Abstract
Aberrations in the cyclin-dependent kinase (CDK) pathways that regulate the cell cycle restriction point contribute to genomic instability and tumor proliferation, and can be targeted by recently developed CDK inhibitors. We therefore investigated the clinical correlates of CDK4/6 and CDKN2A/B abnormalities in diverse malignancies. Patients with various cancers who underwent molecular profiling by targeted next generation sequencing (Foundation Medicine; 182 or 236 cancer-related genes) were reviewed. Of 347 patients analyzed, 79 (22.8%) had aberrant CDK 4/6 or CDKN2A/B. Only TP53 mutations occurred more frequently than those in CDK elements. Aberrations were most frequent in glioblastomas (21/26 patients; 81%) and least frequent in colorectal cancers (0/26 patients). Aberrant CDK elements were independently associated with EGFR and ARID1A gene abnormalities (P < 0.0001 and p = 0.01; multivariate analysis). CDK aberrations were associated with poor overall survival (univariate analysis; HR[95% CI] = 2.09 [1.35–4.70]; p = 0.004). In multivariate analysis, PTEN and TP53 aberrations were independently associated with poorer survival (HR = 4.83 and 1.92; P < 0.0001 and p = 0.01); CDK aberrations showed a trend toward worse survival (HR = 1.67; p = 0.09). There was also a trend toward worse progression-free survival (PFS) with platinum-containing regimens in patients with abnormal CDK elements (3.5 versus 5.0 months, p = 0.13). In conclusion, aberrations in the CDK pathway were some of the most common in cancer and independently associated with EGFR and ARID1A alterations. Patients with abnormal CDK pathway genes showed a trend toward poorer survival, as well as worse PFS on platinum-containing regimens. Further investigation of the prognostic and predictive impact of CDK alterations across cancers is warranted.
Keywords: cancer, CDKN2A/B, CDK4/6, cyclin-Dependent Kinase, next generation sequencing
Introduction
Aberrations in the cyclin D-cyclin-dependent kinase (CDK)-retinoblastoma (Rb) pathway that regulates the cell cycle restriction point is a common feature of human cancer, contributing to tumor proliferation, genomic instability and chromosomal instability.1–3 Cyclin D1 complexes with its catalytic partners, CDK 4/6, leading to Rb protein phosphorylation, and subsequent E2F-mediated transcription of target genes necessary for G1 cell-cycle progression (Fig. S1). This pathway can be altered through multiple mechanisms including mutation or loss of Rb, increased signaling through CDK4 and CDK6 amplification, overexpression of cyclin D1,4 and loss of inhibitors including CDKN2A (p16) and/or CDKN2B (p15).5,6 Alterations in the CDK pathway have also been linked to poorer clinical outcome in multiple cancer types including acute lymphoblastic leukemia,7 ovarian8 and colon cancer,9 and medulloblastoma10; preliminary results in breast cancer suggest that these aberrations may be associated with a better outcome.11
Importantly, there are several inhibitors in clinical trials, with varying selectivity for specific members of the CDK family.2 Some examples include CDK4/6 inhibitors, palbociclib (PD0332991) and LEE011, and the CDK1/2/5/9 inhibitor, dinaciclib (SCH 727965) which all demonstrated clinical activities for patients with advanced malignancies in early phase clinical trials.12–14 Among them, palbociclib (PD0332991), a potent selective CDK4/6 inhibitor, has emerged as an attractive therapeutic option because of its favorable toxicity profile.15–17 Recent trials demonstrate prolonged progression-free survival (PFS) in patients with hormone receptor-positive, Her2-negative metastatic breast cancer when the hormone modulator letrozole was combined with palbociclib as compared to letrozole alone (PALOMA-1 trial, median 20.2 months vs. 10.2 months, respectively).12
Of interest, CDK pathway abnormalities have been reported in numerous cancers,7-10 but the biological implications and landscape of these abnormalities has not been well studied. We therefore used next generation sequencing (NGS) to determine the molecular characteristics and clinical correlates of CDK4/6 or CDKN2A/B abnormalities in 347 patients with diverse malignancies.
Results
CDK4/6 and CDKN2A/B abnormalities
Seventy-nine patients (23%) had an abnormality in CDK4/6 or CDKN2A/B (CDK elements) (Table 1 and Fig. 1): 62 cases (18% of 347) with CDKN2A aberrations (48 patients with CDKN2A loss and 14 with mutations); 37 (11%), CDKN2B loss; 13 (4%), CDK4 amplification; and 8 (2%), CDK6 amplification (Fig. 1). Thirty-eight cases (11%) had more than one abnormality in CDK4/6 or CDKN2A/B.
Table 1.
Clinical characteristics of 347 patients with CDK4/6, CDKN2A/B aberrations (univariate analysis)
| Patient characteristics (N = 347) | Aberrant CDK4/6, DKN2A/B N = 79 (%) | Normal CDK4/6, CDKN2A/B N = 268 (%) | P-value* |
|---|---|---|---|
| Gender | 0.026 | ||
| Women (N = 196) | 36 (18.4%) | 160 (81.6%) | |
| Men (N = 151) | 43 (28.5%) | 108 (71.5%) | |
| Age at diagnosis | 0.13 | ||
| Age ≥ 50 y (N = 212) | 54 (25.5%) | 158 (74.5%) | |
| Age <50 y (N = 135) | 25 (18.5%) | 110 (81.5%) | |
| Types of cancer diagnosis§ | |||
| Breast (N = 74) | 7 (9.5%) | 67 (90.5%) | 0.0021 |
| Glioblastoma (N = 26) | 21 (80.8%) | 5 (19.2%) | < 0.0001 |
| Colorectal (N = 26) | 0 (0%) | 26 (100%) | 0.004 |
| Lung (N = 23) | 6 (26.1%) | 17 (73.9%) | 0.69 |
| Melanoma (N = 23) | 8 (34.8%) | 15 (65.2%) | 0.16 |
| Appendix (N = 20) |
1 (5.0%) |
19 (95.0%) |
0.27 |
|
Site of metastasis (N = 280) |
Aberrant CDK4/6, DKN2A/B N = 50 (%) |
Normal CDK4/6, CDKN2A/B N = 230 (%) |
|
| Lymph node metastasis | 0.73 | ||
| Yes (N = 145) | 27 (18.6%) | 118 (81.4%) | |
| No (N = 135) | 23 (17.0%) | 112 (83.0%) | |
| Liver metastasis | 0.74 | ||
| Yes (N = 101) | 17 (16.8%) | 84 (83.2%) | |
| No (N = 179) | 33 (18.4%) | 146 (81.6%) | |
| Bone metastasis | 0.36 | ||
| Yes (N = 82) | 12 (14.6%) | 70 (85.4%) | |
| No (N = 198) | 38 (19.2%) | 160 (80.8%) | |
| Lung metastasis | 0.23 | ||
| Yes (N = 81) | 11 (13.6%) | 70 (86.4%) | |
| No (N = 199) | 39 (19.6%) | 160 (80.4%) | |
| Omentum/Peritoneum metastasis | 0.92 | ||
| Yes (N = 49) | 9 (18.4%) | 40 (81.6%) | |
| No (N = 231) | 41(17.8%) | 190 (82.2%) | |
| Brain metastasis | 0.16 | ||
| Yes (N = 34) | 9 (26.5%) | 25 (73.5%) | |
| No (N = 246) | 41(16.7%) | 205 (83.3%) | |
| Soft tissue metastasis | 0.21 | ||
| Yes (N = 31) | 3 (9.7%) | 28 (90.3%) | |
| No (N = 249) | 47 (18.9%) | 202 (81.1%) | |
| Adrenal metastasis | 0.51 | ||
| Yes (N = 12) | 3 (25.0%) | 9 (75.0%) | |
| No (N = 268) | 47 (17.5%) | 221 (82.5%) | |
| Site of biopsies | 0.83 | ||
| Primary (N = 125) | 23 (18.4%) | 102 (81.6%) | |
| Metastases (N = 155) | 27 (17.4%) | 128 (82.6%) |
P-values are from Fisher's exact test.
Included characteristics with N ≥ 20 of primary cancer diagnosis. See Table S1 for complete list of characteristics with N < 20.
Excluded patients with hematological malignancy (N = 21) and CNS tumors (N = 46). Reported here with site of metastasis with N ≥ 10.
Figure 1.
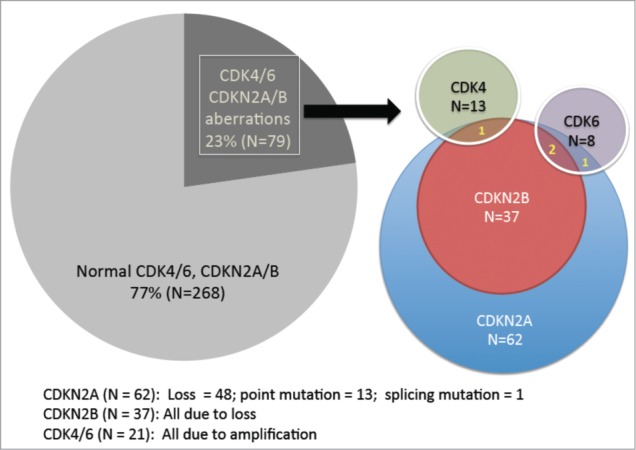
Frequency of CDK-associated genetic aberrations in 347 patients with diverse malignancies. Of 347 patients with diverse malignancies, 23% (N = 79) had an aberration in either CDK4 (N = 13 [3.7% of 347 patients]), CDK6 (N = 8 [2.3%]), CDKN2A (N = 62 [17.9%]) and/or CDKN2B (N = 37 [10.7%]). All cases with CDKN2B aberrations (N = 37) also had aberrant CDKN2A (but the opposite was not true). One case of CDK4 amplification had a co-existing CDKN2A/B aberration. CDK6 amplification occasionally co-existed with CDKN2A/B aberration (N = 2) or a CDKN2A aberration (N = 1).
Clinical and molecular characteristics associated with CDK4/6 or CDKN2A/B abnormalities (univariate analysis) (Tables 1–2 and Table S1)
Table 2.
CDK4/6 or CDKN2A/B abnormality and types of co-existing genetic aberrations (univariate analysis)
| Patient characteristics (N = 347) | Aberrant CDK4/6, CDKN2A/B N = 79 (%) | Normal CDK4/6, CDKN2A/B N = 268 (%) | P-value* |
|---|---|---|---|
| Types of co-existing genetic aberrations§ | |||
| TP53 | 0.28 | ||
| Wild-type (N = 194) | 40 (20.6%) | 154 (79.4%) | |
| Aberrant (N = 153) | 39 (25.5%) | 114 (74.5%) | |
| KRAS | 0.02 | ||
| Wild-type (N = 292) | 73 (25.0%) | 219 (75.0%) | |
| Aberrant (N = 55) | 6 (10.9%) | 49 (89.1%) | |
| FGFR/FGF | 0.11 | ||
| Wild-type (N = 296) | 63 (21.3%) | 233 (78.7%) | |
| Aberrant (N = 51) | 16 (31.4%) | 35 (68.6%) | |
| PIK3CA | 0.01 | ||
| Wild-type (N = 307) | 76 (24.8%) | 231 (75.2%) | |
| Aberrant (N = 40) | 3 (7.5%) | 37 (92.5%) | |
| PTEN | 0.96 | ||
| Wild-type (N = 308) | 70 (22.7%) | 238 (77.3%) | |
| Aberrant (N = 39) | 9 (23.1%) | 30 (76.9%) | |
| MYC | 0.24 | ||
| Wild-type (N = 311) | 68 (21.9%) | 243 (78.1%) | |
| Aberrant (N = 36) | 11 (30.6%) | 25 (69.4%) | |
| EGFR | < 0.0001 | ||
| Wild-type (N = 321) | 59 (18.4%) | 262 (81.6%) | |
| Aberrant (N = 26) | 20 (76.9%) | 6 (23.1%) | |
| CCND1 | 0.52 | ||
| Wild-type (N = 322) | 72 (22.4%) | 250 (77.6%) | |
| Aberrant (N = 25) | 7 (28.0%) | 18 (72.0%) | |
| APC | 0.03 | ||
| Wild-type (N = 324) | 78 (24.1%) | 246 (75.9%) | |
| Aberrant (N = 23) | 1 (4.4%) | 22 (95.7%) | |
| MCL1 | 1.00 | ||
| Wild-type (N = 325) | 74 (22.8%) | 251 (77.2%) | |
| Aberrant (N = 22) | 5 (22.7%) | 17 (77.3%) | |
| NF1 | 1.00 | ||
| Wild-type (N = 325) | 74 (22.8%) | 251 (77.2%) | |
| Aberrant (N = 22) | 5 (22.7%) | 17 (77.3%) | |
| ARID1A | 0.06 | ||
| Wild-type (N = 327) | 71 (21.7%) | 256 (78.3%) | |
| Aberrant (N = 20) | 8 (40.0%) | 12 (60.0%) | |
| BRCA2 | 0.39 | ||
| Wild-type (N = 327) | 76 (23.2%) | 251 (77.2%) | |
| Aberrant (N = 20) | 3 (15.0%) | 17 (85.0%) |
P-values are from Fisher's exact test.
Included characteristics with N ≥ 20 of genetic aberration. See Table S1 for complete list of characteristics with N < 20.
Men more commonly had abnormalities in CDK elements than women (28.5% versus 18.4%; P value = 0.026) (Table 1). Older age at diagnosis (≥50 years old) was associated with a non-significant trend toward greater likelihood of CDK4/6 or CDKN2A/B aberrations (p = 0.13) (Table 1). Frequency of CDK4/6 or CDKN2A/B aberration was similar regardless of the site of biopsies obtained for analysis (18.4% from primary vs. 17.4% from metastatic samples; p = 0.83) (Table 1). Among 155 cases of metastatic samples evaluated for molecular aberration, there were 5 cases evaluated from brain metastasis. Aberrations in CDK elements were associated with a diagnosis of glioblastoma (n = 21 of 26 patients with glioblastoma, 81%; P < 0.0001). CDK4/6 or CDKN2A/B abnormalities were found less frequently (7/74 [9.5%]) in patients with breast cancer (p = 0.0021). We did not observe abnormalities in CDK elements among patients with colorectal cancer (n = 0/0 [0%]; p = 0.004) (Table 1). Regarding sites of metastases, there was a trend (9 of 34 patients [26.5%]; p = 0.16) for CDK4/6 or CDKN2A/B abnormalities in individuals with brain metastases (Table 1). In univariate analysis, CDK4/6 or CDKN2A/B were more likely to be associated with EGFR and ARID1A abnormalities (P < 0.0001 and p = 0.06 [trend]) (Table 2). There was an inverse association with KRAS, PIK3CA and APC aberrations (p = 0.02, p = 0.01 and p = 0.03 respectively).
Multivariate analysis of association between clinical and molecular characteristics and CDK4/6 or CDKN2A/B abnormalities (Table 3):
In multivariate analysis, a diagnosis of glioblastoma was significantly associated with CDK4/6 or CDKN2A/B abnormalities (odds ratio [OR] = 11.2; 95% CI: 3.7–34.5, P < 0.0001) (Table 3). A diagnosis of breast cancer trended to be less frequently associated with abnormal CDK elements (OR = 0.39, 95% CI: 0.15–1.02, p = 0.057).
Table 3.
Multivariate analysis of patient characteristics (N = 347) associated with CDK4/6, CDKN2A/B aberrations
| Characteristics | Odds ratio (95% CI) | P-value* |
|---|---|---|
| Women | 0.92 (0.48–1.76) | 0.80 |
| Breast | 0.39 (0.15–1.02) | 0.057 |
| Glioblastoma | 11.2 (3.7–34.5) | < 0.0001 |
| Colorectal | 0.28 (0.03–2.53) | 0.26 |
| KRAS aberrant | 0.59 (0.22–1.57) | 0.29 |
| PIK3CA aberrant | 0.56 (0.16–1.96) | 0.36 |
| EGFR aberrant | 11.9 (3.92–35.7) | < 0.0001 |
| APC aberrant | 0.19 (0.02–1.83) | 0.15 |
| ARID1A aberrant | 3.98 (1.36–11.8) | 0.01 |
Characteristics selected from Table 1 and 2 with P-value < 0.1.
P-values are from multivariate logistic regression analysis. P < 0.05 was considered to be statistically significant.
Interestingly, EGFR and ARID1A aberrations were significantly associated with CDK4/6 or CDKN2A/B in multivariate analysis (EGFR: OR = 11.9; 95% CI: 3.92–35.7, P < 0.0001) (ARID1A: OR = 3.98; 95% CI: 1.36–11.8, p = 0.01) (Table 3). Co-existing EGFR aberrations included amplification, mutation and EGFR vIII (Table S2). All the co-existing ARID1A aberrations were mutations (Table S3). Indeed, 20 of 26 patients (76.9%) of patients with an EGFR aberration had an abnormality in CDK elements (Table 2). Eight of 20 patients with ARID1A aberrations (40.0%) also had an abnormality in CDK elements (Table 2).
Outcome and abnormalities in CDK4/6 or CDKN2A/B (univariate and multivariate analysis) (Tables 4 and 5):
Table 4.
Clinical outcomes of patients with CDK4/6, CDKN2A/B aberrations (univariate analysis)
| Clinical outcomes | Aberrant CDK4/6, CDKN2A/B Month (range) | Normal CDK4/6, CDKN2A/B Month (range) | P-value* | Comment |
|---|---|---|---|---|
| Median time from diagnosis to metastasis (N = 280) | 17.0 (0–256.7) N = 50 |
24.2 (0–415.1) N = 230 |
0.20 | (a) |
| PFS for first line Therapy | ||||
| Median PFS for all first line therapy (N = 189) | 3.3 (0.6–39.0) N = 41 |
5.0 (0.5–61.0) N = 148 |
0.32 | (b) |
| Subgroup analysis of median PFS for first line therapy | ||||
| Platinum-containing regimen (N = 62) | 3.5 (0.6–11.0) N = 15 |
5.0 (1.0–34.0) N = 47 |
0.13 | (c) |
| 5-FU or capecitabine- containing regimen (N = 50) | 5.0 (0.6–11.0) N = 11 |
5.0 (0.8–21.0) N = 39 |
0.3 | (d) |
| Bevacizumab-containing regimen (N = 33) | 6.5 (2.0–39.0) N = 6 |
6.5 (1.0–34.0) N = 27 |
0.43 | |
| Taxane-containing regimen (N = 32) | 3.0 (0.6–39.0) N = 7 |
9.1 (1.0–38.7) N = 25 |
0.88 | (e) |
P-values are from log-rank test.
(a) Excluded patients with hematological malignancy (N = 21) and CNS tumors (N = 46).
(b) First line therapy referred to first line after metastatic or recurrent disease (N = 189). Excluded patients with hematological malignancy (N = 21). Neoadjuvant/adjuvant therapy was not included (N = 56). Chemotherapy was not initiated on 33 patients and 48 patients were not assessable for accurate PFS for first line therapy.
(c) Platinum-containing regimen (N = 62): Cisplatin (N = 20), carboplatin (N = 21) and oxaliplatin (N = 21).
(d) 5-FU or capecitabine-containing regimen (N = 50): 5-FU (N = 30) and capecitabine (N = 20)
(e) Taxane-containing regimen (N = 32): Paclitaxel (N = 22), docetaxel (N = 5) and abraxane (N = 5).
Table 5.
Univariate and multivariate Cox's regression models predicting duration of overall survival in 347 patients with malignancies
| Characteristics¶ (N = 347) | Hazard Ratio | 95% CI | P-value* |
|---|---|---|---|
| Gender and age | |||
| Women (N = 196) | 0.65 | 0.38–1.04 | 0.08 |
| Age ≥ 50 y (N = 212) | 1.21 | 0.77–1.97 | 0.41 |
| Types of cancer | |||
| Breast (N = 74) | 0.64 | 0.39–1.08 | 0.11 |
| Glioblastoma (N = 26) | 3.01 | 1.73–26.7 | 0.006 |
| Colorectal (N = 26) | 1.73 | 0.71–5.93 | 0.19 |
| Lung (N = 23) | 1.16 | 0.40–3.46 | 0.77 |
| Melanoma (N = 23) | 0.55 | 0.25–1.48 | 0.29 |
| Appendix (N = 20) | 0.89 | 0.30–2.67 | 0.84 |
| Types of genetic aberration | |||
| TP53 aberrant (N = 153) | 2 | 1.27–3.31 | 0.004 |
| CDK4/6, CDKN2A/B aberrant (N = 79) | 2.09 | 1.35–4.70 | 0.004 |
| KRAS aberrant (N = 55) | 1.24 | 0.63–2.53 | 0.51 |
| FGFR/FGF aberrant (N = 51) | 0.67 | 0.39–1.26 | 0.24 |
| PIK3CA aberrant (N = 40) | 0.76 | 0.39–1.57 | 0.49 |
| PTEN aberrant (N = 39) | 4.02 | 5.02–31.9 | <0.0001 |
| MYC aberrant (N = 36) | 1.45 | 0.71–3.33 | 0.27 |
| EGFR aberrant (N = 26) | 2.23 | 1.07–9.71 | 0.04 |
| CCND1 aberrant (N = 25) | 0.48 | 0.27–1.21 | 0.15 |
| APC (N = 23) | 1.75 | 0.65–6.69 | 0.22 |
| MCL1 aberrant (N = 22) | 0.49 | 0.25–1.35 | 0.21 |
| NF1 aberrant (N = 22) | 0.74 | 0.28–2.10 | 0.61 |
| ARID1A aberrant (N = 20) | 0.61 | 0.26–1.65 | 0.38 |
| BRCA2 aberrant (N = 20) | 1.09 | 0.38–3.12 | 0.87 |
| Multivariate Cox's regression model (N = 347) | |||
| Women | 0.6 | 0.35–1.02 | 0.06 |
| Glioblastoma | 1.43 | 0.55–3.72 | 0.46 |
| TP53 aberrant | 1.92 | 1.17–3.14 | 0.01 |
| CDK4/6, CDKN2A/B aberrant | 1.67 | 0.92–3.04 | 0.09 |
| PTEN aberrant | 4.83 | 2.63–8.87 | <0.0001 |
| EGFR aberrant | 1.31 | 0.54–3.16 | 0.55 |
Included characteristics with N ≥ 20.
P-values (univariate) and hazard ratios with 95% CI are from log-rank test or multivariate Cox's regression model as appropriate. P <0.1 from univariate analysis were included for multivariate analysis.
To assess the association between aberrant CDK elements and development of metastases, we first excluded hematological malignancies (n = 21) and CNS tumors (n = 46) (Table 1 and Table S1) from the 347 patients studied. Of 280 patients assessable for development of metastases, 17.9% (n = 50) of patients had abnormalities in CDK elements. Site of biopsies obtained for molecular analysis were similar between the 2 groups (54% [27/50] of patients with aberrant CDK and 56% [128/230] of patients with normal CDK were biopsied from metastatic sites, p = 0.83, Table 1). Numbers of patients who had documented metastases were also similar between the 2 groups (88% [44/50] of patients with aberrant CDK and 90% [206/230] patients with normal CDK eventually had metastatic disease, p = 0.75). The median time from diagnosis to metastases was 17 months versus 24 months in patients with and without abnormalities (p = 0.2) (Table 4).
We next assessed the overall survival for the 347 patients studied. In univariate analysis, the presence of aberrant CDK elements was significantly associated with shorter survival when compared to patients with normal CDK elements (hazard ratio [HR][95% CI] = 2.09 [1,35 – 4.70]; p = 0.004) (Table 5).
In multivariate analysis using the Cox's regression model (variables with P value less than 0.1 from univariate analysis were included [Table 5]: gender [women], diagnosis of glioblastoma, and TP53, CDK4/6 or CDKN2A/B, PTEN, and EGFR aberrations), the only factors independently associated with poorer survival were PTEN and TP53 aberrations (HR = 4.83 and 1.92; P < 0.0001 and p = 0.01 respectively) (Table 5). Patients with abnormalities in CDK elements showed a trend for association with worse survival (HR = 1.67; p = 0.09) (Table 5).
Association of abnormalities in CDK4/6 or CDKN2A/B and PFS for first-line therapy (univariate analysis) (Table 4)
One-hundred-and-8nine patients were assessable for PFS for first-line therapy. (Twenty-one patients with hematological malignancy were not included for the assessment. The other 137 patients were not assessable because PFS for first-line therapy was not accurately available in the electronic medical record [48 patients] and 89 patients had not undergone first-line drug treatment). Among the 189 assessable patients, 22 percent of patients (n = 41) had aberrations in CDK elements. When median PFS for first-line therapy was compared between patients with an aberrant vs. normal CDK pathway, there was no statistically significant difference between the 2 groups (aberrant vs normal CDK pathway; 3.3 versus 5.0 months, p = 0.32) (Table 4). We also assessed whether different types of chemotherapy were associated with PFS in first line. Only platinum-containing regimens had a trend for differences in outcome, with worse PFS (median = 3.5 months) in patients with abnormal CDK elements than in those with normal CDK elements (median PFS = 5 months) (p = 0.13) (Table 4).
Discussion
An aberration in the CDK pathway is an important feature of human cancer, and leads to G1 cell-cycle progression.1–3 In 347 patients with malignancies seen at our center, we found that 79 (23%) had aberrations in CDK4/6 or CDKN2A/B (Table 1 and Fig. 1). Aberrant CDK4/6 or CDKN2A/B was the second most common aberration after TP53. The frequency of abnormalities in our current study of diverse tumors is similar to previous reports where loss of p16 (CDKN2A) expression or strong expression of CDK4 evaluated by immunohistochemistry was seen in 33.9% and 15.3%, respectively, of epithelial ovarian cancer.8 The frequency of CDK4/6 or CDKN2A/B varied by disease, with glioblastoma having the highest frequency, and with no CDK abnormalities found in 26 patients with colorectal cancer (Table 1).
As mentioned, CDK4/6 or CDKN2A/B aberrations were significantly associated with glioblastoma (OR: 11.2, 95% CI: 3.7–34.5, P < 0.0001) (Tables 1 and 3). Indeed, 21 of 26 (81%) patients had abnormalities in CDK elements, making perturbation of this pathway a hallmark of this disease. This result is consistent with previous investigations demonstrating that glioblastoma harbored deletion of CDKN2A in 31–68% of cases18–21 and amplification of CDK4 in 18–50% of cases.18,21 The higher percentages in our study may be due the fact that we examined multiple elements of the CDK pathway. Of clinical interest, in vitro and in vivo studies show that CDK 4/6 inhibition is capable of arresting the growth of glioblastoma.22 Currently phase 2 clinical trials are ongoing to determine the efficacy of the CDK 4/6 inhibitor, palbociclib, in patients with recurrent glioblastoma (NCT01227434).
In multivariate analysis, glioblastoma was independently associated with CDK 4/6 or CDKN2A/B abnormalities (Table 3). CDK4/6 or CDKN2A/B aberrations showed a trend (p = 0.057) toward a lower incidence in breast cancer (7 of 74 cases, 9.5%) (Tables 1 and 3). However, when CCND1 aberrations were included (CCND 1 also being a CDK pathway component), that trend disappeared (18 of 74 patients, 24.3 % positive for CDK4/6, CDKN2A/B or CCND 1 aberrations; p = 0.43). In patients with breast cancer (estrogen or progesterone receptor positivity and Her2 negativity) (n = 37), 12 (32.4%) had an aberration in CDK4/6, CDKN2A/B or CCND1. This frequency may explain the salutary effects in the study of the CDK inhibitor palbociclib plus letrozole vs. letrozole alone in PALOMA-1 trial in this subset of breast cancer (median PFS = 20.2 versus 10.2 months, p = 0.0004).12
Our analysis showed that CDK4/6 or CDKN2A/B aberrations were significantly associated with EGFR aberrations (OR: 11.9, 95% CI: 3.92–35.7, P < 0.0001) (Table 3). Although aberrant EGFR (usually EGFRvIII or EGFR amplification) was frequent in glioblastoma and often co-existed with CDK4/6 or CDKN2A/B (Table S2), the correlation between aberrant EGFR and abnormal CDK elements was found to be independent in multivariate assessment. Indeed, 8 of 14 patients (57%) who had EGFR aberrations with diagnoses other than brain tumors, including lung, breast, esophageal, tongue and appendiceal cancer, also harbored aberrant CDK elements.
There was also a statistically significant association between aberrant ARID1A, an epigenetic regulator that is known to be a tumor suppressor,23 and abnormal CDK elements. In a preclinical study, ARID1A has been shown to be essential for both the induction of p21 and the repression of E2F-responsive genes, which are critical for normal cell cycle arrest.24 Thus having both ARID1A and aberrant CDK elements likely contributes to enhancement of cell cycle progression. However, the clinical significance of and underlying mechanism leading to co-existing aberrations in CDK4/6 or CDKN2A/B and ARID1A are not clear and require further investigation.
In preclinical models, CDKN2A-mutant mice had accelerated tumorigenesis with increased incidence of metastases.25 Also, in patients with head and neck cancer, increased ratios of CCND1: CDKN2A evaluated by polymerase chain reaction was significantly associated with metastases.26 However, in our current study, there was no statistically significant difference in median time from diagnosis to metastasis among patients with aberrant vs. normal CDK elements (17.0 months versus 24.2 months; p = 0.20) (Table 4). Site of metastases were also similar between the 2 groups, except for the observation that metastases to the brain had a non-significant trend toward higher rates of abnormal CDK4/6, CDKN2A/B (9 of 34 [26.5%] brain metastasis cases had abnormal CDK4/6, CDKN2A/B; p = 0.16) (Table 1). Of possible interest in this regard, it has been suggested that CDKN2A is involved in brain development.27 Whether or not the high incidence of CDK abnormalities in glioblastomas and the trend toward brain metastases in their presence has a clinically meaningful connection or implication for understanding tumor migration to the central nervous system would require additional investigation. It is also interesting to note that the frequency of CDK4/6 or CDKN2A/B aberrations was similar regardless of the site of biopsies obtained for molecular analysis (18.4% from primary vs. 17.4% from metastatic samples; p = 0.83) (Table 1). Although conclusion cannot be made from current study, this observation implies that CDK4/6 or CKDN2A/B alterations may be an early event in carcinogenesis.
We also examined association between abnormal CDK elements and PFS on various front-line therapies. We observed a trend toward worse PFS for platinum-containing regimens in CDK-aberrant patients (median = 3.5 months versus 5.0 months; p = 0.13) (Table 4). Interestingly, Kusume et al8 also reported that individuals with epithelial ovarian cancer and an abnormal CDK pathway (p16 [CDKN2A], pRb, CCND1 and CDK4 evaluated by immunohistochemistry) had a trend toward lower complete response rates (55%) compared to those with a normal G1 pathway (81%) (P = 0.1) when treated with surgery followed by platinum based chemotherapy.8 The mechanism by which an aberrant CDK pathway might lead to platinum resistance could be by enabling cancer cells to overcome cell cycle arrest, a generally accepted consequence of DNA damage by platinum agents, thus leading to cell survival.28 Larger, controlled studies would be needed to determine if this relationship to platinum effect is significant.
An altered CDK pathway has also been associated with poorer clinical outcome in multiple cancer types.7–10 Our current study shows that, across diverse cancers, in multivariate analysis, independent factors associated with worse survival in our patients included aberrant TP53 or PTEN (Table 5). CDK4/6 or CDKN2A/B abnormalities showed a trend to correlate with poorer survival, HR = 1.67; 95% CI 0.92–3.04; p = 0.09).
Our study has several limitations. First, it was performed retrospectively in a single institution with a relatively limited number of patients. The small numbers in certain histologies precludes definitive conclusions. Second, multiple comparisons could result in overcalling the implications of positive P values. Third, we included heterogeneous cancers. However, the latter could suggest that the conclusions are generalizable across different histologies. Fourth, molecular analysis was performed on archival tumor tissue, which was obtained at a different time points in relationship to the clinical history. Finally, CDKN2A/B (p16) changes in blood from breast cancer patients undergoing chemotherapy suggests the possibility that treatment could contribute to observed changes in tumor specimens obtained later in the course of disease.29
In conclusion, we have demonstrated that aberrant CDK4/6 or CDKN2A/B are common in various cancer types (79 of 347, 22.8%) (Table 1 and Fig. 1), being second in frequency to only p53 mutations (44.1%). The rate of occurrence of CDK abnormalities varied by type of cancer, with no CDK aberrations seen in colorectal cancer. In contrast, 81% of patients with glioblastomas harbored CDK alterations. CDK4/6 or CDKN2A/B aberrations were associated with poor overall survival in univariate analysis, and a trend was seen in multivariate analysis (Table 5). There was also a trend toward worse PFS with platinum-containing regimens, but not with other chemotherapies, in patients with abnormal CDK elements (Table 4). Targeting specific molecular aberrations in cancer has shown remarkable effects in some patients with advanced malignancies.30–34 Currently, several CDK4/6 inhibitors are in clinical trials, and studies that select patients with aberrant CDK elements for such agents may be warranted. Of interest, aberrant CDK elements were independently associated with abnormalities in EGFR and ARID1A, suggesting that some individuals might need appropriately targeted combination therapy.
Patients and Methods
Patients
We investigated the CDK4/6, CDKN2A/B aberration status of patients with diverse malignancies that were evaluated at UC San Diego Moores Cancer Center from October 2012 through February 2014. To identify the associations between CDK4/6, CDKN2A/B aberration, cancer behavior and response to therapy, we retrospectively reviewed and analyzed the patient demographics, clinical characteristics, response to therapy, time to metastasis and overall survival. This study was performed and patient consents obtained in accordance with UCSD Institutional Review Board guidelines.
Tissue samples and mutational analysis
Available tissue from diagnostic and therapeutic procedures was used to assess molecular aberrations. All histologies were reviewed at Moores Cancer Center. Samples from formalin-fixed paraffin-embedded tissue were sent for targeted next generation sequencing (NGS) at Foundation Medicine (Cambridge, MA). The test sequences the entire coding sequence of 182 (N = 9) or more recently 236 (N = 338 patients) cancer-related genes plus 47 introns from 19 genes often rearranged or altered in cancer to an average depth-of-coverage of greater than 250× (http://foundationone.com/docs/FoundationOne_tech-info-and-overview.pdf).
Endpoints and statistical methods
Descriptive statistics were used to summarize the baseline patient characteristics. The Fisher's exact test was used to assess the association between categorical variables. Time to metastasis was measured from the date of diagnosis to the first date of metastasis reported. Progression-free survival (PFS) was defined as time interval between the start of therapy and the date of disease progression or removal of therapy for any reason, whichever occurred first. Overall survival (OS) was defined as time from diagnosis to last follow up or death. Patients with ongoing therapy without progression and alive at the last follow-up date were censored for progression-free survival. Patients alive at last follow up were censored for survival. The Kaplan-Meier method was used to estimate the probabilities of time to metastasis and PFS. Log-rank test and Cox regression analysis were used to compare subgroups of patients. All tests were 2-sided and P < 0.1 was included for multivariate analysis. P values < 0.05 were considered significant. Statistical analyses were carried out using GraphPad Prism version 6.0 (San Diego, CA, USA) and SPSS version 22.0 (Chicago, IL, USA).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
Funded in part by the Joan and Irwin Jacobs Fund and MyAnswerToCancer philanthropic fund.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 2. Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 2009; 9:153-66; PMID:19238148; http://dx.doi.org/ 10.1038/nrc2602 [DOI] [PubMed] [Google Scholar]
- 3. Rocca A, Farolfi A, Bravaccini S, Schirone A, Amadori D. Palbociclib (PD 0332991): targeting the cell cycle machinery in breast cancer. Expert Opin Pharmacother 2014; 15:407-20; PMID:24369047; http://dx.doi.org/ 10.1517/14656566.2014.870555 [DOI] [PubMed] [Google Scholar]
- 4. Schwaederle M, Daniels GA, Piccioni DE, Fanta PT, Schwab RB, Shimabukuro KA, Parker BA, Kurzrock R. Cyclin alterations in diverse cancers: Outcome and co-amplification network. Oncotarget 2015; 6(5):3033-42; PMID:25596748.16603719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shapiro GI. Cyclin-dependent kinase pathways as targets for cancer treatment. J Clin Oncol 2006; 24:1770-83; PMID:16603719; http://dx.doi.org/ 10.1200/JCO.2005.03.7689 [DOI] [PubMed] [Google Scholar]
- 6. Sheppard KE, McArthur GA. The cell-cycle regulator CDK4: an emerging therapeutic target in melanoma. Clin Cancer Res 2013; 19:5320-8; PMID:24089445; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-0259 [DOI] [PubMed] [Google Scholar]
- 7. Iacobucci I, Ferrari A, Lonetti A, Papayannidis C, Paoloni F, Trino S, Storlazzi CT, Ottaviani E, Cattina F, Impera L, et al. CDKN2A/B alterations impair prognosis in adult BCR-ABL1-positive acute lymphoblastic leukemia patients. Clin Cancer Res 2011; 17:7413-23; PMID:22134481; http://dx.doi.org/ 10.1158/1078-0432.CCR-11-1227 [DOI] [PubMed] [Google Scholar]
- 8. Kusume T, Tsuda H, Kawabata M, Inoue T, Umesaki N, Suzuki T, Yamamoto K. The p16-cyclin D1/CDK4-pRb pathway and clinical outcome in epithelial ovarian cancer. Clin Cancer Res 1999; 5:4152-7; PMID:10632354 [PubMed] [Google Scholar]
- 9. Xing X, Cai W, Shi H, Wang Y, Li M, Jiao J, Chen M. The prognostic value of CDKN2A hypermethylation in colorectal cancer: a meta-analysis. Br J Cancer 2013; 108:2542-8; PMID:23703248; http://dx.doi.org/ 10.1038/bjc.2013.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mendrzyk F, Radlwimmer B, Joos S, Kokocinski F, Benner A, Stange DE, Neben K, Fiegler H, Carter NP, Reifenberger G, et al. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol 2005; 23:8853-62; PMID:16314645; http://dx.doi.org/ 10.1200/JCO.2005.02.8589 [DOI] [PubMed] [Google Scholar]
- 11. Peurala E, Koivunen P, Haapasaari KM, Bloigu R, Jukkola-Vuorinen A. The prognostic significance and value of cyclin D1, CDK4 and p16 in human breast cancer. Breast Cancer Res 2013; 15:R5; PMID:23336272; http://dx.doi.org/ 10.1186/bcr3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finn RS, Crown JP, Lang I, Boer K, Bondarenko IM, Kulyk SO, Ettl J, Patel R, Pinter T, Schmidt M, et al. Final results of a randomized phase II study of PD 0332991, a cyclin-dependent kinase (CDK)-4/6 inhibitor, in combination with letrozole vs letrozole alone for first line treatment of ER+/HER2- advanced breast cancer (PALOMA-1; TRIO-18). 105th Annual Meeting of the American Association for Cancer Research; 2014 Apr 5-9; San Diego, CA. AACR Annual Meeting Abstract # CT101
- 13. Infante JR, Shapiro G, Witteveen P, Gerecitano JF, Ribrag V, Chugh R, Issa I, Chakraborty A, Matano A, Zhao X. A phase I study of the single-agent CDK4/6 inhibitor LEE011 in pts with advanced solid tumors and lymphomas. ASCO Annual Meeting Proceedings, 2014:2528
- 14. Mita MM, Joy AA, Mita A, Sankhala K, Jou YM, Zhang D, Statkevich P, Zhu Y, Yao SL, Small K, et al. Randomized phase II trial of the cyclin-dependent kinase inhibitor dinaciclib (MK-7965) versus capecitabine in patients with advanced breast cancer. Clin Breast Cancer 2014; 14:169-76; PMID:24393852; http://dx.doi.org/ 10.1016/j.clbc.2013.10.016 [DOI] [PubMed] [Google Scholar]
- 15. Flaherty KT, Lorusso PM, Demichele A, Abramson VG, Courtney R, Randolph SS, Shaik MN, Wilner KD, O'Dwyer PJ, Schwartz GK. Phase I, dose-escalation trial of the oral cyclin-dependent kinase 4/6 inhibitor PD 0332991, administered using a 21-day schedule in patients with advanced cancer. Clin Cancer Res 2012; 18:568-76; PMID:22090362; http://dx.doi.org/ 10.1158/1078-0432.CCR-11-0509 [DOI] [PubMed] [Google Scholar]
- 16. Leonard JP, LaCasce AS, Smith MR, Noy A, Chirieac LR, Rodig SJ, Yu JQ, Vallabhajosula S, Schoder H, English P, et al. Selective CDK4/6 inhibition with tumor responses by PD0332991 in patients with mantle cell lymphoma. Blood 2012; 119:4597-607; PMID:22383795; http://dx.doi.org/ 10.1182/blood-2011-10-388298 [DOI] [PubMed] [Google Scholar]
- 17. Schwartz GK, LoRusso PM, Dickson MA, Randolph SS, Shaik MN, Wilner KD, Courtney R, O'Dwyer PJ. Phase I study of PD 0332991, a cyclin-dependent kinase inhibitor, administered in 3-week cycles (Schedule 2/1). Br J Cancer 2011; 104:1862-8; PMID:21610706; http://dx.doi.org/ 10.1038/bjc.2011.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cancer Genome Atlas Research N . Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455:1061-8; PMID:18772890; http://dx.doi.org/ 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jen J, Harper JW, Bigner SH, Bigner DD, Papadopoulos N, Markowitz S, Willson JK, Kinzler KW, Vogelstein B. Deletion of p16 and p15 genes in brain tumors. Cancer Res 1994; 54:6353-8; PMID:7987828 [PubMed] [Google Scholar]
- 20. Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL, Burkhard C, Schuler D, Probst-Hensch NM, Maiorka PC, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res 2004; 64:6892-9; PMID:15466178; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-1337 [DOI] [PubMed] [Google Scholar]
- 21. Schmidt EE, Ichimura K, Reifenberger G, Collins VP. CDKN2 (p16/MTS1) gene deletion or CDK4 amplification occurs in the majority of glioblastomas. Cancer Res 1994; 54:6321-4; PMID:7987821 [PubMed] [Google Scholar]
- 22. Michaud K, Solomon DA, Oermann E, Kim JS, Zhong WZ, Prados MD, Ozawa T, James CD, Waldman T. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res 2010; 70:3228-38; PMID:20354191; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer Dis 2013; 3:35-43; PMID:23208470; http://dx.doi.org/ 10.1158/2159-8290.CD-12-0361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nagl NG, Jr., Patsialou A, Haines DS, Dallas PB, Beck GR, Jr., Moran E. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res 2005; 65:9236-44; PMID:16230384; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-1225 [DOI] [PubMed] [Google Scholar]
- 25. Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 2001; 413:83-6; PMID:11544530; http://dx.doi.org/ 10.1038/35092584 [DOI] [PubMed] [Google Scholar]
- 26. Akervall J, Bockmuhl U, Petersen I, Yang K, Carey TE, Kurnit DM. The gene ratios c-MYC:cyclin-dependent kinase (CDK)N2A and CCND1:CDKN2A correlate with poor prognosis in squamous cell carcinoma of the head and neck. Clin Cancer Res 2003; 9:1750-5; PMID:12738730 [PubMed] [Google Scholar]
- 27. Kawauchi T, Shikanai M, Kosodo Y. Extra-cell cycle regulatory functions of cyclin-dependent kinases (CDK) and CDK inhibitor proteins contribute to brain development and neurological disorders. Genes Cell 2013; 18:176-94; http://dx.doi.org/ 10.1111/gtc.12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 2003; 22:7265-79; PMID:14576837; http://dx.doi.org/ 10.1038/sj.onc.1206933 [DOI] [PubMed] [Google Scholar]
- 29. Sanoff HK, Deal AM, Krishnamurthy J, Torrice C, Dillon P, Sorrentino J, Ibrahim JG, Jolly TA, Williams G, Carey LA, et al. Effect of cytotoxic chemotherapy on markers of molecular age in patients with breast cancer. J Natl Cancer Inst 2014; 106:dju057; PMID:24681605; http://dx.doi.org/ 10.1093/jnci/dju057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Janku F, Hong DS, Fu S, Piha-Paul SA, Naing A, Falchook GS, Tsimberidou AM, Stepanek VM, Moulder SL, Lee JJ, et al. Assessing PIK3CA and PTEN in early-phase trials with PI3K/AKT/mTOR inhibitors. Cell Rep 2014; 6:377-87; PMID:24440717; http://dx.doi.org/ 10.1016/j.celrep.2013.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Janku F, Wheler JJ, Naing A, Stepanek VM, Falchook GS, Fu S, Garrido-Laguna I, Tsimberidou AM, Piha-Paul SA, Moulder SL, et al. PIK3CA mutations in advanced cancers: characteristics and outcomes. Oncotarget 2012; 3:1566-75; PMID:23248156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsimberidou AM, Iskander NG, Hong DS, Wheler JJ, Falchook GS, Fu S, Piha-Paul S, Naing A, Janku F, Luthra R, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res 2012; 18:6373-83; PMID:22966018; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wheler J, Falchook G, Tsimberidou AM, Hong D, Naing A, Piha-Paul S, Chen SS, Heymach J, Fu S, Stephen B, et al. Revisiting clinical trials using EGFR inhibitor-based regimens in patients with advanced non-small cell lung cancer: a retrospective analysis of an MD Anderson Cancer Center phase I population. Oncotarget 2013; 4:772-84; PMID:23800712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wheler JJ, Falchook GS, Tsimberidou AM, Hong DS, Naing A, Piha-Paul SA, Chen SS, Fu S, Stephen B, Fok JY, et al. Aberrations in the epidermal growth factor receptor gene in 958 patients with diverse advanced tumors: implications for therapy. Ann Oncol 2013; 24:838-42; PMID:23139256; http://dx.doi.org/ 10.1093/annonc/mds524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


