Abstract
Spindle assembly checkpoint (SAC) ensures genome stability by delaying anaphase onset until all the chromosomes have achieved proper spindle attachment. Once correct attachment has been achieved, SAC must be silenced. In the absence of mdf-1/MAD1, an essential SAC component, Caenorhabditis elegans cannot propagate beyond 3 generations. Previously, in a dog-1(gk10)/FANCJ mutator background, we isolated a suppressor of mdf-1(gk2) sterility (such-4) which allowed indefinite propagation in the absence of MDF-1. We showed that such-4 is a Cyclin B3 (cyb-3) duplication. Here we analyze mdf-1 such-4; dog-1, which we propagated for 470 generations, with freezing of samples for long time storage at F170 and F270. Phenotypic analysis of this strain revealed additional suppression of sterility in the absence of MDF-1, beyond the effects of such-4. We applied oligonucleotide array Comparative Genomic Hybridization (oaCGH) and whole genome sequencing (WGS) and identified a further amplification of cyb-3 (triplication) and a new missense mutation in dynein heavy chain (dhc-1). We show that dhc-1(dot168) suppresses the mdf-1(gk2), and is the second cloned suppressor, next to cyb-3 duplication, that does not cause a delay in anaphase onset. We also show that amplification of cyb-3 and dhc-1(dot168) cooperate to increase fitness in the absence of MDF-1.
Keywords: Cyclin B3 (cyb-3), dynein heavy chain (dhc-1), mdf-1/MAD1, genome stability, spindle assembly checkpoint (SAC)
Abbreviations
- APC/C
anaphase promoting complex/cyclosome
- CIN
chromosome instability
- EMS
ethyl methanesulfonate
- Him
high incidence of males
- oaCGH
oligonucleotide array Comparative Genomic Hybridization
- SAC
spindle assembly checkpoint
- WGS
whole genome sequencing
Introduction
In order to prevent loss or gain of chromosomes cells employ a surveillance mechanism known as spindle assembly checkpoint (SAC). The checkpoint inhibits the anaphase-promoting complex/cyclosome (APC/C), which stabilizes securin and prevents activation of separase and initiation of anaphase onset until all chromosomes have achieved proper spindle attachment.1,2 The general players in the SAC cascade have been elucidated; however, the precise mechanism of checkpoint activation and checkpoint silencing have yet to be determined.2,3 The core components of the spindle assembly checkpoint [Mad1, Mad2, Mad3 (also named BubR1 in some organisms), Bub1, and Bub3] are conserved in Caenorhabditis elegans.4-7
In the absence of MDF-1, an essential SAC component in C. elegans, genetic errors arise and accumulate leading to extinction of mdf-1(gk2) lines in 3 generations.4 The unique phenotype of mdf-1(gk2) allows for identification of enhancers (strains with enhancer mutations cease propagating before the third generation when MDF-1 is defective)7 and suppressors (strains which propagate beyond the third generation when MDF-1 is defective)8–10 in genetic screens. This is a powerful approach for discovering additional members of the pathway. In an ethyl methanesulfonate (EMS) screen for suppressors of mdf-1(gk2) lethality and sterility, 2 types of suppressors were isolated: 1) suppressors that result in constant anaphase onset delays, and likely suppress MDF-1 requirement by allowing more time for anaphase onset; 2) suppressors that do not affect anaphase onset and partially bypass MDF-1 requirement by an unknown mechanism.9 To date, all of the cloned EMS induced suppressors that result in constant anaphase onset delays are mutations affecting fzy-1/CDC20 and APC/C components.8,9
In addition to the EMS screen for suppressors of mdf-1(gk2) lethality and sterility, we isolated a such-4 suppressor (suppressor of spindle checkpoint defect) from a dog-1(gk10)/FANCJ mutator background.9 dog-1/FANCJ is a conserved gene with the unique task of protecting stretches of guanines from being deleted.11 We showed that such-4 suppresses mdf-1(gk2) sterility as a result of a duplication that amplifies the cyb-3 (Cyclin B3), which is important for proper checkpoint silencing.12 Doubling the dosage of CYB-3 does not result in continuous anaphase onset delays, thus making such-4 the first cloned suppressor of mdf-1(gk2) sterility that does not affect anaphase onset.10
Here we analyzed a mdf-1such-4; dog-1 strain, which was propagated for 470 generations with freezing, for long term preservation, of F170, F270. Phenotypic analysis of this strain revealed a further increase in fitness, independent of such-4 suppression. Using oaCGH analysis we demonstrate another amplification of cyb-3 to three copies, while genetic mapping combined with whole genome sequencing (WGS) analysis identified a missense mutation in dynein heavy chain (DHC-1). We show that the dhc-1 missense allele does not affect anaphase onset under normal conditions which is similar to the effect of the cyb-3 duplication. DHC-1, like CYB-3 is important for silencing SAC, once it is activated, by removing SAC components from properly attached kinetochores.12 Our findings indicate that components of the SAC silencing cascade could partially bypass the MDF-1 checkpoint requirement without constant anaphase onset delays. Finally, we show that amplifications of cyb-3 and base substitution in dhc-1 cooperate to achieve long-term population survival even when MDF-1 is not functioning.
Results
Long-term propagation of unc-46 mdf-1 such-4; dog-1 results in fitness increase
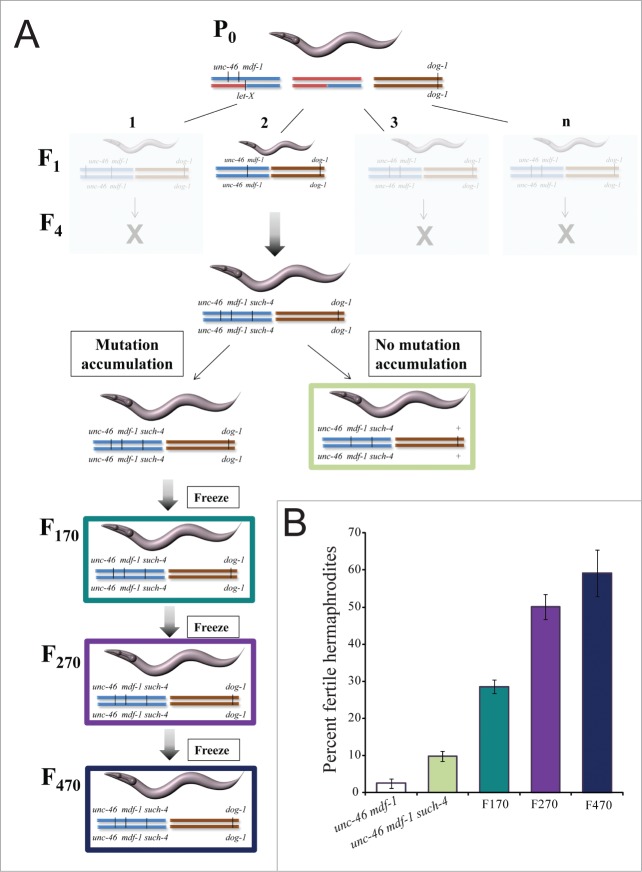
In the absence of MDF-1, C. elegans strains cannot be maintained beyond 3 generations.4 To prevent loss of gk2, the allele was linked to a visible marker unc-46 and balanced over the translocation nT18,9 (Fig. 1A). Previously, using the dog-1(gk10) mutator, we isolated such-4(h2168) as a suppressor of mdf-1(gk2) lethality.9 Briefly, we first constructed a P0 strain (Fig. 1A), then we plated 40 F1 Unc-46 worms (unc-46 mdf-1; dog-1) and analyzed them (Fig. 1).9 One of the 40 worms gave progeny with a different phenotype than the parental phenotype for unc-46 mdf-1 (2% reach fertile F2 adulthood and no fertile F3 adults are recovered). That one strain propagated beyond F3 and 10% of worms on average develop into fertile adults (Fig. 1A).9 One worm from this strain was outcrossed to remove dog-1(gk10) in order to avoid further accumulation of mutations. This strain, KR4233 unc-46(e177) mdf-1(gk2) such-4(h2168), was analyzed further10,13 (Fig. 1A). The second clone unc-46(e177) mdf-1(gk2) such-4(h2168); dog-1(gk10) was maintained for the total of 470 generations, with freezing to preserve genotypes at F170, F270 and F470 (Fig. 1A).
Figure 1.
Long-term propagation of unc-46 mdf-1 such-4; dog-1 homozygotes for 470 generations results in significant fitness recovery. (A) A schematic representation of the long-term propagation experiment. First, we generated a P0 strain of the following genotype: unc-46(e177) mdf-1(gk2) +/+ + nT1[let-X]; dog-1(gk10)/dog-1(gk10). Since mdf-1(gk2) results in lethality, gk2 is kept balanced over nT1 which is a reciprocal translocation between Chromosomes IV (depicted in red) and V (depicted in blue), and serves as an effective recombination suppressor along the translocated portions of each chromosome. dog-1(gk10) on Chromosome I is depicted in brown. We picked F1 unc-46 mdf-1; dog-1 homozygotes (n = 40) and plated them individually. We isolated a single plate containing fertile worms that survived beyond 3 generations as a suppressor candidate and named it such-4.9 The rest of the plates had no surviving progeny after 3 generations (depicted in light gray). We backcrossed one worm from F4 unc-46 mdf-1 such-4; dog-1 to N2 in order to remove dog-1(gk10) and thus avoid further accumulation of dog-1 induced mutations (depicted with light green). A second clone from F4 unc-46 mdf-1 such-4; dog-1 was maintained at 20°C for 470 generations to allow further accumulation of mutations. The worms were frozen for long term storage at the following points: F170 (depicted in dark green), F270 (depicted in purple) and F470 (depicted in blue). (B) Quantification of fitness, measured as the percent of fertile hermaphrodite progeny. Error bars represent SEM (n = 8 trials for each strain and 10 worms per trial). Please note that our initial analysis revealed that F2 unc-46(e177) mdf-1(gk2); dog-1 did not differ significantly from F2 unc-46(e177) mdf-1(gk2) homozygotes (t-test p = 0.845) and F4 unc-46(e177) mdf-1(gk2) such-4(h2168); dog-1 homozygotes did not differ significantly from F4 unc-46(e177) mdf-1(gk2) such-4(h2168) homozygotes (t-test p = 0.357). For that reason we performed multiple trials using F2 unc-46(e177) mdf-1(gk2) (depicted in white) and F4 unc-46(e177) mdf-1(gk2) such-4(h2168) (depicted in light green) as controls.
We previously demonstrated that the such-4 suppressor is a large tandem duplication of CYB-310. Doubling the dosage of CYB-3 alone increases percent of fertile adults from 2% in unc-46 mdf-1 to 10% observed in KR4233.10 During the long-term propagation of unc-46 mdf-1 such-4; dog-1 we detected and quantified significant fitness recoveries (Fig. 1B). We observed that F170 worms produce progeny of which on average 29% develop into fertile hermaphrodites. This is significantly more than unc-46 mdf-1 such-4 (t-test p = 1.16 × 10−4) (Fig. 1B). The longer we grew the strains the better the fitness. After generation F270, we observed that homozygotes produce on average 51% fertile hermaphrodites, which is significantly more than F170 (t-test p = 3.27 × 10−4). Note that this trend did not increase indefinitely as there was no significant difference between the F270 and the F470 unc-46 mdf-1 such-4; dog-1 homozygotes (t-test p = 0.257) (Fig. 1B). Together, these results indicate the presence of additional mutations in these genomes that cooperate to increase fitness when the MDF-1 checkpoint component is not functioning.
Mutation in dynein heavy chain suppresses sterility and lethality in the absence of MDF-1
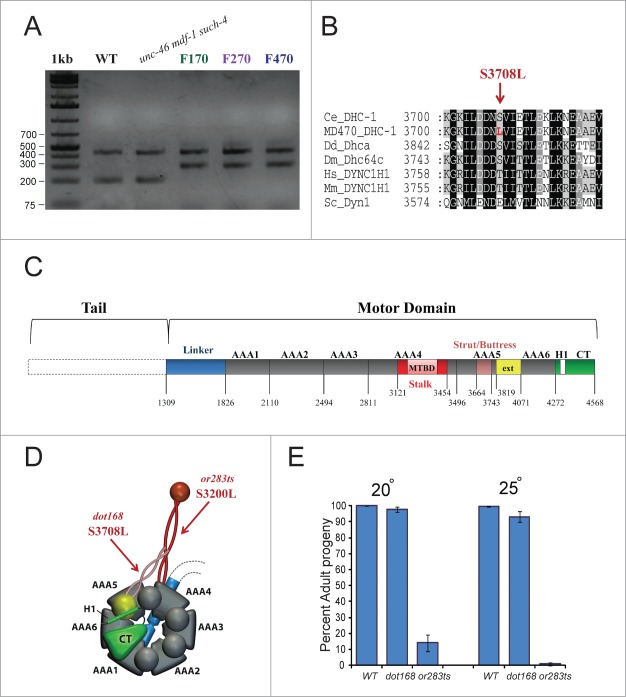
To identify additional mutations responsible for the observed fitness increase in unc-46 mdf-1 such-4; dog-1 worms after multi-generation propagation (Fig. 1), we combined WGS with oaCGH analysis to identify all of the mutations in F170, F270, and F470. Detailed genome analysis of these strains will be published separately. In addition to WGS and oaCGH analysis, we used phenotypic markers to position suppressors to chromosome locations as described previously.9 Our genetic dissection and mapping data for F470 unc-46 mdf-1 such-4; dog-1 revealed 2 chromosomal positions for suppressors: one on Chromosome V and the second one in a gene cluster on Chromosome I tightly linked to the dpy-5. Since such-4 is on Chromosome V,9,10 we focused our suppressor analysis on the dpy-5 region of Chromosome I. The WGS analysis revealed five mutations affecting protein-coding genes in this region: Y110A7A.19, whose function is unknown, has a 50 bp deletion, vacl-14 also of unknown function has a missense mutation, ugt-25 (UDP-Glucuronosyl Transferase) has a missense mutation, D2005.6 (unknown function) has a missense mutation, and a dynein heavy chain homolog (dhc-1), has a missense mutation.
dhc-1 is essential for several cell-cycle processes, including proper silencing of the spindle assembly checkpoint.14-17 The Chromosome I mutation at 4,398,929 (C>T) we identified in the F470 genome was confirmed by Sanger re-sequencing and was later shown to be present in the F170, F270 and F470 genomes (Fig. 2A). This substitution replaces serine with leucine at position 3708, within the strut/buttress motif of the fifth AAA ATPase domain18 (Fig. 2B-D). dhc-1 (also known as let-354 in C. elegans) is an essential gene and the first dhc-1 alleles were isolated in large-scale EMS-screens designed to isolate and maintain mutants having a lethal phenotype.14,19 To determine the consequence of the S3708L substitution, (dot168), we backcrossed unc-46 mdf-1 such-4; dog-1F470 to wild-type (N2) 10 times which isolated dot168 from the F470 genome background. We selected dot168 homozygotes using the tetra-primer ARMS-PCR method.20 After the tenth outcross, we investigated the development of dhc-1(dot168) homozygotes at 20°C and 25°C (Fig. 2E). Our analysis revealed that dhc-1(dot168) animals display subtle developmental arrest phenotype because 97.8 ± 1.8 and 93.3 ± 3.1 percent progeny reach adult stage at 20°C and 25°C, respectively (Fig. 2E) whereas effectively 100% of N2 animals reach adulthood. In C. elegans populations, consisting largely of self-fertilizing hermaphrodites (5A; XX), male progeny (5A; XO) arise as a result of X chromosome nondisjunction in the hermaphrodite germline at a low rate. Mutations in genes that are important for chromosome stability display a Him (high incidence of males) phenotype. To analyze the fidelity of chromosome segregation in the dhc-1(dot168) mutants, we scored the frequency of spontaneous males. At 20°C, we observed 0.3% males (22 males in 7463 adults) in dhc-1(dot168) homozygotes, while 0.05% males (4 males in 7544 adults) were observed in N2. At 25°C, we observed 1.1% males (38 males in 3432 adults) in dhc-1(dot168) homozygotes, while 0.2% males (7 males of 3601 adults) were observed in N2 control. Together, these data indicate a subtle chromosome instability (CIN) phenotype in dhc-1(dot168).
Figure 2.
dhc-1(dot168) suppresses lethality and sterility in the absence of MDF-1. (A) Tetra-primer ARMS-PCR20 analysis of dhc-1(+), unc-46 (e177) mdf-1(gk2) such-4(h2168) and unc-46 (e177) mdf-1(gk2) such-4(h2168); dog-1(gk10) mutation accumulation lines at F170, F270 and F470. The 439 bp product of the 2 outer primers was present in all strains; the 199 bp product of the wild-type allele was present in WT and unc-46 (e177) mdf-1(gk2) such-4(h2168); the 299 bp product of the dhc-1(dot168) mutant allele was present in F170, F270 and F470. (B) dhc-1(dot168) changed serine to leucine at position 3708 within the strut/buttress motif of the fifth AAA ATPase domain. Other organisms also have serine or threonine in this position (or glutamic acid for budding yeast). Organisms: Ce (Caenorhabditis elegans), Dd (Dictyostelium discoideum), Dm (Drosophila melanogaster), Hs (Homo sapiens), Mm (Mus musculus) and Sc (Saccharomyces cerevisiae). (C) Graphic representation of the dynein heavy chain motor domain organization based on the D. discoideum structure.18 We used T-Coffee (http://www.tcoffee.org) to align the C. elegans DHC-1 sequence to D. discoideum Dhca and based on homology extrapolated the location of the important motifs and domains in C. elegans DHC-1. (D) 3D model of the motor domain with arrows pointing to locations of dot168 and or283ts. The model was made based on the previously published model.31 (E) Phenotypic analysis of dhc-1(dot168) and dhc-1(or283ts) at 20°C and 25°C. The graph represents percent of progeny that develop into adults, while error bars represent SEM.
Next we introduced dot168 (outcrossed 10 times) into the mdf-1(gk2) background. We observed that all of the unc-46(e177) mdf-1(gk2); dhc-1(dot168) homozygotes propagated well beyond F3. Phenotypic analysis of unc-46(e177) mdf-1(gk2); dhc-1(dot168) animals revealed that dot168 reduced the percent of embryonic arrests from 22% to 11% and increased the percent of progeny developing into fertile hermaphrodites in the absence of MDF-1 from 2% to 10% (Table 1). These results indicate that dhc-1(dot168) from mdf-1 such-4; dog-1F470 improves fitness in the absence of the MDF-1 checkpoint component.
Table 1.
dhc-1 suppresses mdf-1(gk2) lethality and sterility
| Genotypes1 | Embryonic arrest (%) | Larval arrest (%) | Adult (%) | Fertile2 Hermaphrodites (%) | Males3 (%) |
|---|---|---|---|---|---|
| F2unc-46(e177) mdf-1(gk2) (n = 1985) | 21.8 | 54.2 | 24.0 | 2.0 | 3.6 |
| unc-46(e177) mdf-1(gk2); dhc-1(dot168) (n = 3010) | 10.6 | 54.8 | 34.6 | 10.0 | 2.5 |
| unc-46(e177) mdf-1(gk2); dhc-1(or283ts) (n = 1530) | 13.1 | 51.0 | 35.9 | 15.7 | 2.6 |
Maternal genotypes.
The percent of “Fertile hermaphrodites” was calculated from the total number of progeny.
The percent of “Males” was calculated based on the total number of adult progeny.
Although dhc-1(dot168) had been back-crossed to N2 strain 10 times, it is possible that we were unable to remove all of the background mutations and that one or more of these mutations may account for the observed suppression of the mdf-1(gk2) phenotype. To eliminate this possibility, we investigated whether a dhc-1 allele, isolated in an unrelated study, could suppress mdf-1(gk2) lethality and sterility. dhc-1(or283ts) is a temperature sensitive allele in which a conserved serine was changed to leucine within the microtubule-binding stalk domain (Fig. 2D).21 dhc-1(or283ts) can be propagated at permissive temperature, but shows 100% developmental arrest at 25°C (Fig. 2E). When we introduced dhc-1(or283ts) into mdf-1(gk2) animals, we observed a similar level of suppression as in unc-46(e177) mdf-1(gk2); dhc-1(dot168) at 20°C (Table 1); namely, embryonic arrests were reduced to 13% and percent of fertile hermaphrodites was increased to 16%. This result confirms that mutations in dhc-1 suppress lethality and sterility in the absence of MDF-1.
dhc-1(dot168) Does not result in constant anaphase onset delays
Two types of suppressors of the mdf-1(gk2) phenotype have been described.9 The first type of suppressor delays the onset of anaphase. This type of suppressor compensates for the requirement of MDF-1 by allowing more time for proper attachment of chromosomes to the spindle. The second type of suppressors has no measurable effect on anaphase onset during normal cell division and there is no model for how they suppress.9 To determine whether dhc-1(dot168) suppression has any measurable effect on anaphase onset, we used an integrated histone-GFP transgene (ruIs32) that marks mitotic chromosome behavior22 and measured the length of time to reach anaphase in these animals. Time-lapse fluorescence microscopy of one-cell stage embryos (Fig. 3A) revealed no significant difference in progression from complete nuclear envelope breakdown (NEBD) to anaphase onset between wild-type and dhc-1(dot168) embryos (t-test p = 0.746) (Fig. 3B). We also tested the timing of anaphase onset in the mdf-1(gk2) background (Fig. 3) and have observed a small but significant ∼45 second delay reported previously for mdf-1(gk2)10 (Fig. 3); however, we observed no significant difference in anaphase onset between mdf-1(gk2) and mdf-1(gk2); dhc-1(dot168) embryos (t-test p = 0.859) (Fig. 3B). Therefore, dhc-1(dot168), like the duplication of cyb-3, belongs to the second class of suppressors that have no measurable effect on anaphase onset during normal cell division.
Figure 3.
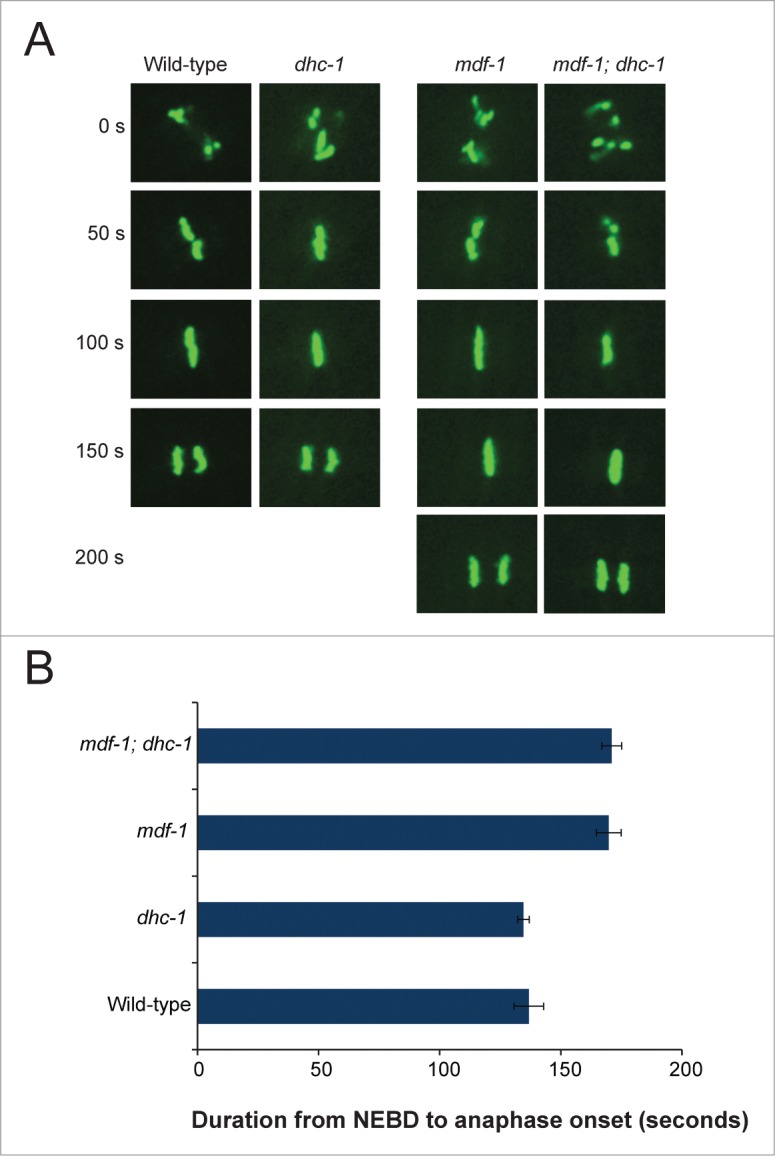
dhc-1(dot168) does not delay anaphase onset. (A) Time-lapse images (in seconds) of one-cell stage embryos that carry ruIs32, an integrated H2B::GFP transgene;22 wild-type [ruIs32], dhc-1 [dhc-1(dot168); ruIs32], mdf-1 [unc-46(e177) mdf-1(gk2); ruIs32] and mdf-1; dhc-1 [unc-46(e177) mdf-1(gk2); dhc-1(dot168); ruIs32] embryos are shown. (B) Summary of the timing measurements of the interval from complete nuclear envelope breakdown (NEBD) to anaphase onset (measured in seconds). Error bars represent standard error of the mean (SEM) for n = 5 measurements for each strain.
Mutations affecting dhc-1 and cyb-3 cooperate to increase fitness in the absence of MDF-1
Doubling the dosage of CYB-3 increases the number of mdf-1(gk2) progeny that develop into fertile hermaphrodites fivefold10 (Fig. 1B). Similarly, dhc-1(dot168) alone allows propagation in the absence of MDF-1 due to a fivefold increase in number of progeny that develop into fertile hermaphrodites (Table 1). However, neither the cyb-3 duplication nor the dhc-1(dot168) substitution alone can explain the level of fitness increase in unc-46 mdf-1 such-4; dog-1 animals after propagation for F170, F270 and F470 (Fig. 1B). When CYB-3 is depleted by RNAi mitotic dynein cannot remove SAC components from kinetochores leading to persistent block in anaphase onset12. Furthermore, genetic experiments revealed that CYB-3 is a positive regulator of dynein functionality.12 Thus, it is possible that increasing the dosage of CYB-3 and reducing function of DHC-1 in combination suppress mdf-1(gk2) lethality. We confirmed that dhc-1(dot168) is present at F170, F270 and F470 but not in KR4233 (unc-46 mdf-1 such-4) (Fig. 2A). If the cyb-3 duplication (Fig. 4A) and dhc-1(dot168) act together to increase fitness in F170 to about 30%, we would expect to see a similar result if we cross dhc-1(dot168) into the KR4233 background. We constructed unc-46(e177) mdf-1(gk2) such-4(h2168); dhc-1(dot168) and indeed observed on average that 33% of progeny developed into fertile hermaphrodites, significantly more than in KR4233 alone (t-test P = 1.08 × 10−4) (Fig. 4C), and comparable to the F170 strain (t-test p = 0.489) (Fig. 1B). This result indicates that increased dosage of CYB-3 together with reduced DHC-1 activity accounts for the observed fitness increase in the F170 strain.
Figure 4.
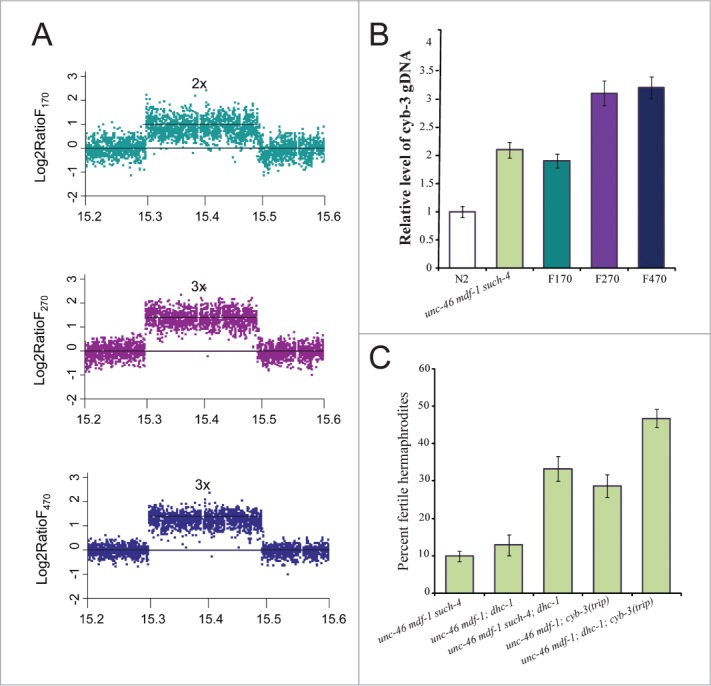
dhc-1(dot168) substitution and cyb-3 amplification work together to increase fitness in the absence of MDF-1. (A) oaCGH plots of 190 kb duplication located on Chromosome V. The y axes represent the log2 ratio of signal intensity of the region in F170 (green) versus N2, F270 (purple) versus N2, and F470 (blue) versus N2. The X axes represent genomic position on Chromosome V. The reference line intersecting at the point 0 reflects wild-type copy number, 2x depicts duplication (F170), while 3x is indicative of triplication (F270 and F470). (B) qRT-PCR was used to confirm the copy number variations affecting the cyb-3 locus in our strains. (C) cyb-3 amplification and dhc-1 act together to increase fitness in the absence of MDF-1. Fitness was measured as the percent of progeny that develop into fertile hermaphrodites. Error bars represent SEM (n = 8 trials for unc-46 mdf-1 such-4 and unc-46 mdf-1 cyb-3trip strains; n = 5 trials for the dhc-1-containing strains).
While this interaction between CYB-3 and reduced DHC-1 activity explains the fitness level at generation F170 it does not explain the further increases in fitness that we observed in later generations. For an explanation of this further increase in fecundity we looked more closely at the suppressor region on chromosome V. Our oaCGH (Fig. 4A) and qRT-PCR (Fig. 4B) analyses revealed that the Chromosome V tandem duplication had amplified again so there are 3 copies (triplication) of this region in the F270 and F470 strains. Previously, using the MosSCI method23 we generated a strain [unc-46(e177) mdf-1(gk2); cyb-3(dotSi100); cyb-3(dotSi110)] that contains three copies of cyb-324 and demonstrated that tripling CYB-3 dosage further increase fitness in the absence of MDF-1 (Fig. 4C).24 Thus, we reasoned that tripling the CYB-3 dosage together with reduced DHC-1 activity offers an explanation for the level of fitness increase we observe in F270 and F470 (Fig. 1B). To test this we constructed unc-46(e177) mdf-1(gk2); cyb-3(dotSi100); cyb-3(dotSi110); dhc-1(dot168) and observed that while on average 29% of progeny in unc-46(e177) mdf-1(gk2); cyb-3(dotSi100); cyb-3(dotSi110) homozygotes develop in fertile hermaphrodites (Fig. 4C), significantly more unc-46(e177) mdf-1(gk2); cyb-3(dotSi100); cyb-3(dotSi110); dhc-1(dot168) homozygotes develop into fertile hermaphrodites (t-test P = 2.09 × 10−4) (Fig. 4C), a level comparable to the fitness observed in the F270 and F470 MA lines (t-test P = 0.529 and P = 0.113 respectively) (Fig. 4C). These experiments indicate that amplification of cyb-3 cooperates with a substitution affecting dhc-1 to increase fitness in the absence of MDF-1. The interplay between the dosage increase in CYB-3 and reduced function of dhc-1 explains the fitness recovery we observed at the various generations we examined. Our experiment in long-term propagation of mdf-1 null animals reveals at least 2 means of compensation for the lack of this important checkpoint protein.
Discussion
MDF-1/Mad1 is a conserved component of the spindle assembly checkpoint that plays an essential role in C. elegans survival and fertility.4 When MDF-1 is absent, C. elegans populations cannot propagate beyond a few generations due to high levels of sterility and lethality.4 Previously, we isolated 10 suppressors of mdf-1(gk2) lethality and sterility from an EMS mutagenesis screen and one suppressor from a dog-1(gk10)/FANCJ mutator background.9 While the majority of the EMS-induced suppressors are alleles of fzy-1/CDC20 and APC/C components that compensate for loss of MDF-1 by delaying anaphase onset,9 the cyb-3 duplication (such-4 suppressor) isolated from the dog-1(gk10) mutator background was shown to be the first suppressor that does not delay anaphase onset.10 In this study, we generated an mdf-1 such-4; dog-1 strain, which we allowed to accumulated mutations for 470 generations, with periodic freezing for long-term storage at F170 and F270. We show further incremental increases in suppression of mdf-1(gk2) lethality and sterility in this strain, much beyond the effects of the initial cyb-3 duplication. This suppression results from further amplification of the cyb-3 locus to three copies and a missense mutation in dhc-1 in this strain. We demonstrate that dhc-1 compensates for the loss of MDF-1 without causing a constant anaphase onset delay. Furthermore, we showed that amplification of cyb-3 and dhc-1 mutation cooperate to increase fitness when MDF-1 absent.
Anaphase onset delay, due to SAC signaling, is essential to allow sufficient time for proper attachment of chromosomes to the spindle, and it is therefore understandable that mutations in fzy-1/CDC20 and APC/C that delay anaphase onset partially compensate for the lack of the checkpoint. Equally important is that SAC signaling is silenced at each kinetochore upon proper attachment of chromosomes to the spindle. SAC silencing occurs on 2 levels, one of which is depletion of essential SAC components, including MDF-1/Mad1, from kinetochores in a dynein-dependent manner.25 In C. elegans, CYB-3 has been shown to promote mitotic dynein function which silences SAC.12 Duplication of cyb-3 and a new dhc-1 missense allele reported here are the only two known suppressors that partially compensate for the lack of the MDF-1 checkpoint without causing anaphase onset delay. This finding opens the exciting possibility that additional components of the SAC silencing pathway may be identified as suppressors of mdf-1(gk2) lethality with normal mitotic timing. Furthermore, the interplay between amplification of cyb-3 and dhc-1(dot168) seems to account for the majority of the striking fitness recovery we observe after long-term propagation of the MDF-1 deficient strain for 470 generations (Fig. 1B). Our data indicate that dhc-1 suppresses mdf-1 lethality; however, the level of suppression is enhanced when cyb-3 amplified from one-to-two-to-three copies. Selection of these events has occurred through long-term propagation. Although we can correlate our findings to fitness recovery observed in unc-46 mdf-1 such-4; dog-1 propagated at F170, F270 and F470 (Figs. 1B and 4C), we acknowledge there may be additional mutation events that influence the phenotype in these genomes.
The discovery that dhc-1, like cyb-3dup suppresses mdf-1(gk2), yet conserves normal mitotic timing is intriguing. In the absence of CYB-3, embryos fail to initiate anaphase onset due to inactive dynein and compromised dynein-dependent removal of the SAC components from the kinetochores.12 Based on these results, one could expect that increased dosage of CYB-3 would result in overactive dynein and precocious anaphase onset, which would result in enhanced lethality in the absence of MDF-1 rather than the observed suppression. However, we showed that cyb-3dup does not result in altered anaphase onset,10 and we also showed here that dhc-1(dot168) embryos progress through anaphase normally. It is important to note that our analysis was performed under normal spindle conditions and did not test anaphase onset timing in the presence of induced spindle defects. One possible hypothesis is that in the presence of spindle defects, increased dosage of CYB-3 results in decreased efficiency and/or precision in promoting activity of dynein to move the activated SAC components away from kinetochores. Proper dynein activity is not only required to remove MAD1 and MAD2, but also other SAC components, such as BUB3 (BUB-3) and BUBR1 (SAN-1) from the kinetochore in order to alleviate SAC signaling.25 Thus, it is possible that increased dosage of CYB-3 combined with a missense allele in dhc-1 compensates for the absence of MDF-1 by causing a progressively more inefficient removal of SAN-1 and BUB-3. There are several lines of evidence that support this hypothesis. First, the analysis of SAC in C. elegans revealed that the checkpoint is composed of 2 largely independent branches, MDF-1/MDF-2 and BUB-3/SAN-1.26 Increasing the dosage of MDF-2/Mad2 bypasses the requirement for BUB-3/Bub3 and SAN-1/Mad3 for the checkpoint activation.26 Second, BUB-3 and SAN-1 are not essential for survival in C. elegans and depletion by RNAi gives only a subtle phenotype in the wild-type worms, but it enhances the lethality of mdf-1(gk2) and mdf-2(tm2190)7. Finally, a slight increase in dosage of these SAC components allows long-term propagation of strains that lack functional MDF-1 (M.T.G, unpublished). To directly test this hypothesis, future analysis using nocodazole to destabilize microtubules and mutants with defective spindles, like zyg-1, should be used to test if increased dosage of CYB-3 combined with reduced activity of DHC-1 could lead to a partial delay in anaphase onset even when MDF-1 is absent.
In summary, our studies have identified a new missense mutation in dynein heavy chain as a suppressor of lethality and sterility when an essential SAC component, MDF-1, is absent. The dhc-1 mutation is the second suppressor, next to cyb-3 duplication, that does not cause a constant anaphase onset delay, indicating that additional components of the SAC silencing cascade could be identified as suppressors with normal cell cycle timing. The mechanism behind this intriguing interplay between cyb-3 amplification and dhc-1 for long-term survival in the absence of MDF-1 should be investigated further for its potential to provide insights into the interdependencies and molecular mechanisms of removal of the SAC components from properly attached kinetochores during SAC silencing.
Materials and Methods
C. elegans strains
The Bristol strain N2 was used as the standard wild-type strain.27 The following mutant alleles were used in this work: unc-46(e177), mdf-1(gk2), dpy-5(s1300), dog-1(gk10), dhc-1(or283ts), dpy-10(e128), dpy-17(e164), dpy-13(e184), unc-119(ed3), ttTi5605, dotSi100, cxTi10882, dotSi110, such-4(h2168) and nT1[let-?(m435)]. The following strains were used in this work: N2 (Bristol strain as a wild-type), KR4233 [unc-46(e177) mdf-1(gk2) such-4(h2168)]; KR3627 [unc-46(e177) mdf-1(gk2) V/nT1[let-?(m435)])]; VC13 [dog-1(gk10)]; EU1385 [dhc-1(or283ts)]; JNC100 [unc-119(ed3) I; dotSi100 II (T06E6.2 + unc-119(+)]]; AZ212 [unc-119(ed3) ruIs32[unc-119(+) pie-1∷GFP∷H2B] III]; and JNC144 [unc-119(ed3) I; dotSi110 IV (T06E6.2 + unc-119(+)]]. Additional strains used in this work were generated in this study. Strains were maintained using standard protocol on nematode growth media (NGM) plates seeded with OP50 bacteria.27 The strains were maintained at 20°C, while the phenotypic analyses were performed at both 20°C and 25°C as noted in the results section.
Mutation accumulation procedure and phenotypic analysis
The first suppressor, such-4, was isolated as previously described.9 One clone from this strain, unc-46 mdf-1 such-4; dog-1 was outcrossed from the dog-1(gk10) background to avoid further accumulation of mutations and the KR4233 [mdf-1(gk2) such-4(h2168)] was generated and analyzed.10,13 The second strain (JNC170) was maintained at 20ºC for 470 generations to allow further accumulation of mutations. Each generation 5 L4 hermaphrodites were transferred to a fresh plate. We also froze the worms at generations 170 (JNC168) and 270 JNC169). Then, at F170, F270 and F470 phenotypic analysis was performed for each strain. Ten L4 hermaphrodites were plated individually and analyzed for number of embryonic arrests, larval arrests, adults and fertile adults, as described previously.9 In total, 5 to eight trials per strain were performed and SEM (standard error of the mean) was calculated and represented as error bars. Note that mdf-1(gk2) is linked to unc-46(e177) which results in an uncoordinated (Unc) phenotype and allows visual identification of mdf-1(gk2) homozygotes.
Genetic analysis of the unc-46 mdf-1 such-4; dog-1F470 genome
To map suppressor mutations to a chromosome, we used Dpy (Dumpy) markers located in the central regions of the autosomes. To identify candidate genes in the mapped regions, we combined WGS and oaCGH analyses. Genomic DNA was prepared from JNC168, JNC169 and JNC170 following a standard protocol (http://www.genetics.wustl.edu/tslab/Protocols/genomic_DNA_prep.htm) originally set up by Andy Fire's Laboratory. For the WGS, using Illumina Solexa technology, libraries were prepared and sequenced at Canada's Michael Smith Genome Sciences Center for JNC170 and Simon Fraser University for JNC168 and JNC169. oaCGH analysis was performed as described by Maydan and colleagues28 using the newly designed 3-plex microarray (120618_Cele_WS230_JK_CGH), manufactured by Roche NimbleGen Inc. Detailed methods on bioinformatics analysis of the WGS and oaCGH data will be published separately.
Phenotypic analysis of dhc-1
dhc-1(dot168) was separated from other accumulated mutations in the unc-46 mdf-1 such-4; dog-1F470 genome by extensive backcrossing to N2 and using the tetra-primer ARMS-PCR20 to select for dhc-1(dot168) homozygotes after each backcross. The following primers were designed and used for tetra-primer ARMS-PCR: C allele: AGCAAAGGAAAGATTCTTGATGATAAGTC; T allele: TCTTCAGTTTTTCGAGAGTTTCAATAAACA; Out_F:CCAGATCTTTATGATCACTCGTGATTC; Out_R:AATCTCATTGAGCTGTTGTAGAGTGTGA. Using these primers, homozygous dhc-1(dot168) is recognizable by 2 PCR-bands, 439 bp of the 2 outer primers and 299 bp of the mutant allele; N2 homozygotes also produce the 439 bp band, but a 199 bp band for the wild-type allele; heterozygotes produce 3 bands (439 bp, 299 bp and 199 bp). First, JNC170 was crossed to N2 and dhc-1(dot168) homozygotes that did not have unc-46(e177) mdf-1(gk2), dog-1(gk10) and such-4(h2168) alleles were selected. Then these dhc-1(dot168) homozygotes were backcrossed to N2 worms an additional 9 times. After each outcross, homozygous dhc-1(dot168) animals were selected. After the final outcross phenotypic analyses and genetic interaction studies were performed.
To assess whether dhc-1(dot168) and dhc-1(or283ts) could suppress the lethality and sterility of mdf-1(gk2), the F1 mdf-1(gk2) homozygotes, segregated from KR3627 (unc-46(e177) mdf-1(gk2) V/nT1[let-?(m435)]) strain were mated to either dhc-1(dot168) or dhc-1(or283ts) males. The phenotypically wild-type l unc-46(e177) mdf-1(gk2)/ + +; dhc-1/ + hermaphrodites were allowed to self-fertilize and Unc-46 progeny were plated individually. All of the worms that propagated for more than 3 generations were genotyped and confirmed to be homozygous for dhc-1, while none of the worms that did not be propagated for longer than 3 generations were homozygous for dhc-1.
To construct unc-46(e177) mdf-1(gk2) such-4(h2168); dhc-1(dot168) we mated dhc-1(dot168) males to KR4233 (unc-46(e177) mdf-1(gk2) such-4(h2168)) hermaphrodites and screened for Unc-46 worms. The Unc-46 worms were then analyzed using PCR and gk2, h2168, dot168 homozygotes were kept. The primers and procedure used to track gk2 and h2168 homozygotes were published previously.9,10 Using the MosSCI method23 we generated a strain (JNC100) that contains 2 copies of cyb-3 gene.10 To eliminate the possibility that potential background mutations in KR4233 would interfer with our results, we also constructed unc-46(e177) mdf-1(gk2); dhc-1(dot168); cyb-3dup(dotSi100). Analysis of this strain revealed similar results to the unc-46(e177) mdf-1(gk2) such-4(h2168); dhc-1(dot168) strain (data not shown). To construct unc-46(e177) mdf-1(gk2); cyb-3(dotSi100); cyb-3(dotSi110); dhc-1(dot168) we mated dhc-1(dot168) males to unc-46(e177); mdf-1(gk2); cyb-3(dotSi100); cyb-3(dotSi110) hermaphrodites then screened for Unc-46 worms. The Unc-46 worms were then analyzed using PCR and gk2, dotSi100, dotSi110, dot168 homozygotes were kept. The primers and procedure used to track dotSi100, dotSi110 homozygotes were published previously.10,24
Phenotypic analysis of the constructed strains was performed as follows; hermaphrodites at L4 stage were grown on fresh OP50 plates at 20⁰C or 25⁰C. The hermaphrodites were transferred to fresh plates every 12 hours. Brood size was calculated based on the total eggs laid by each hermaphrodite. Embryos that did not hatch were scored as embryonic arrests, while the embryos that hatched but did not grow to adult stage were scored as larval arrests. The embryos that developed into adults were analyzed for the presence of males in all the strains analyzed, while all the mdf-1(gk2)-allele containing strains were also analyzed for the percent of the sterile adult progeny by individually plating all the adult progeny and scoring the presence or absence of offspring.
Anaphase onset timing in the early embryo
One-day old gravid adult hermaphrodites were dissected and embryos were mounted onto 3% agarose pads as described previously.29 Embryos were observed using a Quorum WaveFX Spinning Disk system mounted on Zeiss Axioplan microscope. Early embryonic cell division was recorded using time-lapse video microscopy at 400x, with 200 ms fluorescent exposure, one image every 10 seconds. Image acquisition and analysis was performed using the Volocity software package.
qRT-PCR
qRT-PCR analysis was performed on genomic DNA from wild-type (N2), KR4233, JNC168, JNC169 and JNC170. Three internal references were used, eif-3, cdc-42 and mdh-1 following the procedures described previously.30 Mean values and standard deviations of relative ratios from four replicates are shown. Each qRT-PCR reaction contained 50 ng of genomic DNA, 10 μL of 2x SYBR Green Supermix (Biorad), and 1 μM of each primer. qRT-PCR reactions were run in quadruplicate on a Biorad MyIQ Real-time thermocycler. Data were normalized using the 3 internal references and the relative fold change was calculated using the ΔΔCt method. All primers were tested on serial dilutions and primer efficiency was calculated.
Funding Statement
This work was supported by the Canadian Institutes for Health Research (CIHR) and Fanconi Anemia Fellowship to MTG, and the Discovery Grant from the Natural Science and Engineering Research Council (NSERC) to NC. Work in the laboratory of DGM is supported by a grant from CIHR.
Acknowledgments
We thank the C. elegans Gene Knockout Consortium for generating the deletion mutants, the Caenorhabditis Genetics Center (CGC) for providing the strains, and the Michael Smith Genome Sciences Center for Illumina sequencing. We also thank Robert Johnsen, Steven Jones, Harald Hutter and Nancy Hawkins for critical review of the manuscript. NC is a Michael Smith Foundation for Health Research (MSFHR) Scholar and a CIHR New Investigator. DGM is a Senior Fellow of the Canadian Institute for Advanced Research (CIFAR).
References
- 1. Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 2007; 8:379-93; PMID:17426725; http://dx.doi.org/ 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- 2. Hauf S. The spindle assembly checkpoint: progress and persistent puzzles. Biochem Soc Trans 2013; 41:1755-60; PMID:24256287 [DOI] [PubMed] [Google Scholar]
- 3. Wang Y, Jin F, Higgins R, McKnight K. The current view for the silencing of the spindle assembly checkpoint. Cell Cycle Georget Tex 2014; 13:1694-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kitagawa R, Rose AM. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat Cell Biol 1999; 1:514-21; PMID:10587648; http://dx.doi.org/ 10.1038/70309 [DOI] [PubMed] [Google Scholar]
- 5. Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol 2001; 153:1209-26; PMID:11402065; http://dx.doi.org/ 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nystul TG, Goldmark JP, Padilla PA, Roth MB. Suspended animation in C. elegans requires the spindle checkpoint. Science 2003; 302:1038-41; PMID:14605367; http://dx.doi.org/ 10.1126/science.1089705 [DOI] [PubMed] [Google Scholar]
- 7. Tarailo M, Tarailo S, Rose AM. Synthetic lethal interactions identify phenotypic “interologs” of the spindle assembly checkpoint components. Genetics 2007; 177:2525-30; PMID:18073444; http://dx.doi.org/ 10.1534/genetics.107.080408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kitagawa R, Law E, Tang L, Rose AM. The Cdc20 homolog, FZY-1, and its interacting protein, IFY-1, are required for proper chromosome segregation in Caenorhabditis elegans. Curr Biol CB 2002; 12:2118-23; http://dx.doi.org/ 10.1016/S0960-9822(02)01392-1 [DOI] [PubMed] [Google Scholar]
- 9. Tarailo M, Kitagawa R, Rose AM. Suppressors of spindle checkpoint defect (such) mutants identify new mdf-1/MAD1 interactors in Caenorhabditis elegans. Genetics 2007; 175:1665-79; PMID:17237515; http://dx.doi.org/ 10.1534/genetics.106.067918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tarailo-Graovac M, Wang J, Tu D, Baillie DL, Rose AM, Chen N. Duplication of cyb-3 (cyclin B3) suppresses sterility in the absence of mdf-1/MAD1 spindle assembly checkpoint component in Caenorhabditis elegans. Cell Cycle Georget Tex 2010; 9:4858-65; http://dx.doi.org/ 10.4161/cc.9.24.14137 [DOI] [PubMed] [Google Scholar]
- 11. Cheung I, Schertzer M, Rose A, Lansdorp PM. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat Genet 2002; 31:405-9; PMID:12101400 [DOI] [PubMed] [Google Scholar]
- 12. Deyter GMR, Furuta T, Kurasawa Y, Schumacher JM. Caenorhabditis elegans cyclin B3 is required for multiple mitotic processes including alleviation of a spindle checkpoint-dependent block in anaphase chromosome segregation. PLoS Genet 2010; 6:e1001218; http://dx.doi.org/ 10.1371/journal.pgen.1001218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao Y, Tarailo-Graovac M, O’Neil NJ, Rose AM. Spectrum of mutational events in the absence of DOG-1/FANCJ in Caenorhabditis elegans. DNA Repair 2008; 7:1846-54; PMID:18708164; http://dx.doi.org/ 10.1016/j.dnarep.2008.07.011 [DOI] [PubMed] [Google Scholar]
- 14. Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 2001; 155:1159-72; PMID:11756470; http://dx.doi.org/ 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Griffis ER, Stuurman N, Vale RD. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 2007; 177:1005-15; PMID:17576797; http://dx.doi.org/ 10.1083/jcb.200702062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schmidt DJ, Rose DJ, Saxton WM, Strome S. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol Biol Cell 2005; 16:1200-12; PMID:15616192; http://dx.doi.org/ 10.1091/mbc.E04-06-0523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivaram MVS, Wadzinski TL, Redick SD, Manna T, Doxsey SJ. Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint. EMBO J 2009; 28:902-14; PMID:19229290; http://dx.doi.org/ 10.1038/emboj.2009.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. The 2.8Å crystal structure of the dynein motor domain. Nature 2012; 484:345-50; PMID:22398446; http://dx.doi.org/ 10.1038/nature10955 [DOI] [PubMed] [Google Scholar]
- 19. Mains PE, Sulston IA, Wood WB. Dominant maternal-effect mutations causing embryonic lethality in Caenorhabditis elegans. Genetics 1990; 125:351-69; PMID:2379819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 2001; 29:E88-88; PMID:11522844; http://dx.doi.org/ 10.1093/nar/29.17.e88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell 2002; 3:673-84; PMID:12431374; http://dx.doi.org/ 10.1016/S1534-5807(02)00327-1 [DOI] [PubMed] [Google Scholar]
- 22. Vanoosthuyse V, Hardwick KG. Overcoming inhibition in the spindle checkpoint. Genes Dev 2009; 23:2799-805; PMID:20008930; http://dx.doi.org/ 10.1101/gad.1882109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Praitis V, Casey E, Collar D, Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 2001; 157:1217-26; PMID:11238406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen S-P, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 2008; 40:1375-83; PMID:18953339; http://dx.doi.org/ 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tarailo-Graovac M, Chen N. Proper cyclin B3 dosage is important for precision of metaphase-to-anaphase onset timing in Caenorhabditis elegans. G3 Bethesda Md 2012; 2:865-71; http://dx.doi.org/ 10.1534/g3.112.002782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 2013; 14:25-37; PMID:23258294; http://dx.doi.org/ 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Essex A, Dammermann A, Lewellyn L, Oegema K, Desai A. Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol Biol Cell 2009; 20:1252-67; PMID:19109417; http://dx.doi.org/ 10.1091/mbc.E08-10-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brenner S. The genetics of Caenorhabditis elegans. Genetics 1974; 77:71-94; PMID:4366476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maydan JS, Flibotte S, Edgley ML, Lau J, Selzer RR, Richmond TA, Pofahl NJ, Thomas JH, Moerman DG. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res 2007; 17:337-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 1983; 100:64-119; PMID:6684600; http://dx.doi.org/ 10.1016/0012-1606(83)90201-4 [DOI] [PubMed] [Google Scholar]
- 31. Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol 2008; 9:9; PMID:18211699; http://dx.doi.org/ 10.1186/1471-2199-9-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Höök P, Vallee R. Dynein dynamics. Nat Struct Mol Biol 2012; 19:467-9; PMID:22551707; http://dx.doi.org/ 10.1038/nsmb.2290 [DOI] [PubMed] [Google Scholar]