Abstract
Mitochondria are key players in aging and cell death. It has been suggested that mitochondrial fragmentation, mediated by the Dnm1/Fis1 organelle fission machinery, stimulates aging and cell death. This was based on the observation that Saccharomyces cerevisiae Δdnm1 and Δfis1 mutants show an enhanced lifespan and increased resistance to cell death inducers. However, the Dnm1/Fis1 fission machinery is also required for peroxisome division. Here we analyzed the significance of peroxisome fission in yeast chronological lifespan, using yeast strains in which fission of mitochondria was selectively blocked. Our data indicate that the lifespan extension caused by deletion of FIS1 is mainly due to a defect in peroxisome fission and not caused by a block in mitochondrial fragmentation. These observations are underlined by our observation that deletion of FIS1 does not lead to lifespan extension in yeast peroxisome deficient mutant cells.
Keywords: chronological aging, Fis1, fission, mitochondria, peroxisome, yeast
Introduction
Cells continuously have to cope with factors that affect their viability. Important threats are reactive oxygen species (ROS), which can damage important macromolecules such as proteins, lipids and DNA. Intracellular accumulation of damaged components contributes to aging, a process that can be defined as the deterioration of cells in time, accompanied by a loss of viability. However, ROS can also have positive effects by acting as second messengers that activate pathways aimed at saving the cell from demise.1
Eukaryotic cells contain two types of organelles that produce ROS as byproduct of oxidative metabolism, namely mitochondria and peroxisomes. It is generally accepted that the mitochondrion is a key player in aging.2,3 Recent data however indicate that peroxisomes play a role in this process as well.4-7
In respiratory active yeast cells mitochondria form a large network of tubular organelles. However, during aging this network fragments resulting in the formation of multiple spherical organelles.8 In Saccharomyces cerevisiae this process depends on the organelle fission machinery consisting of the GTPase Dnm1, the tail-anchored mitochondrial outer membrane protein Fis1 and the accessory proteins Mdv1 and Caf4. It has been suggested that mitochondrial fragmentation contributes to aging, because deletion of genes encoding proteins of the mitochondrial fission machinery (DNM1, FIS1 or MDV1) enhanced the cellular lifespan.8,9
However, the Dnm1/Fis1 machinery is not only responsible for mitochondrial fragmentation, but also involved in peroxisome fission.10,11 So far, the effect of DNM1, FIS1 or MDV1 deletion on yeast aging were supposed to be the consequence of a defect in mitochondrial fission, but a possible role for peroxisomal fission in aging has not been investigated to date.
Here we reanalyzed the role of the Fis1/Dnm1 organelle fission machinery in S. cerevisiae chronological aging focusing on a possible contribution of peroxisome fission to the reported effects. Our data indicate that a defect in peroxisome fission is the major cause of yeast lifespan extension caused by the absence of the Fis1/Dnm1 machinery.
Results
Construction of strains specifically affected in mitochondrial fission
Because in S. cerevisiae two dynamin-like proteins, Vps1 and Dnm1, are involved in peroxisome fission,11 this process is more severely blocked in a Δvps1Δdnm1 double deletion strain relative to Δvps1 and Δdnm1 single deletion strains.10 Dnm1 is only involved in mitochondrial fission, therefore deletion of VPS1 does not affect this process (Fig. 3A, compare Fig. S1B). In order to be able to selectively assess the role of the Fis1/Dnm1-containing fission machinery in yeast chronological aging, we performed all experiments in a Δvps1 background. Because Hughes and Gottschling recently showed that vacuolar defects may affect yeast mitochondrial function and lifespan,12 we first examined the effect of deletion of VPS1 on the chronological lifespan yeast. As shown in Fig. S1A, the chronological lifespan (CLS) of Δvps1 cells did not significantly differ from that of wild-type (WT) cells. Also, Δvps1 cells were capable to grow on the non-fermentable carbon source glycerol, indicating that mitochondrial function is not strongly compromised (data not shown).
Figure 3.
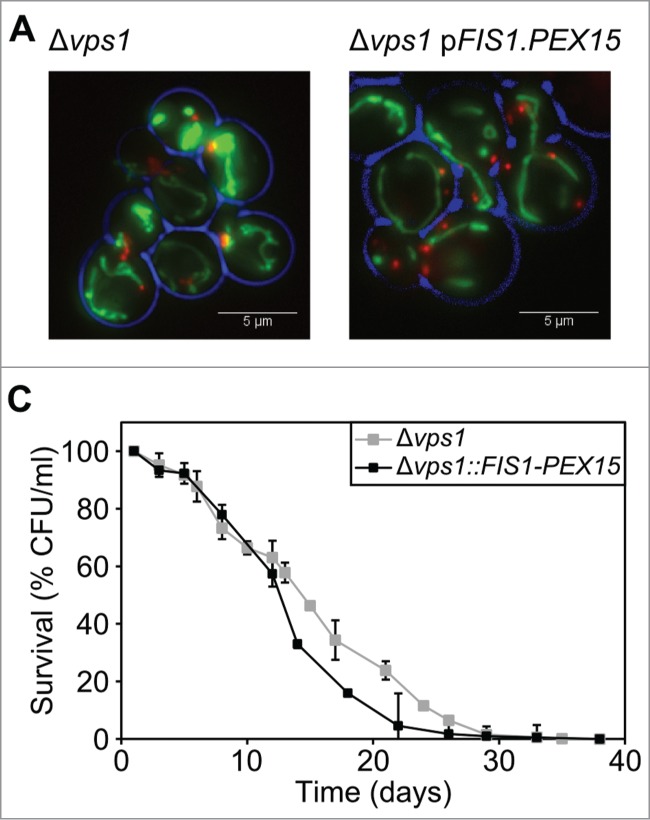
Increased peroxisome fission does not affect the CLS. (A) Mitochondrial and peroxisomal morphology in Δvps1 and Δvps1 FIS1-PEX15 cells. Peroxisomes were labeled with DsRED-SKL and mitochondria by mitoGFP. (B) Chronological lifespan analysis of Δvps1 and Δvps1 FIS1-PEX15 cells. Data represent mean ± SEM from at least 2 experiments.
Previous reports indicated that deletion of FIS1 in S. cerevisiae can result in the acquisition of a secondary mutation in the stress-response gene WHI2.13 To exclude the occurrence of such secondary mutations, all Δfis1 strains used in this study were checked for the absence of mutations in WHI2.
In line with our previous observations,10 fluorescence microscopy analysis of Δvps1Δfis1 cells revealed the presence of very low peroxisome numbers relative to Δvps1 control cells (Fig. 1A,Table 1). In addition, these cells harbor a collapsed mitochondrial network, which is characteristic for mutants defective in mitochondrial fission (Fig. 1A). As expected, upon reintroduction of FIS1 in Δvps1Δfis1 cells (strain Δvps1Δfis1::FIS1), the WT mitochondrial morphology was restored and the number of peroxisomes increased to those observed in Δvps1 cells (Table 1, Fig. 1B).
Figure 1.
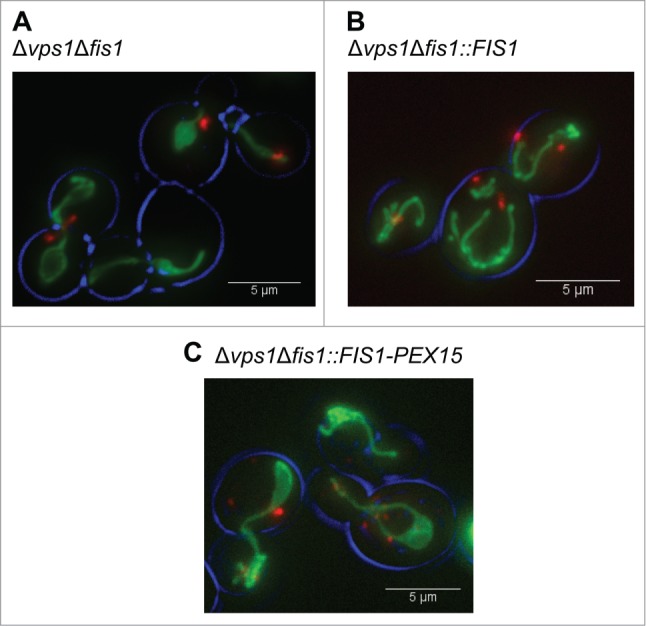
Peroxisome and mitochondrial fission defects in various yeast mutant strains. Fluorescence microscopy images showing mitochondrial and peroxisome morphology in Δvps1Δfis1 (A), Δvps1Δfis1::FIS1 (B) and Δvps1Δfis1::FIS1-PEX15 (C) cells. Cells were grown until the mid-exponential growth phase on MM containing 2% glucose. Peroxisomes are marked by DsRED-SKL and mitochondria by mitoGFP.
Table 1.
Summary of the results of the fluorescence microscopy analyses.
| Strain | mitochondria morphology | peroxisome number* |
|---|---|---|
| Δvps1 | Normal | 1.30 ± 0.02 |
| Δvps1Δfis1 | Collapsed | 0.85 ± 0.05 |
| Δvps1Δfis1::fis1 | Normal | 1.27 ± 0.01 |
| Δvps1Δfis1::fis1-PEX15 | Collapsed | 3.32 ± 0.18 |
| Δvps1fis1-PEX15 | Normal | 2.90 ± 0.08 |
| Δpex3 | Normal | ND |
| Δpex3Δfis1 | Collapsed | ND |
Mean peroxisome numbers (± standard error of mean).
In order to assess the role of peroxisome fission in yeast chronological aging, we constructed a yeast strain that is selectively blocked in mitochondrial fission. Previously, Halbach et al. showed that a fusion protein consisting of the N-terminal, soluble domain of S. cerevisiae Fis1 and the C-terminal peroxisomal membrane anchor of Pex15 exclusively sorts to peroxisomes.14 Moreover, Motley et al. showed that this Fis1-Pex15 fusion protein is able to recruit the Dnm1 fission machinery to yeast peroxisomes.11 Using the identical construct we confirmed that upon introduction of this Fis1-Pex15 fusion protein in Δvps1Δfis1 cells (produced under control of the FIS1 promoter), the cells still showed collapsed mitochondria, indicative for a mitochondrial fission defect (Fig. 1C). However, the number of peroxisomes increased to an average of 3.3 per cell, indicating that peroxisome fission is not blocked anymore (Fig. 1C;Table 1). The enhanced peroxisome number in this strain relative to the Δvps1 control is in line with previous observations11 and most likely is due to the fact that the entire cellular Fis1 pool is localized to peroxisomes instead of being distributed over both peroxisomes and mitochondria.
The lifespan extension observed in Δfis1 cells is restored upon sorting of Fis1 to peroxisomes
Next, we compared the CLS of strains in which fission of mitochondria and peroxisomes was blocked (Δvps1Δfis1), unaffected (Δvps1 and Δvps1Δfis1::FIS1) or in which only mitochondrial fission was defective (Δvps1Δfis1::FIS1-PEX15). CLS experiments revealed no significant differences between the mean lifespans of these 4 strains (Fig. 2B). However, deletion of FIS1 in Δvps1 cells (Δvps1Δfis1) significantly increased the maximum lifespan relative to the Δvps1 control (p = 3.10−4; Fig. 2B;Table 2). Upon FIS1 reintroduction (Δvps1Δfis1::FIS1) this effect was abolished (Fig. 2A; Table 2).
Figure 2.
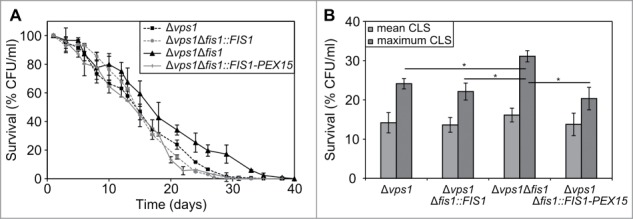
Exclusive sorting of Fis1 to peroxisomes does not result in lifespan extension. (A) Chronological lifespans of Δvps1, Δvps1Δfis1, Δvps1Δfis1::FIS1 and Δvps1Δfis1::FIS1-PEX15 cells. Data represent mean ± SEM from at least 2 experiments. (B) Statistical analysis for mean and maximum lifespans of strains presented in panel A. *, p < 0.001.
Table 2.
Mean and maximal lifespans.
| Strain | Mean CLS (days) | Maximum CLS (days) |
|---|---|---|
| Δvps1 | 14.2 ± 1.45 | 24.4 ± 1.06 |
| Δvps1Δfis1 | 16.0 ± 1.22 | 31.1 ± 0.36 |
| Δvps1Δfis1::FIS1 | 13.6 ± 1.34 | 22.1 ± 1.52 |
| Δvps1Δfis1::fis1-PEX15 | 13.9 ± 1.35 | 20.3 ± 1.91 |
| Δvps1fis1-PEX15 | 12.3 ± 0.35 | 23.0 ± 2.83 |
| Δpex3 | 9.9 ± 0.18 | 16.0 ± 0.00 |
| Δpex3Δfis1 | 9.6 ± 0.88 | 16.0 ± 0.00 |
The mean chronological lifespan is defined as the time point where 50% of the cells are viable. The maximum lifespan is the time point where 10% of the cells are viable. Mean values (± standard error of mean) are presented of at least 2 independent cultures.
Interestingly, no statistically different changes in mean or maximum lifespans were observed when mitochondrial fission was selectively blocked (Δvps1 versus Δvps1Δfis1::FIS1-PEX15 cells) (mean CLS, p = 0.829; maximum CSL, p = 0.064), whereas the maximum CLS of Δvps1Δfis1 was significantly higher than that of Δvps1Δfis1::FIS1-PEX15 cells (mean CLS, p = 0.208; maximum CLS, p = 0.001) (Fig. 2; Table 2). These data suggest that the extension of the maximum lifespan caused by FIS1 deletion is mainly caused by a block in peroxisome fission (Fig. 2B; Table 2).
Enhanced levels of Fis1 on the peroxisomal membrane do not negatively affect the CLS
The number of peroxisomes is enhanced in Δvps1Δfis1::FIS1-PEX15 relative to Δvps1Δfis1::FIS1 (Fig. 1B,Table 1),11 most likely due to increased levels of Fis1 protein at the peroxisomal membrane. In order to test whether this influences the CLS, we expressed FIS1-PEX15 in Δvps1 cells and compared the CLS with Δvps1 control cells. Indeed, peroxisome numbers increased (2.9 per cell in Δvps FIS1-PEX15 relative to 1.3 in Δvps1 cells; Fig. 3A; Table 1) to a value similar as observed in Δvps1Δfis1::FIS1-PEX15 cells (3.3; Table 1). CLS analysis revealed no significant changes in mean or maximum lifespan (mean CLS, p = 0.129; maximum CLS, p = 0.215) (Fig. 3B; Table 2), indicating that enhanced peroxisomal Fis1 levels or fission do not affect yeast CLS.
Deletion of FIS1 has no impact on lifespan in Δpex3 cells
To further substantiate that a block in mitochondrial fission does not result in an increased CLS, we deleted FIS1 in Δpex3 cells. Pex3 is a peroxisomal membrane protein that is essential for peroxisome biogenesis. Δpex3 cells fully lack normal peroxisomal structures and all peroxisomal matrix proteins are mislocalized to the cytosol.15
Fluorescence microscopy analysis revealed that Δpex3 cells show normal branched mitochondria, which collapse upon deletion of FIS1 (Fig. 4A; Table 1). As expected, the peroxisomal matrix marker DsRed-SKL is mislocalized to the cytosol in both Δpex3 and Δpex3Δfis1 cells (Fig. 4A). Chronological aging experiments revealed that both strains show similar mean and maximum lifespans (Fig 4B; Table 2). This observation indicates that FIS1 deletion only results in a decreased CLS when peroxisome are present, demonstrating that blocking mitochondrial fission does not increase the CLS in S. cerevisiae.
Figure 4.
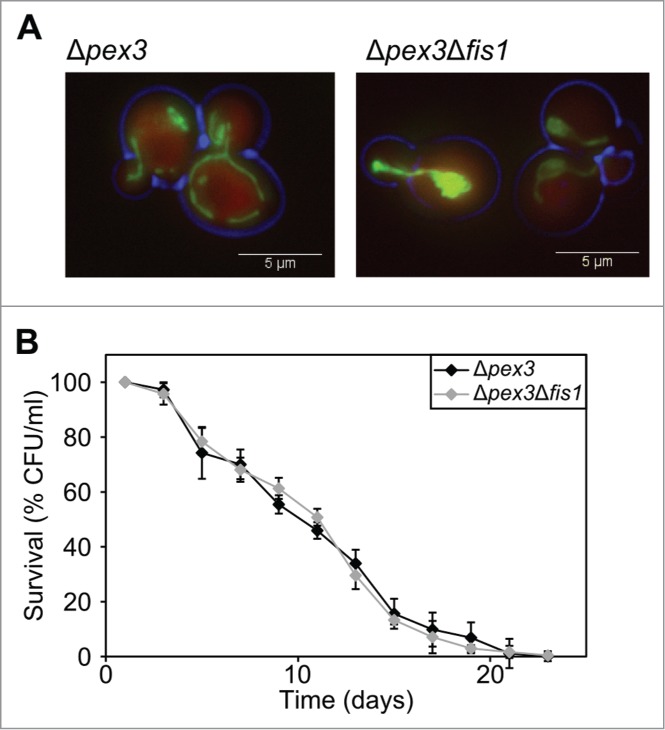
FIS1 deletion in Δpex3 cells has no effect on the CLS. (A) Fluorescence microscopy of Δpex3 and Δpex3Δfis1 cells producing DsRED-SKL or mitoGFP. (B). Chronological lifespan experiment of Δpex3 and Δpex3Δfis1 cells. Data represent mean ± SEM from at least 2 experiments.
Discussion
Here we show that a block in peroxisome fission, but not a block in mitochondrial fission, results in an increased chronological lifespan of S. cerevisiae.
First, we showed that, like reported before for WT cells, deletion of FIS1 in a Δvps1 background results in an increase in chronological lifespan.9 This allowed us to specifically study the role of Fis1/Dnm1 mediated peroxisome and mitochondrial fission in yeast aging. In Δfis1 Δvps1 cells both peroxisome and mitochondrial fission are blocked. By re-introducing a Fis1 variant that is selectively targeted to peroxisomes, we were able to create a strain that was selectively blocked in mitochondrial fragmentation, whereas peroxisomes still divided. Because in these cells the CLS was not extended anymore, we conclude that blocking peroxisomes fission, but not mitochondrial fission increases yeast CLS. Our observation that deletion of FIS1 does not increase the lifespan of cells lacking peroxisomes (Δpex3) supports the conclusion that the positive effect of FIS1 deletion on yeast CLS is mainly related to peroxisomes and not to mitochondria.
Our data reveal that only the maximum CLS and not the mean CLS is enhanced by blocking peroxisome fission. This suggests that peroxisome fission has a pro-aging effect mainly at later stages of chronological aging. We speculate that this may be related to the block in pexophagy which accompanies peroxisome fission defects.16-18 We previously showed that peroxisomal β-oxidation is important for energy supply during yeast chronological aging,15 because a reduction in β-oxidation caused by either the absence of an enzyme of this pathway or a defect in peroxisome biogenesis resulted in a decrease in chronological lifespan. Conversely a defect in pexophagy is likely to result in enhanced peroxisomal β-oxidation and thus a higher capacity of the cells to generate energy during chronological aging especially at the later stages.
An alternative explanation may be related to the effect of peroxisome fission on ROS homeostasis. Peroxisomes play an important role in ROS homeostasis and oxidative stress. This is illustrated by the observation that deletion of S. cerevisiae PEX6, a crucial gene for peroxisome biogenesis, resulted in enhanced ROS levels during chronological aging ultimately leading to necrotic cell death.19 Also, the absence of the peroxisomal peroxiredoxin PMP20 in the yeast Hansenula polymorpha caused increased oxidative stress and necrotic cell death.20 These results suggest that intact peroxisomes containing sufficient levels of peroxisomal antioxidant enzymes may prevent necrosis. The block in pexophagy and/or the presence of enlarged peroxisomes in Δvps1Δfis1 cells may result in altered ROS homeostasis, thereby reducing the induction of necrosis and an extension of the CLS in Δvps1Δfis1 cells.
Mitochondrial fragmentation has been suggested to be important for selective mitochondrial autophagy (mitophagy). Studies in mammals suggested that fission, fusion and autophagic degradation of mitochondria are important processes to prevent accumulation of dysfunctional mitochondria.21-24 According to this model mitochondrial fission results in uneven daughter mitochondria thereby separating dysfunctional and functional parts. Subsequently, the dysfunctional organelles are degraded by autophagy whereas the functional ones fuse to form healthy mitochondrial networks. If this model is correct, one would assume that in mammalian cells a block in mitochondrial fission (by DNM1 or FIS1 deletion) results in the accumulation of dysfunctional organelles leading to a decreased instead of an enhanced lifespan. In yeast this issue is still controversial, because data have been presented indicating that mitochondrial fission is important for autophagy,25 whereas other studies suggest that fission is not required for mitophagy.26
Summarizing our data indicate that a block in peroxisome fission, but not in mitochondrial fission, enhances yeast chronological lifespan.
Materials and Methods
Strains and growth conditions
The S. cerevisiae strains, all derived from wild-type BY4742, are listed in Table 3. We sequenced the WHI2 gene in all strains and found no mutations (data not shown). Cells were grown in batch cultures at 30°C, 200 rpm, on mineral media (MM)27 containing 0.25% ammonium sulfate, 0.05% yeast extract and 2% or 0.5% glucose. MM was supplemented with the required amino acids to a final concentration of 20 μg/ml (histidine) or 30 μg/ml (leucine, lysine, and uracil). For growth on agar plates YPD medium (1% yeast extract, 1% peptone, 1% glucose) was supplemented with 2% agar. Escherichia coli DH5α was used for plasmid constructions and cultured at 37°C on LB medium (1% trypton, 0.5% yeast extract, 0.5% NaCl) supplemented with 100 μg/ml ampicillin, when required.
Table 3.
Yeast strains used in this study.
| Strains | Genotype | Reference |
|---|---|---|
| BY4742 WT | MATα his3A1 leu2A0 lys2A0 ura3A0 | Euroscarf |
| Δvps1 | vps1::KanB | 10 |
| Δvps1Δfis1 | vps1::loxP fis1::loxP | 10 |
| Δvps1Δfis1::FIS1 | vps1::loxP fis1::FIS1 | this study |
| Δvps1Δfis1::FIS1-PEX15 | vps1::loxP fis1::FIS1-PEX15 | this study |
| Δvps1 FIS1-PEX15 | vps1::KanB FIS1/FIS1-PEX15 | this study |
| Δpex3 | pex3::KanMX4 | Euroscarf |
| Δpex3Δfis1 | pex3::KanMX4 fis1::NatMX4 | this study |
Construction of strains
Plasmids and primers used in this study are listed in Tables 4 and 5. Yeast were transformed according to the lithium acetate transformation protocol described by Ito et al. 198327 and modified by Hill et al. 1991 and Gietz et al. 1992.28,29
Table 4.
Plasmids used in this study.
| Plasmid | Description | Reference |
|---|---|---|
| mitoGFP* | pVT100U-mtGFP*, PADH1-Su9(1–69)DsRed.T1/URA3, 2μ | 30 |
| mitoDsRED* | pVT100U-mtDsRED, PADH1-Su9(1–69)DsRed.T1/URA3, 2μ | 30 |
| DsRED-SKL | pUG34-DsREDskl, PMET25-DsRED-T1.SKL/HIS3, 2μ | 10 |
| pEH107** | PFIS1-FIS1-TFIS1/URA3, 2μ** | 11 |
| pEH111.2** | PFIS1-FIS1-PEX15-TPEX15/URA3, 2μ** | 11 |
| pFIS1 | pSL35, PFIS1-FIS1-TFIS1/HghMX4, integrative | this study |
| pFIS1.PEX15 | pSL36, PFIS1-FIS1-PEX15-TPEX15/ HghMX4, integrative | this study |
gift from Dr. Jodi Nunnari (University of California, Davis, USA).
gift from Dr Alison M. Motley (University of Sheffield, Sheffield UK)
Table 5.
Primers used in this study.
| Primer | Sequence (5′−3′) |
|---|---|
| FIS1.A | GCATACAGTTCATCCCAGTATTTTT |
| FIS1.D | CATGCGTAGTTAAACCTTGACTGTA |
| FIS1up | CACATAGAAGCACAGATCAGAGCACAGCCATACAACATAAGTATGCTTAACTATGCGGCATCAGAG |
| FIS1dw | ATTCTTATGTATGTACGTATGTGCTGATTTTTTATGTGCTTGTTAGCGCCCAATACGCAAACC |
| FIS1.6 | CCGGCTTCCAGTCACCACTA |
| FIS1.9 | CGTCCGCGGTGGGTCTAAACCGTATGA |
| NATrev | GTAAGCCGTGTCGTCAAGAG |
| POT1.6 | AATTCAACGCGTCTGTGAGG |
| WHI2.1 | CGCAAGAAGACAACTCCTTCA |
| WHI2.2 | ACCGTTTTGCCAGTTTCTTG |
| WHI2.3 | AGGGGTCCAATTCTTCTTCAAAT |
| WHI2.4 | TGTGTCTTTGGCCCGATCT |
pEH107 (FIS1 ORF) and pEH111.2 (FIS1-PEX15 hybrid gene) are a gift from A. Motley.11 Both are 2μ plasmids allowing expression of either full-length FIS1 or the gene encoding the fusion protein Fis1-Pex15361–383 in which the C-terminal anchor domain of Fis1 is replaced by the one of Pex15.14 Both genes are under control of the FIS1 promoter. Because integrative plasmids are more accurate while investigating chronological aging, we subsequently cloned both constructs into an integrative plasmid containing an antibiotic resistance marker. pFIS1 (pSL35) and pFIS1-PEX15 (pSL36) were constructed by digestion of pEH107 and pEH111.2 with EcoRI/HindIII and cloned into pBluescript SK+. The hygromycin resistance gene from pAG3230 was then introduced using SpeI/BamHI. pSL35 and pSL36 were then linearized using BstBI and transformed into yeast cells allowing genomic integration into the FIS1 promoter. Positive clones were selected on YPD plates containing 300 μg/ml hygromycin B (Invitrogen). Successful integration by homologous recombination was checked using FIS1.6 + FIS1.D and POT1.6 + FIS1.9 primers (Table 5).
FIS1 deletion in Δpex3 cells was obtained by replacement of the FIS1 open reading frame by NatMX4 gene from pAG2531 using primers FIS1up and FIS1dw. For the selection of positive clones, YPD plates containing 100 μg/ml nourseothricin (Invitrogen, Carlsbad, CA, USA) was used. Correct integration was checked using Euroscarf primers FIS1.A and FIS1.D and the NATrev primer (Table 5).
Fluorescence microscopy
Cells from fresh plates were grown overnight on MM medium containing 2% glucose. Cultures were diluted at OD600 nm = 0.1 in fresh MM 2% glucose and grown for 4 hours before imaging. Fluorescence images were captured by an inverted microscope (Observer Z1; Carl Zeiss) using AxioVision software (Carl Zeiss) and a digital camera (CoolSNAP HQ2; Photometrics). All fluorescence images were acquired using a 100x 1.30 NA Plan-Neofluar objective (Carl Zeiss). GFP signal was visualized with a 470/40-nm band pass excitation filter, a 495-nm dichromatic mirror, and a 525/50-nm band pass emission filter. DsRed fluorescence was visualized with a 546/12-nm bandpass excitation filter, a 560-nm dichromatic mirror, and a 575–640-nm bandpass emission filter. Fluorescence image were analyzed using ImageJ software software (US National Institutes of Health, Bethesda, MD, USA). Pictures presented here are z-projection of stacks resulting in complete overview of organelles morphology/number. The number of peroxisomes per cell was calculated from 2 different clones grown at the same conditions and the results presented are mean values and standard error of mean.
Chronological lifespan measurements
Overnight cultures were grown in MM medium containing 0.5% glucose and diluted twice at OD600 nm = 0.1 in fresh MM and grown for 8 hours. After the last pre-cultivation step, cells were diluted in final fresh MM. Survival was assayed by determining the colony forming units (CFUs) after 2 d of incubation at 30°C on YPD agar plates. Twenty-four hours after the last dilution (D1) was considered as 100% of survival. The results shown are mean values and standard error of mean. At the end of each aging experiments, the WHI2 gene was amplified using primers WHI2.1 and WHI2.4 and sequenced with primers WHI2.2 and WHI2.3 to check appearance of a random mutation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1.Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Rad Biol Med 2010; 48:749–62; PMID:20045723; http://dx.doi.org/ 10.1016/j.freeradbiomed.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban RS, Nemoto S, Finkel T. Mitochondria, Oxidants, and Aging. Cell 2005; 120:483–95; PMID:15734681; http://dx.doi.org/ 10.1016/j.cell.2005.02.001 [DOI] [PubMed] [Google Scholar]
- 3.Bratic A, Larsson N-G. The role of mitochondria in aging. J Clin Invest 2013; 123:951–7; PMID:23454757; http://dx.doi.org/ 10.1172/JCI64125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beach A, Titorenko VI. Essential roles of peroxisomally produced and metabolized biomolecules in regulating yeast longevity. Subcell Biochem 2013; 69:153–67; PMID:23821148; http://dx.doi.org/ 10.1007/978-94-007-6889-5_9 [DOI] [PubMed] [Google Scholar]
- 5.Fransen M, Nordgren M, Wang B, Apanasets O, Van Veldhoven PP. Aging, age-related diseases and peroxisomes. Subcell Biochem 2013; 69:45–65; PMID:23821142; http://dx.doi.org/ 10.1007/978-94-007-6889-5_3 [DOI] [PubMed] [Google Scholar]
- 6.Kumar S, Kawałek A, van der Klei IJ. Peroxisomal quality control mechanisms. Curr Opin Microbiol 2014; 22:30–7; PMID:25305535; http://dx.doi.org/ 10.1016/j.mib.2014.09.009 [DOI] [PubMed] [Google Scholar]
- 7.Manivannan S, Scheckhuber CQ, Veenhuis M, van der Klei IJ. The impact of peroxisomes on cellular aging and death. Front Oncol 2012; 2(5); PMID:22662318; http://dx.doi.org/ 10.3389/fonc.2012.00050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nyström T, Osiewacz HD. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol 2007; 9:99–105; PMID:17173038; http://dx.doi.org/ 10.1038/ncb1524 [DOI] [PubMed] [Google Scholar]
- 9.Palermo V, Falcone C, Mazzoni C. Apoptosis and aging in mitochondrial morphology mutants of S. cerevisiae. Folia Microbiol (Praha) 2007; 52:479–83; PMID:18298044; http://dx.doi.org/ 10.1007/BF02932107 [DOI] [PubMed] [Google Scholar]
- 10.Kuravi K, Nagotu S, Krikken AM, Sjollema K, Deckers M, Erdmann R, Veenhuis M, van der Klei IJ. Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisae. J Cell Sci 2006; 119:3994–4001; PMID:16968746; http://dx.doi.org/ 10.1242/jcs.03166 [DOI] [PubMed] [Google Scholar]
- 11.Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci 2008; 121:1633–40; PMID:18445678; http://dx.doi.org/ 10.1242/jcs.026344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature 2012; 492:261–5; PMID:23172144; http://dx.doi.org/ 10.1038/nature11654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng WC, Teng X, Park HK, Tucker CM, Dunham MJ, Hardwick JM. Fis1 deficiency selects for compensatory mutations responsible for cell death and growth control defects. Cell Death Differ 2008; 15:1838–46; PMID:18756280; http://dx.doi.org/ 10.1038/cdd.2008.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbach A, Landgraf C, Lorenzen S, Rosenkranz K, Volkmer-Engert R, Erdmann R, Rottensteiner H. Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J Cell Sci 2006; 119:2508–17; PMID:16763195; http://dx.doi.org/ 10.1242/jcs.02979 [DOI] [PubMed] [Google Scholar]
- 15.Lefevre SD, van Roermund CW, Wanders RJA, Veenhuis M, van der Klei IJ. The significance of peroxisome function in chronological aging of Saccharomyces cerevisiae. Aging Cell 2013; 12:784–93; PMID:23755917; http://dx.doi.org/ 10.1111/acel.12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao K, Liu X, Feng Y, Klionsky DJ. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy 2014; 10:652–61; PMID:24451165; http://dx.doi.org/ 10.4161/auto.27852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nowikovsky K, Reipert S, Devenish RJ, Schweyen RJ. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ 2007; 14:1647–56; PMID:17541427; http://dx.doi.org/ 10.1038/sj.cdd.4402167 [DOI] [PubMed] [Google Scholar]
- 18.Manivannan S, de Boer R, Veenhuis M, van der Klei IJ. Lumenal peroxisomal protein aggregates are removed by concerted fission and autophagy events. Autophagy 2013; 9:1044–56; PMID:23614977; http://dx.doi.org/ 10.4161/auto.24543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jungwirth H, Ring J, Mayer T, Schauer A, Buttner S, Eisenberg T, Carmona-Gutierrez D, Kuchler K, Madeo F. Loss of peroxisome function triggers necrosis. FEBS letters 2008; 582:2882–6; PMID:18656474; http://dx.doi.org/ 10.1016/j.febslet.2008.07.023 [DOI] [PubMed] [Google Scholar]
- 20.Bener Aksam E, Jungwirth H, Kohlwein SD, Ring J, Madeo F, Veenhuis M, van der Klei IJ. Absence of the peroxiredoxin Pmp20 causes peroxisomal protein leakage and necrotic cell death. Free Rad Biol Med 2008; 45:1115–24; PMID:18694816; http://dx.doi.org/ 10.1016/j.freeradbiomed.2008.07.010 [DOI] [PubMed] [Google Scholar]
- 21.Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen W-L, et al.. A Genomic Screen for Yeast Mutants Defective in Selective Mitochondria Autophagy. Mol Biol Cell 2009; 20:4730–8; PMID:19793921; http://dx.doi.org/ 10.1091/mbc.E09-03-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou J-C. Preventing Mitochondrial Fission Impairs Mitochondrial Function and Leads to Loss of Mitochondrial DNA. PLoS ONE 2008; 3:e3257-8; PMID:18806874; http://dx.doi.org/ 10.1371/journal.pone.0003257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ 2005; 12:1613–21; PMID:15947785; http://dx.doi.org/ 10.1038/sj.cdd.4401697 [DOI] [PubMed] [Google Scholar]
- 24.Twig G, Elorza A, Molina AJA, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, et al.. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J 2008; 27:433–46; PMID:18200046; http://dx.doi.org/ 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendl N, Occhipinti A, Müller M, Wild P, Dikic I, Reichert AS. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J Cell Sci 2011; 124:1339–50; PMID:21429936; http://dx.doi.org/ 10.1242/jcs.076406 [DOI] [PubMed] [Google Scholar]
- 26.Dijken LPV, Otto R, Harder W. Growth of Hansenula polymorpha in a methanol-limited chemostat. Arch Microbiol 1976; 111:137–44; PMID:1015956; http://dx.doi.org/ 10.1007/BF00446560 [DOI] [PubMed] [Google Scholar]
- 27.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol 1983; 153:163–8; PMID:6336730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 1999; 15:1541–53; PMID:10514571; http://dx.doi.org/ 10.1002/(SICI)1097-0061(199910)15:14%3c1541::AID-YEA476%3e3.0.CO;2-K [DOI] [PubMed] [Google Scholar]
- 29.Hill J, Donald KA, Griffiths DE, Donald G. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res 1991; 19:5791; PMID:1945859; http://dx.doi.org/ 10.1093/nar/19.23.6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westermann B, Neupert W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 2000; 16:1421–7; PMID:11054823; http://dx.doi.org/ 10.1002/1097-0061(200011)16:15%3c1421::AID-YEA624%3e3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- 31.Gietz D, Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 1992; 20:1425; PMID:1561104; http://dx.doi.org/ 10.1093/nar/20.6.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]


