was unchanged in siB23 cells (ratio of NCL/ß-actin of 106 % and a B23/ßactin ratio of 19% in siB23 cells, against 100% in control cells, derived from WB quantification shown in Fig. S3A) and that nucleolin staining was still present at the centrosome (compare Fig. S3D with Fig. S3C). In addition, siRNA induced depletion of nucleolin did not change B23 expression, nor its centrosomal localization, thereby supporting the fact that these 2 nucleolar proteins are not mutually required for their respective localization at centrosomes.
Altogether, our results provide evidence that centrosomal localization of nucleolin is maintained in the absence of microtubules and is independent of B23.
Non-random distribution of nucleolin toward the mature centriole
Since nucleolin localizes at a single centriole (Fig. 1), we then determined whether it was randomly distributed on the 2 centrioles, or if it was specifically associated with the centriole carrying mature or immature markers. Nucleolin localization was assessed by performing co-immunodetections with distal (CEP164),33 subdistal (ninein)34 or both distal and subdistal (cenexin)35 appendage proteins (Fig. 3). These analyzes confirmed that among cells exhibiting 2 centrioles visualized with centrin-1-GFP (C1G), a single centriole co-distributed with ninein (blue-green inset on Fig. 3A), cenexin (Fig. 3B, blue-green inset) or CEP164 (Fig. 3C, blue-green inset). More than 95% of G1/S cells had a single centriole positive for ninein (Fig. 3F). 80% of these G1/S cells had both nucleolin and ninein labeling (Fig. 3G, gray bars). Furthermore, 100% of the centrioles labeled with nucleolin were also positive for ninein (Fig. 3G black bars and Fig. 3A). A negligible percentage of cells showed both or no centrioles positive for nucleolin, in agreement with our previous quantifications (see above and Fig. 1C).
Figure 3 (See previous page).
Non-random distribution of nucleolin toward the mature centriole. (A–C) Co-visualization of nucleolin and centrin-1-GFP (C1G) together with ninein (A), cenexin (B) or CEP164 (C) in U2OS-centrin-1-GFP cells. Enlarged images of the centrosome area (displayed on the 3 color merged images) are presented in the insets as 2 color merged images to facilitate colocalization visualization. (D, E) Co-visualization of nucleolin (NCL) and centrin-1-GFP (C1G) together with ninein (NIN, D) and with CEP164 (C164, E) in U2OS-centrin-1-GFP cells, exhibiting 4 centrin dots, characteristic of G2 cells. The arrow on the left image shows the centrosome area. Enlarged images of the centrosome area (displayed on the 3 color merged images) are presented in the upper insets for a single channel and in the lower insets as 2 color merged images to facilitate colocalization visualization. In A-E, nucleolin was detected with a monoclonal antibody (detected with a secondary antibody coupled to Alexa647) [red]; centrin-1-GFP detection was enhanced with a GFP booster [green], while ninein, cenexine and CEP164 were detected with polyclonal antibodies (detected with a secondary antibody coupled to Alexa555) [in white on the unmerged image and in blue on merged images]. Outline of the nuclei, counterstained with DAPI (not shown) is highlighted on the merged images. Scale bars represent 5 μm on full size images and 1 μm on enlarged insets. (F) Quantification of the number of centrioles (centrin-1-GFP label) positive for ninein [dark bars] and nucleolin [gray bars] per cell in G1/S cell cycle phase, from experiment in A. Cells exhibiting 0/2, 1/2 and 2/2 centrioles labeled for each marker are reported as percentages of the number n of cells studied. ND stands for not determinable and mainly corresponds to cases where the centrosomal signal of nucleolin is superimposed to the strong nucleolar signal. (G) Quantification of co-distribution between both centrosomal markers, recorded in F. The presence of ninein on nucleolin positive centrioles is shown on the left, while the presence of nucleolin on ninein positive centrioles is shown on the right, expressed as percentages of the number n of cells studied. Similarly as in F, ND stands for “Not Determinable." (H) Quantification of the number of centrioles (centrin-1-GFP label) positive for ninein [dark bars] and nucleolin [gray bars] per cell in G2 cell cycle phase, from experiment in D and E. Cells exhibiting 0/4, 1/4 and >1/4 centrioles labeled for each marker are reported as percentages of the number n of cells studied. As in F, ND stands for “Not Determinable." (I) Quantification of co-distribution between both centrosomal markers, recorded in H. The presence of ninein on nucleolin positive centrioles is shown on the left, while the presence of nucleolin on ninein positive centrioles is shown on the right, expressed as percentages of the number n of cells studied. As in F, ND stands for “Not Determinable."
To assess whether the same remained true in G2 cells, ninein and CEP164 were further analyzed and quantified in cells exhibiting 4 centrin-1-GFP signals, separated or not (Fig. 3D and E respectively). As observed for non-duplicated cells, ninein and CEP164 labeled a single centriole out of 4 (Fig. 3D and E, respectively) in 100% of the analyzed G2 cells (Fig. 3H). 85% of G2 cells had both nucleolin and ninein labeling (Fig. 3I, gray bars) and 100% of the centrioles labeled with nucleolin were also positive for ninein (Fig. 3I black bars and Fig. 3D). The association of nucleolin with the mature centriole was not restricted to U2OS cells, since this was also observed in several cell lines such as RPE1 cells, where nucleolin co-distributed with ninein in 84% of the cell population (Fig. S2B). Therefore, our results highlight that nucleolin is widely found at the mature centriole, co-distributing with ninein, cenexin and CEP164, throughout the whole interphase.
Nucleolin silencing leads to amplification of centriole immature markers
One characteristic phenotype associated with nucleolin silencing is the presence of supernumerary centrosome-like structures, as identified by γ-tubulin staining together with multipolar spindle formation.28,29 The presence of these supernumerary structures in interphase cells was analyzed in regards to PCM components like γ-tubulin (Figs. 4A-E and K), pericentrin (Figs. 4F and G), as well as with specific markers of mature (Fig. 4H, I, and L, ninein) or immature (Fig. 4H, I, and M, centrobin) centrioles. A siRNA transfection approach was used to decrease nucleolin protein level down to 20% in whole cell extracts (Fig S3 A and B). The percentage of interphase cells with an abnormal number of centrin-1-GFP labeled structures increased in nucleolin-silenced cells (compare Figs. 4D–E with Figs. 4A–C). Indeed, cells exhibiting more than 4 centrin-1-GFP signals reached about 80% in nucleolin-silenced cells, compared to less than 5% in control cells, while the percentage of cells exhibiting 2 centrin-1-GFP signals dropped from 80% in control cells down to about 5% in nucleolin-silenced cells (Fig. 4J). This is specific of silencing as when nucleolin is overexpressed we do not see any difference in the centrosome cycle and in the cell proliferation rate (data not shown). In these silenced cells, the elongated form of the γ-tubulin signal can appear as 2 (80% of the cells) or 4 (20% of the cells) individual signals (Figs. 4D, E, and K) colocalizing with centrin-1-GFP, revealing that, contrary to centrin-1-GFP, the number of γ-tubulin signals is not significantly increased in these interphase silenced nucleolin cells. We next investigated whether the fluorescence intensity of γ-tubulin is increased. The local fluorescence ratio of γ-tubulin vs. centrin-1 was unchanged (Fig. 4N). Altogether, our results indicate that absence of nucleolin increases specifically the number of centrin-1-GFP signals without affecting the number or fluorescence intensity of the γ-tubulin signal, thereby ruling out that nucleolin is involved in the maintenance of γ-tubulin at centrosome, but supporting a role for nucleolin in the initial recruitment of γ-tubulin on newly formed centrioles.
Figure 4 (See previous page).
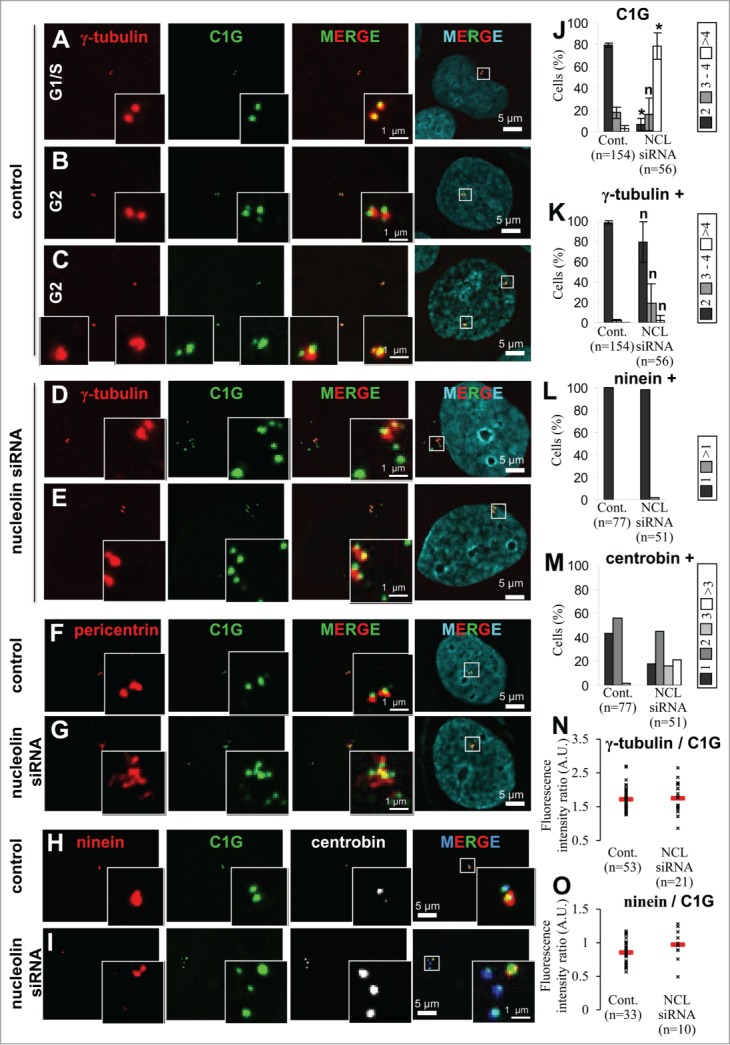
Nucleolin depletion leads to amplification of immature centriole markers. (A–E) Presence of centrosome amplification in nucleolin-silenced cells during interphase. Co-visualization of γ-tubulin, centrin-1-GFP (C1G) and nuclear counterstain (DAPI) shown as individual projections and as a 2- and 3-color merged projections in untransfected control (A, B and C) or nucleolin siRNA transfected (D, E) U2OS-centrin-1-GFP cells. Cells were harvested 4 days after transfection. Centrin-1-GFP detection was enhanced with a GFP booster [green] and γ-tubulin was detected with a primary antibody directly coupled to TRITC [red]. Nuclei were counterstained with DAPI [cyan]. Asynchronously growing cells were selected under the microscope with reference to the number of centrin dots (2 dots for G1/S cells and 4 dots for G2 cells). Early G2 and late G2 control cells are shown in B and C respectively. Enlarged projections of the centrosome area (boxed on the 3-color image) are shown as insets. Scale bars represent 5μm on full size images and 1 μm on enlarged insets. (F, G) Nucleolin silencing leads to an amplification of pericentrin. Co-visualization of centrin-1-GFP (C1G) and pericentrin in untransfected control (F) or in nucleolin-silenced (G) U2OS-centrin-1-GFP cells. Cells were harvested 4 days after transfection. Centrin-1-GFP detection was enhanced with a GFP booster [green] and pericentrin was detected with a monoclonal antibody (detected with a secondary antibody coupled to Alexa555) [red]. Nuclei were counterstained with DAPI [cyan]. Enlarged images of the centrosome area (displayed on the 3 color merged images) are presented in the insets. Scale bars represent 5 μm on full size images and 1 μm on enlarged insets. (H, I) Nucleolin silencing leads to a specific amplification of centriole immature mark. Co-visualization of centrin-1-GFP (C1G) with ninein and centrobin in untransfected control (H) or in nucleolin-silenced (I) U2OS-centrin-1-GFP cells. Cells were harvested 4 days after transfection. Centrin-1-GFP detection was enhanced with a GFP booster [green], ninein was detected with a polyclonal antibody (detected with a secondary antibody coupled to Alexa 555) [red] and centrobin was detected with a monoclonal antibody (detected with a secondary antibody coupled to Alexa 647) [in white on the unmerged images and in blue on the merge images]. Nuclei were counterstained with DAPI [cyan]. Enlarged images of the centrosome area (displayed on the 3 color merged images) are presented in the insets. Scale bars represent 5 μm on full size images and 1 μm on enlarged insets. (J, K) Quantification of the number of centrin-1-GFP dots (C1G, J), and of γ-tubulin positive centrin-1-GFP dots (γ-tubulin +, K) per cell in untransfected control (Cont., left) or nucleolin siRNA transfected (NCL siRNA, right), for A-E experiments. Cells exhibiting 2 dots [dark bars], 3 or 4 dots [gray bars] and more than 4 dots (>4) [white bars] are expressed as percentages of the number n of cells studied. The error bars correspond to standard deviations calculated for 3 independent experiments. *: Significant difference with a p value < 0.01 with control populations (left columns). n: non-significant difference with a p value < 0.01 with control populations (left columns). (L, M) Quantification of the number of ninein positive centrin-1-GFP dots (ninein +, L) and of centrobin positive centrin-1-GFP dots (centrobin +, M) per cell in untransfected control (Cont., left) or nucleolin siRNA transfected (NCL siRNA, right), for H and I experiments. Cells are distributed in different classes according to their number of dots (see individual legends), whose values are expressed as percentages of the number n of cells studied. (N, O) Quantification of γ-tubulin and ninein at the centrosome in absence of nucleolin. γ-tubulin (N) or ninein (O) fluorescence intensity were measured at the centrosome in untransfected control or nucleolin siRNA transfected U2OS-centrin-1-GFP cells. Fluorescence intensities of γ-tubulin or ninein were measured on the centrin-1-GFP dots positive for γ-tubulin or ninein respectively. These intensities were normalized to that of centrin-1-GFP. For each condition, the mean value is reported on the graph with standard deviation error bars.
In contrast to γ-tubulin, another PCM marker, pericentrin, was also found to be amplified in nucleolin-silenced cells and displayed a high number of dispersed dots associated with numerous centrin-1-GFP signals (Fig. 4F and G compare merged insets).
As far as mature markers were concerned, the ninein signal did not appear to be amplified and was always associated with a single centrin-1-GFP signal in 100% of nucleolin-silenced cells (Fig. 4H–I, red signals, and Fig 4L, black bars). This unique ninein signal likely corresponds to the ancient mature centriole, which exhibits a constant fluorescence ratio of ninein vs. centrin-1 in nucleolin-silenced cells compared to control cells (Fig. 4O). These results illustrate that the supernumerary centrin-1 containing structures observed in nucleolin-silenced cells are not able to acquire maturation markers following nucleolin depletion.
Finally, centrobin was used to investigate the status of these extra-centriolar markers within nucleolin-silenced cells. In about 40% of the control cells, centrobin appeared associated with a single centrin-1-GFP dot deprived of ninein (Fig. 4H, white and blue signals, Fig. 4M left, black bar). In agreement with previous publication,36 centrobin was found associated with both centrioles in more than 50% of the cells (Fig. 4M, left, gray bar). This distribution is consistent with the U2OS cell cycle (data not shown). In nucleolin-silenced cells, the percentage of 1 and 2 dots labeling diminished for centrobin, while 2 new classes appeared: a class with 3 dots representing 15% of the cells (Fig. 4H, blue and white signals; Fig. 4M, right, light gray bars) and a class with more than 3 dots representing 20% of the cells (Fig. 4M, right, white bars). Therefore, the number of centrobin dots was also amplified in nucleolin-silenced cells. Altogether, our results bring conclusive evidence for an amplification of immature centriole markers and for a disorganization of the PCM marker pericentrin, upon nucleolin silencing.
To gain some clues about the role of nucleolin in the centrosome cycle, we performed co-immunoprecipitation with several centrosomal proteins. Immunoprecipitation experiments with nucleolin antibodies on whole cell lysates, were validated with a nucleolar protein partner of nucleolin, B23 (Fig. 5A). We first found that a major PCM protein, γ-tubulin, could readily be immunoprecipitated with nucleolin antibodies (Fig. 5A). This result was specific to γ−tubulin, since this was not true for α or β tubulin (data not shown). Since nucleolin was found to specifically localize at the mature centriole (Fig. 3), we next investigated whether nucleolin might also interact with the mature centriole appendage protein, ninein. Using cell extracts, ninein antibodies were able to immunoprecipitate nucleolin (Fig. 5B). Therefore, our results demonstrate that cytoplasmic nucleolin associates with both soluble γ-tubulin and ninein complexes. If these interacting complexes need to be recruited together at immature centrioles this could explain why supernumerary centrosomes do not acquire mature markers in nucleolin-silenced cells.
Figure 5.
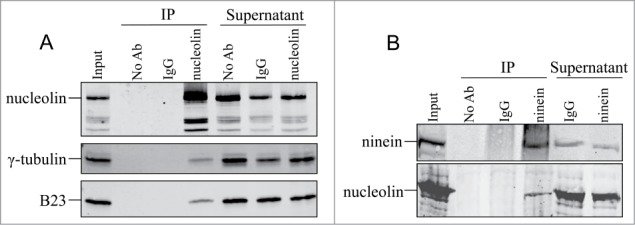
Nucleolin belongs to protein complexes containing centrosomal proteins. (A) γ-tubulin coimmunoprecipitates with nucleolin. Immunoprecipitation (IP) assay was performed in HeLa cells with anti nucleolin polyclonal antibody (polyclonal antibody 5567). Following immunoprecipitation, proteins were separated on a SDS-PAGE gel and probed with an anti nucleolin polyclonal antibody (polyclonal antibody 134, first row), detected with a secondary antibody coupled to IRdye800, and with anti γ-tubulin (second row) and B23 (third row) monoclonal antibodies, detected with a secondary antibody coupled to Alexa680. (B) Nucleolin coimmunoprecipitates with ninein. Immunoprecipitation (IP) assay was performed in HeLa cells with anti ninein monoclonal antibody. Following immunoprecipitation, proteins were separated on a SDS-PAGE gel and probed with an anti nucleolin polyclonal antibody (polyclonal antibody 5567, lower row), detected with a secondary antibody coupled to IRdye800, and with anti ninein monoclonal antibody (upper row), detected with a secondary antibody coupled to Alexa680. IP without antibody (No Ab) and preimmune serum (IgG) were used as control. Input: input pre-immunoprecipitation fractions; IP: immunoprecipitated proteins; Supernatant: supernatant after immunoprecipitation.
Microtubule network is disorganized in nucleolin depleted cells
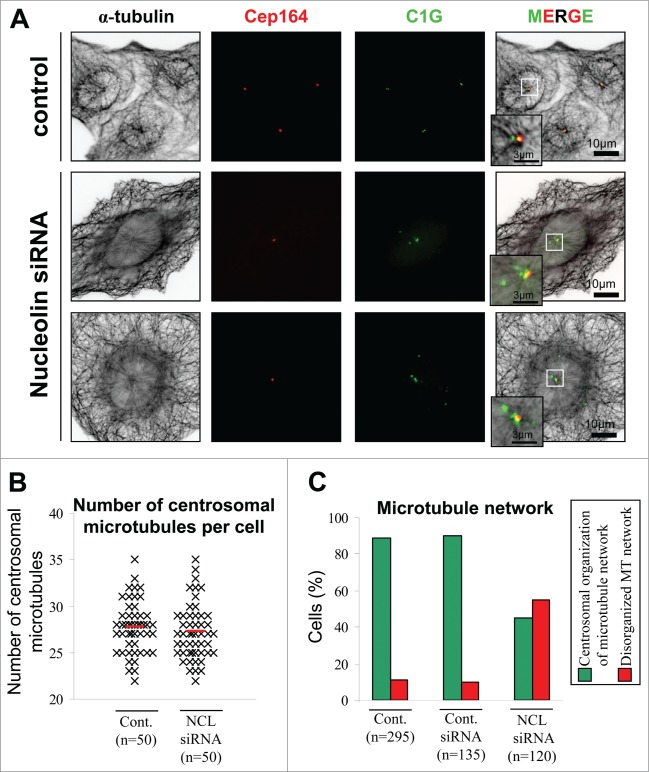
As nucleolin was associated with the mature centriole (Fig. 3), investigation of the functional consequences of nucleolin silencing was focused on microtubule organization. The microtubule network was first analyzed using α-tubulin staining in control and nucleolin-silenced cells (Fig. 6A). Nucleolin-silenced cells exhibited the typical phenotype of centrin-1-GFP amplification without any changes in CEP164 labeling (Fig. 6A). In nucleolin-silenced cells, a single aster of microtubules arising from the remaining mature centriole was observed despite numerous centrin-1-GFP signals (Fig. 6A), as observed above (Figs. 3D and E). Therefore, the amplified centrin-1GFP structures are not able to nucleate microtubules.
Figure 6.
Microtubule network is disorganized in nucleolin depleted cells. (A) Co-visualization of α-tubulin, CEP164 and centrin-1-GFP (C1G) shown as individual projections and as 3-color merged projections in control or nucleolin siRNA transfected U2OS-centrin-1-GFP cells. Cells were harvested 4 days after transfection. Centrin-1-GFP detection was enhanced with a GFP booster [green], CEP164 was detected with a polyclonal antibody (detected with a secondary antibody coupled to Alexa647) [red] and α-tubulin was detected with a monoclonal antibody (detected with a secondary antibody coupled to Alexa555) [black]. Enlarged projections of the centrosome area (boxed on the 3-color image) are shown as insets. Scale bars represent 10 μm on full size images and 3 μm on enlarged insets.(B) Quantification of the number of microtubules emanating from the centrosome of untransfected control (cont.) and nucleolin siRNA transfected (NCL siRNA) U2OS-centrin-1GFP cells. Each black mark corresponds to a cell (cont. n = 50 and NCL siRNA n = 50) and the red lines correspond to the mean (cont. 27.72 +/− 2.74 and NCL siRNA 27.24 +/− 2.97). (C) Quantification of the changes in the interphase microtubule network organization following nucleolin depletion from experiment in A. The histogram shows the percentage of the number n of U2OS-centrin-1-GFP cells analyzed for untransfected control (cont., left), control siRNA transfected (Cont. siRNA, middle), or nucleolin siRNA transfected (NCL siRNA, right), harboring a microtubule network mainly organized from the centrosome [green bars, see example on first row of A], or disorganized [red bars, see examples on second and third row of A].
In control and silenced cells the number of microtubules forming a well-organized aster centered on the centriole doublets was very similar (Figs. 6A and B). We next looked at the general organization of microtubules outside the centrosomal area and we observed that 55% of silenced cells exhibited a dense network of microtubules surrounding the nucleus which were not connected to the centrosomal aster (Figs. 6A and C).
Inhibition of microtubule nucleation and alteration of microtubule anchoring in nucleolin-silenced cells following depolymerization
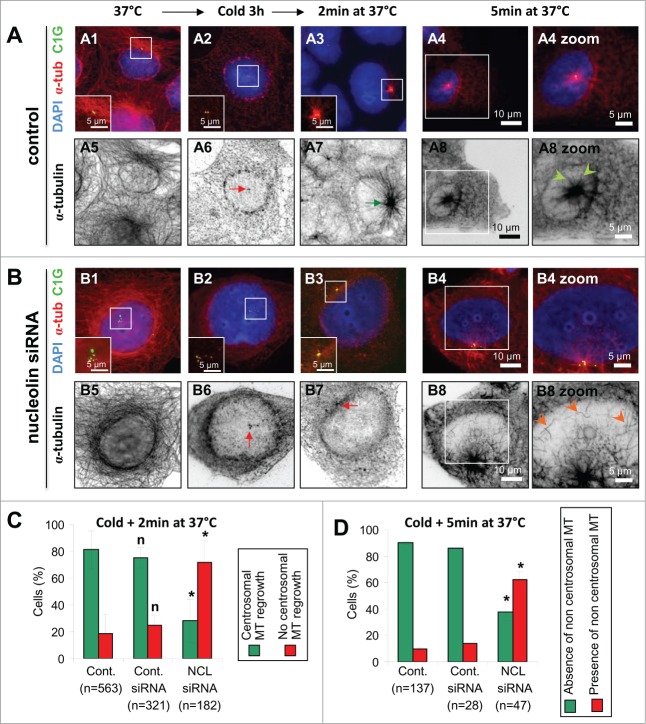
To characterize microtubule network formation in nucleolin-silenced cells, we looked at the kinetics of microtubule initiation following cold-induced depolymerization. Cells expressing centrin-1-GFP were used to analyze microtubule regrowth after 2 minutes recovery at 37°C to assess microtubule nucleation (Fig. 7A3–B3) and after 5 minutes at 37°C to assess centrosomal microtubule anchoring (Fig. 7A4-B4). Microtubules were detected by immuno-staining of α-tubulin (Figs. 7A and B, black and white pictures) in control and nucleolin-silenced cells. Complete depolymerization (Fig. 7A2 and red arrow on 7A6) was observed in 100% of control and nucleolin-silenced cells after a 3h incubation time at 4°C. Following a 2 minute incubation at 37°C a clear microtubule aster centered on the centrosome was observed (Fig. 7A3 and green arrow in A7) in more than 80% of the control cells (Fig. 7C, green bar, n = 563). Comparable results were observed in control siRNA transfected cells (Fig. 7C, cont. siRNA). In contrast, centrosomal microtubule regrowth was significantly reduced down to less than 30% in nucleolin-silenced cells, after 2 minutes at 37°C (Fig. 7B3, 7B7 and 7C -red bar, n = 182). Similar results were obtained when experiment was performed with acetylated tubulin detection, showing microtubule regrowth in only 15% of nucleolin siRNA cells against 65% in control (data not shown). In addition, the percentage of cells with centrosomal microtubule regrowth at short times (2 minutes) was decreased by an additional factor of 2, in the sub-population of nucleolin-silenced cells exhibiting supernumerary centrioles (less than 20% in cells with >4 centriole dots, against 40% in cells with ≤ 4 centriole dots, see Fig. S4B). Therefore, these results reveal that, in the absence of nucleolin, the kinetics of microtubule nucleation at centrosome is either inhibited or delayed.
Figure 7 (See previous page).
Alteration of microtubule regrowth in nucleolin-silenced cells after depolymerization. (A-B) Microtubule regrowth after microtubule induced depolymerization in untransfected control (A) and nucleolin siRNA transfected (B) U2OS-centrin-1-GFP cells. Co-visualization of α-tubulin (α-tub), centrin-1-GFP (C1G) and nuclear counterstain (DAPI) shown as 3-color merged projections (first row) and as individual sections for α-tubulin (inverted dynamics, second row) before depolymerization (A1, A5, B1 and B5), immediately after depolymerization (A2, A6, B2 and B6), after 2 minutes at 37°C (A3, A7, B3 and B7), and finally after 5 minutes at 37°C (A4, A8, B4, B8, and zooms A4, A8, B4, and B8). Centrin-1-GFP detection was enhanced with a GFP booster [green]; α-tubulin was detected with a monoclonal antibody (detected with a secondary antibody coupled to Alexa555) [red on the 3-color merged images and black on individual projections]. Enlarged projections of the centrosome area (boxed on the 3-color images) are shown as insets (A1-A3 and B1-B3) or as full sized pictures (A4, A8, B4, and B8 zooms). Arrows pointing to the centrosomal area appear red when no microtubule regrowth is observed (A6, B6 and B7) or green when microtubule regrowth is observed (A7). Arrowheads pointing to microtubules appear light green when microtubules are directed to the centrosomal area (A8 zoom) or orange when microtubules are not pointing to the centrosomal area (B8 zoom). Scale bars represent 10 μm on full size images, 5 μm on the zoomed images and the insets. (C–D) In C, quantification of the cells incubated for 2 minutes at 37°C (A7 and B7) exhibiting either microtubules regrowth (green bars on C, see also green arrows in A7) or no microtubule regrowth (red bars on C, see also red arrows in B7). In D, quantification of the cells incubated for 5 minutes at 37°C (A8 and B8) exhibiting either absence (green bars on D, see also cells with only green arrowheads on A8 zoom) or presence of cytoplasmic microtubules (orange bars on D, see also cell with orange arrowheads on B8 zoom). These classes are displayed for untransfected control cells (Cont., left), cells transfected with control siRNA (Cont. siRNA, middle) and nucleolin siRNA transfected cells (NCL siRNA, right), whose values are expressed as percentages of the total number n of cells studied. Error bars represent the standard deviation from 4 independent experiments. *: Significant difference with a p value < 0.01 with control populations (left columns). n: non-significant difference with a p value < 0.01 with control populations (left columns).
To discriminate between these 2 hypotheses, a longer incubation at 37°C was performed (Fig. 7A4–A8 and B4-B8). In control cells, the α-tubulin labeling formed an even better defined aster (Fig. 7A8), centered on the centrosome area (Fig. 7A4, centrin-1-GFP labeled in green), compared to that obtained at 2 minutes (Fig. 7A7). A closer view of the aster showed that microtubules were well focus on the centrosome area (Fig. 7A8, zoom, green arrowheads). In nucleolin-silenced cells incubated for 5 minutes following complete depolymerization, the centrosome area appeared labeled with α-tubulin (Fig. 7B8), resembling aster-like structures observed in control cells (Fig. 7A8). This observation was in agreement with the fact that microtubules could nucleate from centrosomes without nucleolin. However, a difference was clearly visible between control and silenced cells. Indeed, the presence of transverse microtubules, that were not converging toward the centrosomal area, was observed in more than 60% of nucleolin-silenced cells (Fig. 7B8, Fig. 7D – red bars – and Fig. S5), while only 40% of these nucleolin-silenced cells had the characteristic centrosomal anchored microtubules (vs. 90% in control cells and 85 % in siRNA control cells). Altogether, our results point to a role for nucleolin in restricting microtubule nucleation and anchoring at centrosomes.
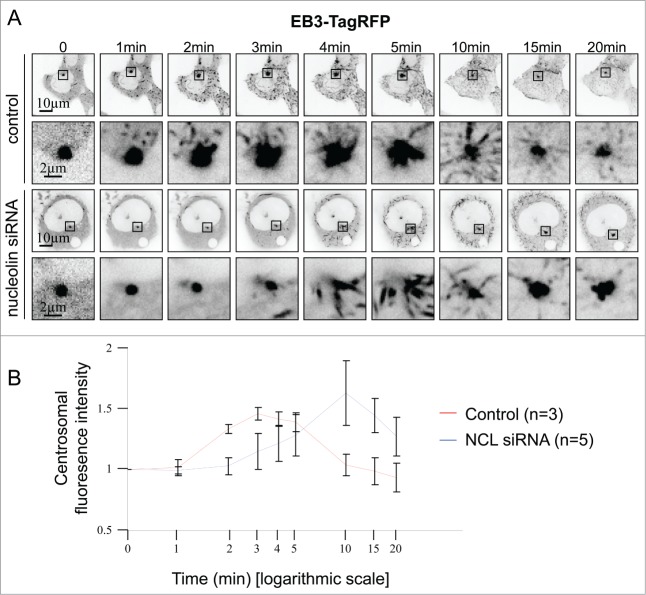
To explore in more detail microtubule polymerization after complete depolymerization in nucleolin-silenced cells, we followed microtubule regrowth in live cells. To this end, microtubules were visualized using the plus tip tracker protein EB3 fused to the tagRFP (Fig. 8 and Videos S1 and C2). After microtubule depolymerization (cold and nocodazole treatment), EB3-tagRFP staining was focused on the centrosome (Fig. 8A, time 0). In control cells, one minute after nocodazole washout, EB3-tagRFP dots were observed around the centrosome area suggesting that short microtubules were already nucleating from the centrosome (Fig. 8A, time 1 min). Several EB3-tagRFP dots were also observed in the cytoplasm showing that microtubule nucleation was also effective in non-centrosomal sites. In nucleolin-silenced cells, centrosomal and non-centrosomal microtubule nucleation were only observed after 3 minutes following nocodazole washout (Fig. 8A time 3 min), suggesting a delay in microtubule nucleation.
Figure 8.
Analysis of microtubule regrowth after depolymerization using the microtubule plus-end tracking protein EB3-tagRFP. (A) Still frames from time-lapse experiments show EB3-tagRFP expressing untransfected control and nucleolin siRNA transfected U2OS-centrin-1-GFP cells, after microtubule depolymerization. Enlarged images of the centrosome area (displayed on the full size images) are shown under the full size images. For each time point (except for t = 0min), a projection of 10 temporal images prior to the image matter was performed. Scale bars represent 5 μm on full size images and 2 μm on enlarged images. Times are given in minutes (full videos are available in supplementary material as video S1 and S2). (B) Quantification of centrosomal fluorescence intensity at the indicated times in untransfected control and nucleolin siRNA transfected U2OS-centrin-1-GFP cells, after microtubule depolymerization from A. Mean values of 3 control [red line] and 5 nucleolin depleted (NCL siRNA) [blue line] cells were plotted.
In order to quantify microtubule nucleation, EB3-tagRFP fluorescence intensity was measured around the centrosome area (Fig. 8B). In control cells, centrosomal EB3-tagRFP fluorescence intensity increased starting 1 minute after nocodazole washout to reach a maximum value at 3 minutes (Fig. 8B, red line). Next, EB3-tagRFP fluorescence intensity decreased to reach the initial value at 20 minutes suggesting a stabilization of microtubule polymerization (Fig. 8B, red line). In nucleolin-silenced cells, EB3-tagRFP centrosomal fluorescence intensity increased starting 2 or 3 minutes following nocodazole washout, showing that centrosomal microtubule nucleation was delayed compared to control cells. Next, fluorescence intensity reached a maximal value at 10 minutes in siNCL cells before decreasing. Altogether, these results clearly demonstrate a delay in microtubule nucleation in live nucleolin-silenced cells, thereby supporting a role for nucleolin in the regulation of centrosomal microtubule nucleation timing.
Discussion
In this report, we characterized the centrosomal localization of nucleolin and explored the functions of nucleolin at centrosome. For the first time, we provided evidences to show that nucleolin is a centrosomal component of interphase cells. Indeed, by immunofluorescence, nucleolin was detected at the centrosome (Fig. 1) and specifically associated with the mature centriole throughout interphase (Fig. 3). Biochemically, we also proved that nucleolin is a core component of centrosome, as it remains associated with the centrosomal structure after centrosome purification. Furthermore, the centrosomal fraction of nucleolin does not require microtubule activity since microtubule depolymerization by nocodazole or cold treatment did not affect its localization (Fig. 2 D–I). Even though an amplification of centrin-1 containing structures was highly frequent in nucleolin-silenced cells, microtubule nucleation was not found on these supernumerary centrin1-structures, which colocalized with centrobin, an immature centriole marker and with pericentrin, a PCM marker, but did not colocalized with mature markers like ninein or CEP164 (Fig. 4). Using co-immunoprecipitation experiments, we showed that nucleolin interacts with γ-tubulin and ninein, 2 key centrosomal proteins involved in microtubule nucleation and anchoring (Fig. 5). Even though the number of microtubules emanating from the single mature centriole in silenced cells was not decreased compared to control, we frequently observed a highly dense microtubule network surrounding the nucleus of silenced cells not connected with the centrosomal area (Fig. 6), suggesting that nucleolin is involved in restricting microtubule nucleation and anchoring at centrosomes. In agreement with that, we show that both in fixed and live silenced cells (Figs. 7 and 8), microtubule nucleation and anchoring steps were delayed or disrupted after microtubule depolymerization treatment, compared to control cells.
Within the centrosome, ninein localizes at the subdistal appendages of the mature centriole 37 and is involved in microtubule anchoring to the mature centriole.34,38 Ninein localizes to the centriole via its C-terminus and interacts with γ-tubulin containing complexes via its N-terminus, linking microtubule nucleation and anchoring at the centrosome.14 Nucleolin localization at the mature centriole (Fig. 3) and its interaction with ninein and γ-tubulin containing complexes (Fig. 5) suggest a role for nucleolin in microtubule nucleation and/or anchoring at the centrosome. The centrosomal microtubule nucleation delay observed in absence of nucleolin (Fig. 7 and 8) is not a consequence of γ-tubulin displacement from the centrosome (Fig. 4), as it was shown to be the case after overexpressing a dominant negative form of ninein.14 This nucleation delay is neither a consequence of NCL silenced cells accumulating in G2,29 since the rate of microtubule nucleation is unchanged in G2 compared to G1.39 Since purified nucleolin did not favor microtubule nucleation by itself (data not shown), we concluded that nucleolin might act as an activator of microtubule nucleation when bound to centrosomal γTuRC, thereby playing a role in the acquisition of mature markers on immature centrioles.
In a microtubule regrowth experiment, we also observed a microtubule anchoring defect at the centrosome in nucleolin-silenced cells (Fig. 7 and Fig. S4C). Thus, as ninein,14 nucleolin is involved in both microtubule nucleation and anchoring at the centrosome. Nucleolin could anchor microtubules directly or via other anchoring protein localizing at the mature centriole such as ninein14 since nucleolin antibodies immunoprecipitated ninein, or through p150glued, APC, FOP, CAP350 and EB1,40-42 Kif3a,43 nudel 44 or 4.1R-135.45
Previous studies have shown the presence of extra centrosomes markers containing structure in mitotic cells silenced for nucleolin.28,29 Thus, it was suggested that nucleolin may act as a negative regulator of centriole duplication as it is the case for B23,46 p53,47 cdk1,48 BRCA149 and CDK5RAP2.50 However, the link between nucleolin and centrosome duplication regulation is unknown. In this report, we observed an amplification of centrin dots in nucleolin-silenced cells during interphase (Fig. 4). Interestingly, these extra centrin dots contained centriole immature markers such as centrobin (Fig. 4I and M) but were devoid of mature markers such as ninein (Fig. 4I and L) and CEP164 (Fig. 6). This observation is in agreement with the fact that these extra centrin dots appear during interphase, suggesting that in nucleolin-silenced cells, centrosome cycle and cell cycle are uncoupled. In the absence of nucleolin, cells accumulate in G2 phase.29 Centriole amplification has been previously shown to occur as a consequence of a prolonged window in G2 that is permissive for centriole duplication.51 Moreover, no transmission of maturation markers is observed in nucleolin-silenced cells on the extra centrin-1 structures indicating that their presence could not be explained by cytokinesis failure. Thus, the centriole amplification observed in nucleolin-silenced cells might be an indirect effect of nucleolin depletion due to a prolonged G2 phase.
On the mother centriole, whereas subdistal appendages are involved in microtubule anchoring, distal appendages are important during ciliogenesis. Cenexin is localized on these 2 types of appendages35 and is involved in both ciliogenesis and the stabilization of centrosomal microtubules.13 It remains to be determined if nucleolin could also have a function related to ciliogenesis.
Several key centrosomal proteins have been observed in the nucleoli: γ-tubulin,52 cenexin53 and centrin.54 On the other hand, nucleolar proteins such as B23,32 HCA6655 and fibrillarin 15 have also been detected at the centrosome. Nucleolin and B23 have numerous functions in common. Both proteins interact 56 and are involved in ribosome biogenesis within the nucleoli. In the same manner as nucleolin, B23 is localized to the centrosome during interphase57 and its absence leads to microtubule polymerization defects.58 All these observations suggest a cross talk between centrosome and nucleoli. A molecular communication between the 2 compartments may be essential to coordinate nucleolar and centrosomal functions.
Materials and Methods
Antibodies and reagents
Nucleolin was detected with a mouse monoclonal antibody (4E2 Assay designs #ADI-KAM-CP100-E / Immunogen: human full length nucleolin / dilution: IF 1/300 and WB 1/1000), a rabbit polyclonal antibody raised against purified human nucleolin (number 134, developed in our laboratory / dil. WB 1/1000),29 a rabbit polyclonal antibody raised against 3 peptides of human nucleolin (Covalab, pab0971-P / dil. WB 1/1000),59,60 or a rabbit polyclonal antibody raised against the first hundred amino acids of human nucleolin (Abcam ab#22758 / dil. IF 1/300). γ-tubulin was detected with a mouse monoclonal antibody (GTU-88 Sigma-Aldrich #T6557 / dil. WB 1/1000) and a rabbit polyclonal antibody coupled to TRITC (C-20 Santa-Cruz #SC-7396 / dil. IF 1/100). B23 was detected with a mouse monoclonal antibody (FC82291 Sigma-Aldrich #B0556 / dil. WB 1/1000) and a rabbit polyclonal antibody (kind gift from K. Fukasawa / dil. IF 1/100).57 Ninein was detected with a rabbit polyclonal antibody (kind gift from M. Bornens / dil. IF 1/5000) and a mouse monoclonal antibody (Millipore #MABT29 / dil. WB 1/1000). Cenexin was detected with a rabbit polyclonal antibody (Rockland #600-401-A46 / dil. IF 1/100). CEP164 was detected with a rabbit polyclonal antibody (Novus NBP1-81445, dil. IF 1/200). Pericentrin was detected with a mouse monoclonal antibody (Abcam #ab28144 / dil. IF 1/300). Centrobin (Abcam #ab70448 / dil. IF 1/1000). α-tubulin was detected with a mouse monoclonal antibody (DM1A Sigma-Aldrich #T9026 / dil. IF 1/300). β-actin was detected with a mouse monoclonal antibody (AC-15 Sigma-Aldrich #A5441 / dil. WB 1/1000). GFP detection was boosted with an anti-GFP antibody directly coupled to Atto488 (Chromotek / dil. IF 1/200).
For IF, secondary antibodies used were coupled to Alexa555 (Molecular Probes DaMAlexa555 #A31570 dil. 1/2000 and DaRAlexa555 #A31572 dil. 1/1000), Alexa647 (Molecular Probes DaMAlexa647 #A31571 dil. 1/200) and Alexa488 (Invitrogen GaMA488 #A11025 dil. 1/1000). For WB, secondary antibodies used were coupled to IRdye800 (Li-Cor #92632211 dil. 1/2500) and Alexa680 (Li-Cor #92632220 dil. 1/15000).
Nocodazole was obtained from Sigma-Aldrich (#M1404) and cytochalasin D from Tocris (#1233).
Cell culture
U2OS-centrin-1-GFP (kind gift from M. Bornens) 38 were grown in Dulbecco's modified Eagles medium DMEM (PAA #E15-883) supplemented with 10% fetal calf serum (FCS) (PAA #A15-151), 1% of non essential amino acids (PAA #M11-003), 1% of penicillin-streptomycin (PAA #P11-010) and 1mM of Sodium Pyruvate (PAA #S11-003).
HTERT-RPE1 cells were grown in a mixture of DMEM and Ham's F12 (PAA # E15-890) medium supplemented with 10% FCS, 1% of non-essential amino acids and 1% of penicillin-streptomycin.
Instead of trypsin, we used a mixture of collagenases referred to as accutase for cell detachment (PAA).
KE37 cells were grown in RPMI (PAA #E15-039) supplemented with 10% fetal calf serum, 1% of penicillin-streptomycin and 2mM of glutamax (PAA #M11-006). Cells were maintained at 37°C in a 5% CO2-humidified incubator.
siRNA transfection
For nucleolin siRNA, a mixture of functional small interfering RNAs (siRNAs) specific for human nucleolin was used as previously described.29,61,62 For B23 silencing, a mixture of siRNAs specific for human B23 was used (Invitrogen NPM1-HSS143152 GAUGGAACUCCACCCUUUGCUUGGU and NPM1-HSS143153 UGUAUGGAAUGUUAUGAUAGGACAU).63 All siRNAs were reconstituted at a concentration of 100μM and stored at −20°C. As a control siRNA, we used stealth high GC siRNA (Invitrogen). Cells were transfected in a 6-well dishes using siRNA at 20nM final concentration. SiRNAs were diluted in 200μl of OptiMEM and plated in a well. 80μl of INTERFERin (Polyplus) diluted 1/10 in RNase-free water were added. After 10 min incubation, 2 ml of medium containing 3.105 cells were added. After 2 days, cells were detached and plated in 24-well dishes onto glass coverslips for further immunofluorescence or plated in 10cm dishes for western blot analysis. Cell fixation and cell lysis were performed 96 h after siRNA transfection. For EdU incorporation and detection, immediately before fixation, cells were incubated with 10 μM EdU (5-ethynyl-2′-deoxyuridine) for 20 min (Click-iT EdU kit from Invitrogen). EdU detection was performed using an Alexa647 fluorophore according to the manufacturer's instructions.
Immunofluorescence
Cells were plated 5.104 cells/well in 24-well dishes onto glass coverslips. 2 days after plating, cells were usually fixed in cold methanol for 3 min at −20°C and then permeabilized with 0.1% Triton X-100 in PBS (PBS-T) for 2 × 10 min (more fixation procedures are described in Fig. S1A). All subsequent incubations were performed in a humidified chamber maintained at 37°C. Non specific binding of antibodies was blocked by 10% FCS, 3% BSA and 0.1% Triton X-100 in PBS (blocking buffer) for 30 min. Coverslips were next incubated with primary antibodies diluted in the blocking buffer for 30 min. After 3 washes at room temperature in PBS-T, they were incubated with secondary antibodies also diluted in the blocking buffer, for 30 min. After 3 more washes in PBS-T, coverslips were washed in PBS, rinsed in ddH2O and briefly dipped in absolute ethanol. After a quick dry, coverslips were mounted on a slide with Fluoromount G (FMG Southern biotech #0100-01) containing 400 ng/ml 4′,6′-diamidino-2-phenylindole (DAPI).
Microscopic image acquisition and treatment
12-bit images were acquired using a Cool Snap HQ charge-coupled-device (CCD) camera mounted on a Zeiss Axio-Imager Z1 equipped with a 63x oil-immersion objective lens (numerical aperture [NA] = 1.4 / working distance 0.19 mm) and fluorescence filters suited for the visualization of DAPI, Alexa488 and Atto488, Alexa555 and Alexa647. For each field of view, z-stacks of about 25 images with a pixel size of 102 nm were obtained by setting the z-step at 200 nm. Image stacks were processed using a 3D constrained iterative deconvolution module running under Metamorph (Meinel Algorithm on Metamorph [iteration: 7x / σ: 0.7 / frequency: 5 / without auto background]), using the Point Spread Functions (PSF) measured for the different channels under similar acquisition conditions for PS-speck beads (Molecular probes) mounted in the same mounting medium. For each analyzed cell, the optical section in which the centrosome was the most in focus was chosen and a projection of 5 consecutive sections around this focus section was performed (for DAPI and DIC, images correspond to a single section). The x, y and z shifts between individual channels were corrected on the 3D stack by imaging 100 nm multi fluorescent microspheres under similar acquisition conditions (translation of red channel x-1 and z-1).
Western blot
Cells were detached, lysed in 2% SDS, 10% glycerol and 20% β-mercapto-ethanol for a final concentration of 1.104 cells/μl, and boiled at 95°C for 5 min. 1.105 cells were loaded onto a 10% SDS poly acrylamide gel electrophoresis. The proteins were then transferred to Protan membranes (Protran BA85, Whatman, /GE Healthcare/ #/10 401 196). Membranes were blocked in 5% milk and incubated with the primary antibodies over night at 4°C in PBS containing 1% milk. Membranes were washed in PBS and secondary antibody incubations were performed for 30 min in PBS containing 1% milk at room temperature. Monoclonal antibodies were generally detected with a secondary antibody coupled to Alexa680, while polyclonal antibodies were detected with a secondary antibody coupled to IRdye800. After PBS washes, protein gel blot imaging was performed with an Odyssey Infrared Imaging System (Li-Cor Biosciences).
Co-immunoprecipitation
For Nucleolin immunoprecipitation 5.106 cells were resuspended in 400 μl of lysis buffer (150 mM NaCl, 10 mM Tris -HCl pH 8, 1% NP-40 and a cocktail of protease inhibitor) and incubated on ice for 10min. Lysates were then clarified (12 000g for 10 min at 4°C). To avoid unspecific binding to the beads, the supernatants were incubated with protein A sepharose beads (Sigma-Aldrich #P3391) at 4°C for 3h. Beads were then eliminated by centrifugation. Meanwhile, 5μl of the antibody (rabbit polyclonal anti-nucleolin 5567 antibody, no antibody or pre-immune serum for controls) was incubated with beads at 4°C for 3h. Equal amounts of lysates were then added on beads and incubated over night at 4°C. Supernatants (supernatant unbound fraction) were then mixed with 5x protein loading buffer (250 mM Tris-HCl pH 6.8, 2% SDS, 50% glycerol, 500 mM β-mercapto-ethanol and 0.5% Bromophenol Blue) and beads (IP fraction) were extensively washed and eluted at 95°C for 5 min in protein loading buffer for western blot analysis.
For Ninein immunoprecipitation 5.106 cells were resuspended in 1ml of homogenization buffer (250mM sucrose, 10 mM Tris-HCl pH 7.4 and protease inhibitors) and centrifuged (2000rpm for 5min at 4°C). Pellet was resuspended in 200 μl of homogenization buffer and was homogenized on ice. Nuclei were eliminated by centrifugation (4000rpm for 5 min at 4°C) and the supernatant containing the cytosolic fraction was used for immunoprecipitation as described before for nucleolin immunoprecipitation using the ninein monoclonal antibody (Millipore #MABT29).
Centrosome isolation
Centrosomes were isolated from KE37 cells as previously described.31 One liter of cultured KE37 cells was incubated with 0.2 μM nocodazole and 1μg/ml cytochalasin D for 1h at 37°C. All subsequent steps were performed at 4°C. Cells were recovered by low centrifugation, and after one wash step in 1xPBS, they were resuspended in 8% sucrose, 0.1xPBS solution. Cells were then extracted for 5min in lysis buffer (1 mM HEPES, 0.5 mM MgCl2, 0.5% NP-40, 1mM PMSF and containing a mixture of protease inhibitor). After centrifugation (10 min at 2500 g), supernatant was filtered; HEPES was added to have a 10mM final concentration and then incubated for 30 min with 2units/ ml of DNase (Roche #10 776 785). Centrosomes were sedimented on 12.5 ml of 60% sucrose solution (30 min at 10 000 g). Pellet (25 ml) was then transferred to a sucrose gradient (3ml 70%, 3 ml 50% and 3ml 40% sucrose solutions) and centrifuged for 1h15 min at 40 000 g. 12 fractions of 0.5 ml were collected from the bottom of the tube and immediately frozen in liquid nitrogen. Aliquot of each fraction was collected to analyze centrosome enrichment by IF and WB. For Immunofluorescence analysis of sucrose gradient fractions, 10μl centrosome fraction aliquots were resuspended in 4 ml of kPIPES, sedimented on glass coverslips (10 min 20 000 g) and fixed 6 min in cold methanol before IF experiment. For Western blot analysis, centrosome fractions were resupended in 1 ml of kPIPES and centrifuged 20 min at 15 000 g. Pellets were then resupended in protein loading buffer and boiled 5min at 95°C for protein gel blot analysis.
Establishment of U2OS-centrin-1-GFP cells stably expressing EB3-tagRFP
EB3-tagRFP construct was extracted from a pTagRFP-EB3 plasmid (Evrogen #FP365), using the SalI and NotI restriction sites. To obtain a resistance to blasticidin, the construct was then cloned in the pbos-H2B-GFP plasmid (kind gift of T. Kanda),64 in which the H2B-GFP construct was previously removed using the same restriction sites.
A U2OS-centrin-1-GFP cell line stably expressing the new EB3-tagRFP construct was then established (for protocol see 61).
Time-lapse analysis of microtubule regrowth after depolymerization using EB3-tagRFP
150 000 cells (U2OS-centrin-1-GFP cells stably expressing EB3-tagRFP) were plated on 35mm Ibidi dishes (μ-dish high Ibidi treat, Biovalley) 2 days before imaging. Before live cell imaging, cells were incubated in a pre cold medium containing 10 μM nocodazole for 2 hours. Next, cells were incubated at 37°C for 20 min and then washed once in PBS and imaged in FluoroBrite DMEM medium without phenol red (Gibco #A18967) and supplemented with 10% FCS and 2 mM of glutamax. 16 bits images were acquired with a spinning disk confocal microscope setup on a Leica-inverted microscope equipped with an EMCCD camera (iXon3 897 / Andor). Using the 491 and 561 nm laser lines and a 100x oil immersion objective lens (NA = 1.4), individual sections were acquired every 5s during 20min in a 37°C thermo regulated atmosphere. For each time point (except for T = 0 min), a projection of 10 temporal images prior to the image matter (sum intensity projection of 10 temporal images from Ti to Ti-45s) was performed. Centrosomal fluorescence intensity was measured using a 30 pixel squared ROI (20.79μm2). In order to compensate that T = 0 min is not a projected image; the intensity value was multiplied by 10. Centrosomal fluorescence intensity was setup to 1 at T = 0 min. For each time point, the relative fluorescence intensity compared to T = 0 min was measured.
Quantification of the number of microtubules emanating from the centrosome
U2OS-centrin-1-GFP cells were labeled for α-tubulin and analyzed. The fluorescence intensity profile was measured on the perimeter of a circle (diameter = 10.24 μm) centered on the centrosome using the Oval Profile Plot plugin on ImageJ freeware. For each pixel, the fluorescence intensity value was an average of the 6 surrounding pixels. To measure the number of fluorescent peaks (corresponding to the number of microtubules), the fluorescence intensity profile curve was next derived. Between the 2 conditions, the mean values were compared using a Student test.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank PLATIM (PLAteau Technique d’Imagerie et de Microscopie, UMS3444, Lyon, FRANCE) for assistance in the acquisition and analysis of microscopic data and Christophe Place for critical reading of the manuscript.
Funding
This project was funded thanks to the Agence Nationale de la Recherche [ANR-07-BLAN-0062-01], the Association pour la Recherche sur le Cancer n_ECL2010R01122, Ligue contre le Cancer (Rhône), FINOVI, the CNRS and Ecole Normale Supérieure de Lyon.
References
- 1. Bornens M. The centrosome in cells and organisms. Science 2012; 335:422-6; PMID:22282802; http://dx.doi.org/ 10.1126/science.1209037 [DOI] [PubMed] [Google Scholar]
- 2. Gonczy P. Towards a molecular architecture of centriole assembly. Nat Rev Mol Cell Biol 2012; 13:425-35; PMID:22691849; http://dx.doi.org/ 10.1038/nrm3373 [DOI] [PubMed] [Google Scholar]
- 3. Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol 2007; 17:215-21; PMID:17383880; http://dx.doi.org/ 10.1016/j.tcb.2007.03.003 [DOI] [PubMed] [Google Scholar]
- 4. Nigg EA, Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 2011; 13:1154-60; PMID:21968988; http://dx.doi.org/ 10.1038/ncb2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meraldi P, Nigg EA. The centrosome cycle. FEBS Lett 2002; 521:9-13; PMID:12067716; http://dx.doi.org/ 10.1016/S0014-5793(02)02865-X [DOI] [PubMed] [Google Scholar]
- 6. Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer 2006; 119:2717-23; PMID:17016823; http://dx.doi.org/ 10.1002/ijc.22245 [DOI] [PubMed] [Google Scholar]
- 7. Lingle WL, Lukasiewicz K, Salisbury JL. Deregulation of the centrosome cycle and the origin of chromosomal instability in cancer. Adv Exp Med Biol 2005; 570:393-421; PMID:18727509; http://dx.doi.org/ 10.1007/1-4020-3764-3_14 [DOI] [PubMed] [Google Scholar]
- 8. Zyss D, Gergely F. Centrosome function in cancer: guilty or innocent? Trends Cell Biol 2009; 19:334-46; PMID:19570677; http://dx.doi.org/ 10.1016/j.tcb.2009.04.001 [DOI] [PubMed] [Google Scholar]
- 9. Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet 2011; 27:307-15; PMID:21680046; http://dx.doi.org/ 10.1016/j.tig.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell 2009; 139:663-78; PMID:19914163; http://dx.doi.org/ 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- 11. Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol 1992; 108:107-28; PMID:1486002; http://dx.doi.org/ 10.1016/1047-8477(92)90011-X [DOI] [PubMed] [Google Scholar]
- 12. Ishikawa H, Kubo A, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol 2005; 7:517-24; PMID:15852003; http://dx.doi.org/ 10.1038/ncb1251 [DOI] [PubMed] [Google Scholar]
- 13. Tateishi K, Yamazaki Y, Nishida T, Watanabe S, Kunimoto K, Ishikawa H, Tsukita S. Two appendages homologous between basal bodies and centrioles are formed using distinct Odf2 domains. J Cell Biol 2013; 203:417-25; PMID:24189274; http://dx.doi.org/ 10.1083/jcb.201303071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci 2005; 118:1565-75; PMID:15784680; http://dx.doi.org/ 10.1242/jcs.02302 [DOI] [PubMed] [Google Scholar]
- 15. Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 2003; 426:570-4; PMID:14654843; http://dx.doi.org/ 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- 16. Jakobsen L, Vanselow K, Skogs M, Toyoda Y, Lundberg E, Poser I, Falkenby LG, Bennetzen M, Westendorf J, Nigg EA, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J 2011; 30:1520-35; PMID:21399614; http://dx.doi.org/ 10.1038/emboj.2011.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luders J. The amorphous pericentriolar cloud takes shape. Nat Cell Biol 2012; 14:1126-8; PMID:23131920; http://dx.doi.org/ 10.1038/ncb2617 [DOI] [PubMed] [Google Scholar]
- 18. Mennella V, Agard DA, Huang B, Pelletier L. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol 2014; 24:188-97; PMID:24268653; http://dx.doi.org/ 10.1016/j.tcb.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat Cell Biol 2012; 14:1159-68; PMID:23086239; http://dx.doi.org/ 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol 2012; 14:1148-58; PMID:23086237; http://dx.doi.org/ 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- 21. Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open 2012; 1:965-76; PMID:23213374; http://dx.doi.org/ 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol 2000; 2:352-7; PMID:10854326; http://dx.doi.org/ 10.1038/35014045 [DOI] [PubMed] [Google Scholar]
- 23. Raynaud-Messina B, Merdes A. Gamma-tubulin complexes and microtubule organization. Curr Opin Cell Biol 2007; 19:24-30; PMID:17178454; http://dx.doi.org/ 10.1016/j.ceb.2006.12.008 [DOI] [PubMed] [Google Scholar]
- 24. Murphy SM, Preble AM, Patel UK, O'Connell KL, Dias DP, Moritz M, Agard D, Stults JT, Stearns T. GCP5 and GCP6: two new members of the human gamma-tubulin complex. Mol Biol Cell 2001; 12:3340-52; PMID:11694571; http://dx.doi.org/ 10.1091/mbc.12.11.3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J Cell Biol 2006; 172:505-15; PMID:16461362; http://dx.doi.org/ 10.1083/jcb.200510028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol 2002; 14:25-34; PMID:11792541; http://dx.doi.org/ 10.1016/S0955-0674(01)00290-3 [DOI] [PubMed] [Google Scholar]
- 27. Cong R, Das S, Bouvet P. The multiple properties and functions of nucleolin. In: Olson MOJ, ed. The nucleolus, 2011:185-212; PMID:NOT_FOUND; http://dx.doi.org/ 10.1007/978-1-4614-0514-6_9 [DOI] [Google Scholar]
- 28. Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K. Nucleolin functions in nucleolus formation and chromosome congression. J Cell Sci 2007; 120:2091-105; PMID:17535846; http://dx.doi.org/ 10.1242/jcs.008771 [DOI] [PubMed] [Google Scholar]
- 29. Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, et al. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol 2007; 8:66; PMID:17692122; http://dx.doi.org/ 10.1186/1471-2199-8-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sauer G, Korner R, Hanisch A, Ries A, Nigg EA, Sillje HH. Proteome analysis of the human mitotic spindle. Mol Cell Proteomics 2005; 4:35-43; PMID:15561729; http://dx.doi.org/ 10.1074/mcp.M400158-MCP200 [DOI] [PubMed] [Google Scholar]
- 31. Moudjou M, Bornens M. Method of centrosome isolation from cultured animal cells. In: Celis JE, ed. Cell Biology: a Laboratory Handbook: Academic Press, London, 1994:595-604 [Google Scholar]
- 32. Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 2000; 103:127-40; PMID:11051553; http://dx.doi.org/ 10.1016/S0092-8674(00)00093-3 [DOI] [PubMed] [Google Scholar]
- 33. Graser S, Stierhof YD, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. CEP164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol 2007; 179:321-30; PMID:17954613; http://dx.doi.org/ 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci 2000; 113 (Pt 17):3013-23; PMID:10934040 [DOI] [PubMed] [Google Scholar]
- 35. Nakagawa Y, Yamane Y, Okanoue T, Tsukita S. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol Biol Cell 2001; 12:1687-97; PMID:11408577; http://dx.doi.org/ 10.1091/mbc.12.6.1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, Gao Q. Centrobin: a novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol 2005; 171:437-45; PMID:16275750; http://dx.doi.org/ 10.1083/jcb.200506185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bouckson-Castaing V, Moudjou M, Ferguson DJ, Mucklow S, Belkaid Y, Milon G, Crocker PR. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J Cell Sci 1996; 109 (Pt 1):179-90; PMID:8834802 [DOI] [PubMed] [Google Scholar]
- 38. Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol 2000; 149:317-30; PMID:10769025; http://dx.doi.org/ 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Piehl M, Tulu US, Wadsworth P, Cassimeris L. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc Natl Acad Sci U S A 2004; 101:1584-8; PMID:14747658; http://dx.doi.org/ 10.1073/pnas.0308205100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Askham JM, Vaughan KT, Goodson HV, Morrison EE. Evidence that an interaction between EB1 and p150(Glued) is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol Biol Cell 2002; 13:3627-45; PMID:12388762; http://dx.doi.org/ 10.1091/mbc.E02-01-0061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Louie RK, Bahmanyar S, Siemers KA, Votin V, Chang P, Stearns T, Nelson WJ, Barth AI. Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J Cell Sci 2004; 117:1117-28; PMID:14970257; http://dx.doi.org/ 10.1242/jcs.00939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yan X, Habedanck R, Nigg EA. A complex of two centrosomal proteins, CAP350 and FOP, cooperates with EB1 in microtubule anchoring. Mol Biol Cell 2006; 17:634-44; PMID:16314388; http://dx.doi.org/ 10.1091/mbc.E05-08-0810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kodani A, Salome Sirerol-Piquer M, Seol A, Garcia-Verdugo JM, Reiter JF. Kif3a interacts with Dynactin subunit p150 Glued to organize centriole subdistal appendages. EMBO J 2013; 32:597-607; PMID:23386061; http://dx.doi.org/ 10.1038/emboj.2013.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guo J, Yang Z, Song W, Chen Q, Wang F, Zhang Q, Zhu X. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol Biol Cell 2006; 17:680-9; PMID:16291865; http://dx.doi.org/ 10.1091/mbc.E05-04-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krauss SW, Spence JR, Bahmanyar S, Barth AI, Go MM, Czerwinski D, Meyer AJ. Downregulation of protein 4.1R, a mature centriole protein, disrupts centrosomes, alters cell cycle progression, and perturbs mitotic spindles and anaphase. Mol Cell Biol 2008; 28:2283-94; PMID:18212055; http://dx.doi.org/ 10.1128/MCB.02021-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang W, Budhu A, Forgues M, Wang XW. Temporal and spatial control of nucleophosmin by the Ran-Crm1 complex in centrosome duplication. Nat Cell Biol 2005; 7:823-30; PMID:16041368; http://dx.doi.org/ 10.1038/ncb1282 [DOI] [PubMed] [Google Scholar]
- 47. Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science 1996; 271:1744-7; PMID:8596939; http://dx.doi.org/ 10.1126/science.271.5256.1744 [DOI] [PubMed] [Google Scholar]
- 48. Vidwans SJ, Wong ML, O'Farrell PH. Anomalous centriole configurations are detected in Drosophila wing disc cells upon Cdk1 inactivation. J Cell Sci 2003; 116:137-43; PMID:12456723; http://dx.doi.org/ 10.1242/jcs.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ko MJ, Murata K, Hwang DS, Parvin JD. Inhibition of BRCA1 in breast cell lines causes the centrosome duplication cycle to be disconnected from the cell cycle. Oncogene 2006; 25:298-303; PMID:16170356; http://dx.doi.org/ 10.1038/sj.onc.1209683 [DOI] [PubMed] [Google Scholar]
- 50. Barrera JA, Kao LR, Hammer RE, Seemann J, Fuchs JL, Megraw TL. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev Cell 2010; 18:913-26; PMID:20627074; http://dx.doi.org/ 10.1016/j.devcel.2010.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. EMBO J 2004; 23:3864-73; PMID:15359281; http://dx.doi.org/ 10.1038/sj.emboj.7600393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Horejsi B, Vinopal S, Sladkova V, Draberova E, Sulimenko V, Sulimenko T, Vosecká V, Philimonenko A, Hozák P, Katsetos CD, et al. Nuclear gamma-tubulin associates with nucleoli and interacts with tumor suppressor protein C53. J Cell Physiol 2012; 227:367-82; PMID:21465471; http://dx.doi.org/ 10.1002/jcp.22772 [DOI] [PubMed] [Google Scholar]
- 53. Rivkin E, Tres LL, Kierszenbaum AL. Genomic origin, processing and developmental expression of testicular outer dense fiber 2 (ODF2) transcripts and a novel nucleolar localization of ODF2 protein. Mol Reprod Dev 2008; 75:1591-606; PMID:18398819; http://dx.doi.org/ 10.1002/mrd.20911 [DOI] [PubMed] [Google Scholar]
- 54. Keryer G, Di Fiore B, Celati C, Lechtreck KF, Mogensen M, Delouvee A, Lavia P, Bornens M, Tassin AM. Part of Ran is associated with AKAP450 at the centrosome: involvement in microtubule-organizing activity. Mol Biol Cell 2003; 14:4260-71; PMID:14517334; http://dx.doi.org/ 10.1091/mbc.E02-11-0773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fant X, Gnadt N, Haren L, Merdes A. Stability of the small gamma-tubulin complex requires HCA66, a protein of the centrosome and the nucleolus. J Cell Sci 2009; 122:1134-44; PMID:19299467; http://dx.doi.org/ 10.1242/jcs.035238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li YP, Busch RK, Valdez BC, Busch H. C23 interacts with B23, a putative nucleolar-localization-signal-binding protein. Eur J Biochem 1996; 237:153-8; PMID:8620867; http://dx.doi.org/ 10.1111/j.1432-1033.1996.0153n.x [DOI] [PubMed] [Google Scholar]
- 57. Shinmura K, Tarapore P, Tokuyama Y, George KR, Fukasawa K. Characterization of centrosomal association of nucleophosmin/B23 linked to Crm1 activity. FEBS Lett 2005; 579:6621-34; PMID:16297385; http://dx.doi.org/ 10.1016/j.febslet.2005.10.057 [DOI] [PubMed] [Google Scholar]
- 58. Amin MA, Matsunaga S, Uchiyama S, Fukui K. Depletion of nucleophosmin leads to distortion of nucleolar and nuclear structures in HeLa cells. Biochem J 2008; 415:345-51; PMID:18729828; http://dx.doi.org/ 10.1042/BJ20081411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cong R, Das S, Ugrinova I, Kumar S, Mongelard F, Wong J, Bouvet P. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res 2012; 40:9441-54; PMID:22859736; http://dx.doi.org/ 10.1093/nar/gks720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Das S, Cong R, Shandilya J, Senapati P, Moindrot B, Monier K, Delage H, Mongelard F, Kumar S, Kundu TK, et al. Characterization of nucleolin K88 acetylation defines a new pool of nucleolin colocalizing with pre-mRNA splicing factors. FEBS Lett 2013; 587:417-24; PMID:23353999; http://dx.doi.org/ 10.1016/j.febslet.2013.01.035 [DOI] [PubMed] [Google Scholar]
- 61. Gaume X, Monier K, Argoul F, Mongelard F, Bouvet P. In vivo study of the histone chaperone activity of nucleolin by FRAP. Biochem Res Int 2011; 2011:187624; PMID:21403913; http://dx.doi.org/ 10.1155/2011/187624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Greco A, Arata L, Soler E, Gaume X, Coute Y, Hacot S, Callé A, Monier K, Epstein AL, Sanchez JC, et al. Nucleolin interacts with US11 protein of herpes simplex virus 1 and is involved in its trafficking. J Virol 2012; 86:1449-57; PMID:22130536; http://dx.doi.org/ 10.1128/JVI.06194-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Murano K, Okuwaki M, Hisaoka M, Nagata K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol Cell Biol 2008; 28:3114-26; PMID:18332108; http://dx.doi.org/ 10.1128/MCB.02078-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol 1998; 8:377-85; PMID:9545195; http://dx.doi.org/ 10.1016/S0960-9822(98)70156-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.