Abstract
Fishing and climate change impact the demography of marine fishes, but it is generally ignored that many species are made up of genetically distinct locally adapted populations that may show idiosyncratic responses to environmental and anthropogenic pressures. Here, we track 80 years of Atlantic cod (Gadus morhua) population dynamics in West Greenland using DNA from archived otoliths in combination with fish population and niche based modeling. We document how the interacting effects of climate change and high fishing pressure lead to dramatic spatiotemporal changes in the proportions and abundance of different genetic populations, and eventually drove the cod fishery to a collapse in the early 1970s. Our results highlight the relevance of fisheries management at the level of genetic populations under future scenarios of climate change.
Global change impacts the abundance and distribution of biodiversity in the world´s oceans, which in turn affects the services provided by marine ecosystems1,2. Marine fisheries are an invaluable resource supporting human welfare worldwide3. Historically, dramatic changes in both the abundance and distribution of many important fish stocks have been observed4,5,6,7,8. However, the relative impact of fishing and climate on stock dynamics is still under debate9, as few studies have been able to assess the integrated responses of fish stocks to these pressures.
Many marine fish species are made up of genetically distinct populations that often do not match traditional fisheries management units, “stocks”10,11. Stocks are defined geographically rather than biologically, and can potentially lead to overexploitation of more vulnerable populations, in a fishery consisting of a mixture of different fish populations12, a so-called “mixed-stock fishery”. Genetic populations may exhibit unique adaptations and tolerances to specific environments13, and might display distinct responses to fisheries and climate change14,15. Disentangling how different populations have responded to intense fishing pressure and climate variability could hold key answers for understanding historical fish distribution and abundance patterns and for improved future management of marine fish resources.
Here, we used a large archive of fish earstones (otoliths) to study the population dynamics of Atlantic cod (Gadus morhua) during the historical commercial fishery in West Greenland, which displayed a dramatic collapse similar to a number of other cod fisheries16,17,18,19. From peak landings that ranged between 400 K and 500 K tons in the 1960’s, the catch dropped dramatically through the 1970’s to a complete collapse in the early 1990s20,21 (see Fig. 1), and have since remained low21. Traditionally, biomass increases and large catches were partly ascribed to a generally warmer climate and the influx of cod eggs and larvae from Iceland22,23, but no clear explanation of the population dynamics has emerged. Recent work has shown that at least four genetically distinct cod populations occur along the coasts of Greenland24, presenting an opportunity to examine fishery dynamics within distinct populations.
Figure 1. Historical Atlantic cod (Gadus morhua) biomass (dotted line) and commercial catch (solid line) in West Greenland (readapted after20,21).
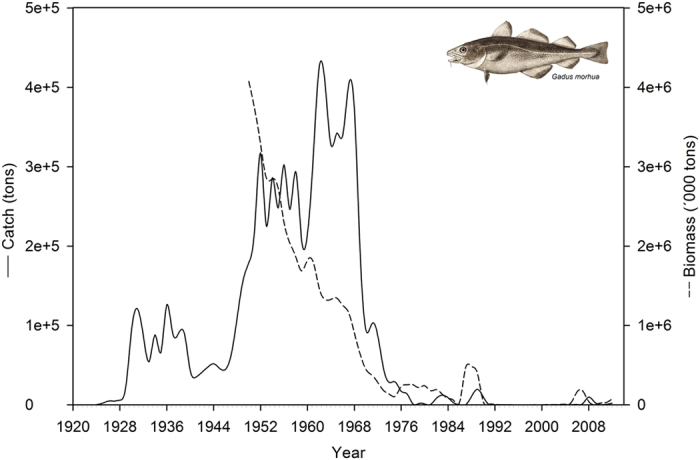
This figure has been drawn by S.B. using SigmaPlot 12 software.
Over the last century, otoliths have been collected for age and growth determination, and now also provide an opportunity for recovering historical DNA25,26. In this study, DNA was extracted and analyzed from 872 cod otoliths and a panel of 81 gene-associated Single Nucleotide Polymorphisms (SNPs)27 was applied to allow the assignment of each cod individual to its genetic populations of origin among those described within Greenlandic waters24: West Greenland offshore, West Greenland inshore, Iceland offshore (also known as East Greenland/Iceland offshore population28) and Iceland inshore, (population names re-adapted from previous study24). We generated a probability distribution of the contribution of the different genetic populations to the total catch over time and related temporal variation in catch composition within fisheries management areas to historical sea surface temperatures using ecological niche modeling29.
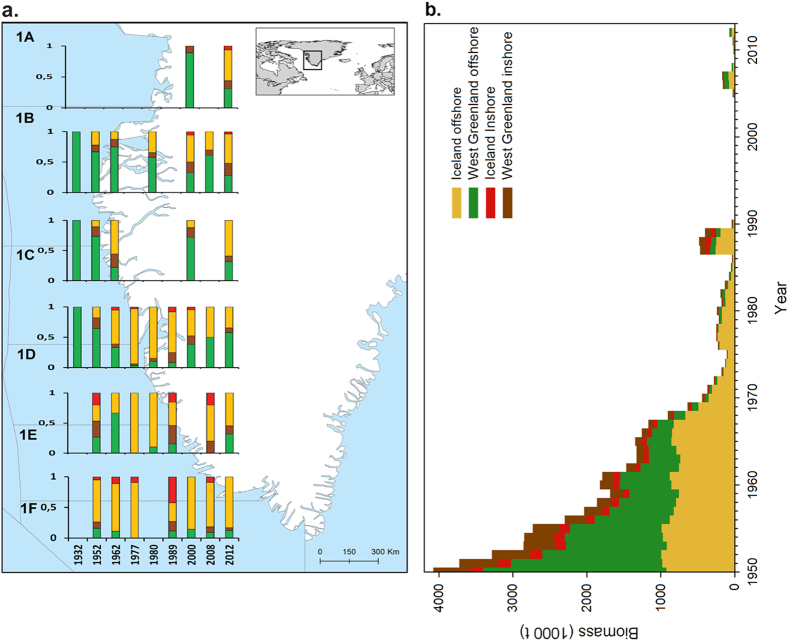
Genetic assignment of archived otoliths revealed different spatiotemporal contributions of populations to the historical commercial fishery (Fig. 2a). Catches during the 1930s consisted almost exclusively of cod from the local West Greenland offshore population (98.8%). The contribution from this population decreased dramatically during the peak fishing in the 1950s (46.5%) and 1960s (34%), but in the 1970s and 1980s this population had almost disappeared from the catch in the southwest Greenland fishing areas (NAFO divisions 1D to1 F, 1.1%, Fig. 2a), but still appeared to be present in the northern areas. In contrast, the contribution from the Icelandic offshore population with spawning areas offshore Iceland and offshore East Greenland23 increased from the start of the major fishing boom in the 1950s (29.1%) to constitute the vast majority of the catch during and immediately following the stock collapse around the 1970s (91.6%).
Figure 2.
(a) Spatiotemporal development in the proportions of different Atlantic cod (Gadus morhua) populations in the historical West Greenland fishery (NAFO divisions from 1A to 1F): West Greenland offshore (green), West Greenland inshore (brown), Iceland offshore (dark yellow), Iceland inshore (red).The Greenland map and the small-scale world map have been drawn by S.B. using ArcGIS and R software respectively. (b) Estimated stock biomass composition of cod along West Greenland 1950–2012 (NAFO divisions 1A–1F). Biomass is estimated based on catch proportions (Supplementary Figure 1) and the biomass of 3+ years old cod in the stock.
The collapse of the West Greenland offshore population was predicted from the fishing intensity and productivity estimates of the two main populations. The average spawning stock biomass needed to produce one recruit (3 years old) was 5.56 kg for the West Greenland offshore cod compared to 1.06 kg for the Iceland offshore cod, which corresponds to equilibrium fishing mortalities (Feq) of 0.14 and 0.82 respectively30 (see details in Methods). These estimates are sensitive to errors in the estimation of spawning stock biomass and recruitment, where overestimation of spawning stock biomass and underestimation of recruitment will lead to inflated Feq. A sensitivity analysis showed that the West-Greenland spawning stock biomass during 1955–1972 would have been overestimated by a factor of five to allow for a Feq corresponding to the Icelandic Feq. Within all reasonable scenarios of errors, the West-Greenlandic Feq is considerably lower than the Icelandic Feq (see details in Supplementary Information). Accordingly, historically observed intermediate fishing intensities (between the two Feq estimates)30 would sustain the Icelandic offshore cod population while leading to the collapse of the West Greenland offshore cod population. This pattern was observed during 1950–1968 when the biomass of the West Greenland offshore population plummeted while the biomass of the Iceland offshore population remained stable (Fig. 2b). Following the crash and the post-1990 absence of commercial landing in the northern and central NAFO divisions31, the local West Greenland offshore population has rebounded in terms of contribution to the mixed stock (Fig. 2a). This increase has primarily been seen in the central areas (division 1D; Fig. 2a). The contribution from the Icelandic offshore population has generally decreased but remained very high in the southern NAFO divisions. Throughout the period, West Greenland inshore and Icelandic inshore populations were present, but never constituted a major part of the catch.
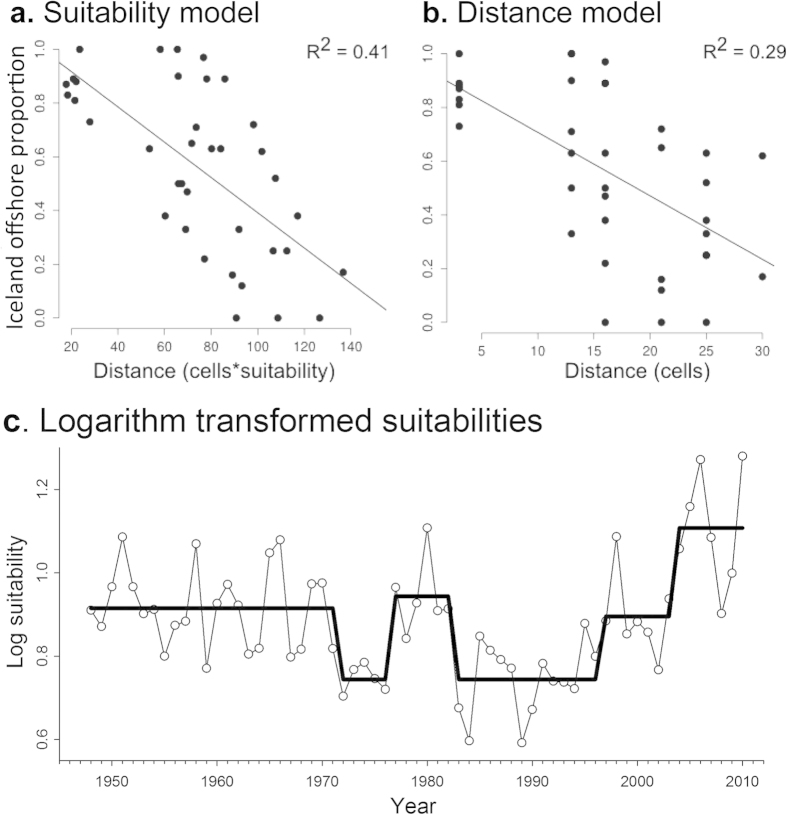
We related the shift of the proportion of the two major stock components, (i.e. West Greenland offshore relative to the Iceland offshore populations) to the historical climatic suitability for cod along West Greenland area (see details in Methods). We estimated yearly cod climate suitability using species distribution models, hindcasted to 1948–2011 hydrographic conditions. Historical climate suitability was estimated both at the species level (mean model accuracy measured with Area Under the Curve (AUC) = 0.83), as well as for distinct populations assuming population-level genetic adaptations (AUC = 0.78). For each year and each 50 × 50 km pixel, we computed the least-cost path distance along the coast through climatically suitable conditions to the southern tip of Greenland (presumed entry point of Icelandic offshore cod into West Greenlandic waters). We found that the spatiotemporal shift in the proportion of the Iceland offshore population relative to that from offshore West Greenland was better explained by a population level climatic niche assuming distinct genetic adaptation for the population (general linear model with a binomial distribution, coefficient of determination R2 = 0.41, slope = −0.04, Wald-z test p = 0.001; Fig. 3a), than considering a general climatic requirement for the stock as a whole (R2 = 0.30, slope = −0.05, p = 0.03) or static geographic distance (R2 = 0.29, slope = −0.06, p = 0.03, Fig. 3b). Moreover, the proportion of Icelandic cod catches was best explained by the least cost path distance to spawning areas via oceanographic routes with suitable conditions (e.g. surface temperatures) in contrast to the shortest geographic route to spawning areas (Fig. 3a,b). We also observed that the West Greenland offshore population occured predominantly in colder sea surface temperatures, while the Iceland offshore population occupied a relatively broader range of temperature conditions including warmer temperatures (see Supplementary Figure S2). The climatic niche modelling documented an increased suitability of the West Greenland area for the Icelandic offshore population during the warm periods, facilitating the distribution at higher latitude areas. A regime shift detection analysis applied to the habitat suitability data showed a significant decrease concurrent with the collapse of the fishery during the cold period beginning in 1970s (see Methods). This low suitability regime persisted until 1997 only interrupted by a short improvement in the period 1977–1982 (Fig. 3c).
Figure 3. Climatic suitability for the Iceland offshore population linked to cod population structure in NAFO divisions.
(a) Proportion of cod from the Iceland offshore population in a given NAFO division in relation to of least-cost path distance to nearest spawning areas weighted by suitability. (b) Proportion of cod from the Iceland offshore population as a function of the shortest sea distance to nearest spawning area. (c) Yearly averaged logarithm transformed random forest generated habitat suitability for Icelandic offshore cod along West-Greenland. Solid line is mean value from a regime shift detection analysis40 (cut-off: 5 years, significance level 0.1).
This study demonstrates the value of using DNA from archived fish in order to identify vital processes at the population level for understanding the impact of exploitation and climate change on the historical distribution of marine fish. First, we showed that the high and indiscriminate fishing pressure resulted in a collapse of the local, cold-adapted, West Greenland offshore population in the 1970s. Concurrent with this collapse, the colder conditions prevented the Icelandic cod population from increasing in abundance, and the flourishing West Greenland cod fishery vanished.
Our results suggest that tracking the genetic origin of harvested fish could support spatially differentiated management plans and help avoid a disproportional impact on the most vulnerable populations. This will have important consequences for fisheries management in West Greenland and require that separate quotas are set for the different biological populations. However, we expect that maintenance of this biocomplexity will lead to more stable ecosystem services in the future due to the portfolio effect13, whereby today’s strained cod populations may thrive under future conditions. A number of previous studies have shown that species are not equal in the face of climate change, with some geographic ranges expanding while others contract22. Our results suggest that within species, different, locally-adapted genetic pools may win or lose disproportionally under climate change. If not counteracted through population based fisheries management, exploitation may lead to the collapse of populations needed to adapt to future environmental conditions.
Methods
Sampling
Archived tissue samples (otoliths and scales) of Atlantic cod (Gadus morhua) were originally collected off west Greenland through a combination of commercial fishing, during annual surveys (with R/V Paamiut) and as part of the cod tagging program conducted by the Greenland Institute of Natural Resources in Nuuk, Greenland. 872 samples were collected between late June and January in: 1932, 1952, 1962, 1977, 1980, 1989, 2000, 2008 and 2012 (see Supplementary Table S1 and Supplementary Table S2). Sampling was stratified according to the Northwest Atlantic Fisheries Organization Convention Area in West Greenland (NAFO Subarea 1; from 1A to 1F divisions). Further details about seasonal composition of sampling are outlined in Supplementary Information.
SNPs selection and genotyping
81 SNPs (Single Nucleotide Polymorphisms) were selected as the most informative for population assignment out of a panel of 935 SNPs that was recently screened in spawning population samples collected throughout Greenlandic and Icelandic waters24. The reduced panel was selected to achieve the minimum assays with maximum power and the selected loci showed the highest FCT(differentiation between groups) in pairwise comparisons of the four distinct spawning groups identified. For this study, a total of 867 cod individuals were genotyped for the 81 SNP panel using the Fluidigm 96.96 Dynamic Arrays system (BioMark HD System) following the instructions from the manufacturer and standard methods32.
Individual assignment tests
Two different genetic assignment approaches were used to estimate the historical contribution of different cod populations to the fished stock. Individual assignment test was first conducted with the program GENECLASS233. These tests were based on the Bayesian probability approach34 and a Monte-Carlo resampling method for probability computation35 with 10000 simulated individuals (α = 0.01) to evaluate the probability that a certain multilocus genotype originated from one of the four baseline populations previously identified24 (populations names were changed according to defined spawning grounds28). Furthermore, Discriminant Analysis of Principal Components (DAPC36) implemented in the adegenet R package was also employed. In this multivariate framework, the mixed stock individuals were assigned with the predict.dapc() function that projects multilocus genotypes onto discriminant functions (i.e. synthetic variables that maximize differences between and minimize differences within a priori defined reference groups), thereby deriving posterior membership probabilities to each reference group (spawning population) for each individual. In both approaches, the baseline genetic signature (allele frequencies) of each spawning population was defined based on individual samples collected at the spawning time for a previous study24, further details are outlined in Supplementary Information. Only mixed stock individuals that could be assigned with >0.90 probability to a particular population with both methods were included in the following analysis. Individuals assigned with lower probability or with inconsistent results for the two methods were discarded. However, individual assignment test with GENECLASS2 was generally highly consistent with that of DAPC, and 85% of individuals were assigned to one of the genetic populations inhabiting Greenlandic waters using the above criteria. Out of 872 individuals analyzed, 103 were discarded.
Probability distribution of total catch composition over time
At each observation time ti, i = 1 … n, the total number of investigated fish Ni was categorized into four genetically distinguished spawning groups: West Greenland offshore (1), West Greenland inshore (2), Iceland offshore (3), and Iceland inshore (4) (Supplementary Figure S1). These observations are naturally described by a multinomial distribution with sample size Ni. To describe the development of the stock proportions over time it is assumed that the probability vector: pt = (p1, p2, p3, p4) follows an unobserved process. The process is set up such that the sum of ptis 1 and such that the elements of pteach are between 0 and 1. The following transformation defines the process: let αt = (αt1, αt2, αt3) follow a three dimensional random walk, such that: αt = αt−1 + εt, where εt ~ N (0, σ2I3×3). Then, we defined 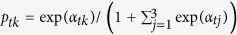 , for k = 1 … 3, and
, for k = 1 … 3, and 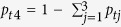 .
.
The catch probability distributions were multiplied by the total biomass of fish age 3 years and older (30) in order to estimate the biomass contribution of each population during 1950–2012 (Fig. 1).
Equilibrium fishing mortalities
The equilibrium fishing mortality (Feq) is the fishing mortality that balances mortality and reproduction, ensuring that enough fish survive to reproduce to exactly replenish them via recruitment37. One approach to determine Feq is to calculate the spawning biomass needed to produce one new recruit from time series of spawning stock biomasses and recruitment, and subsequently use an exponential decay function (1) to search for the equilibrium fishing mortality (Feq) that would lead to this equilibrium biomass. Actual fishing mortalities can then be evaluated against Feq to reveal whether a current or historical fishing pattern was sustainable. This analysis was performed for the West Greenland offshore and Icelandic offshore populations. The recruitment per spawning stock biomass (R/SSB) index of fish stock productivity was obtained from30 for both the West Greenlandic offshore and Icelandic offshore populations. For the period 1924 to 1973 the average SSB needed to produce 1 new recruit, and hence ensure replacement of the adult population by new recruits, was 5.56 g for West Greenland offshore cod compared to 1.06 kg for the Icelandic offshore cod during 1955–2002. The period 1924–1973 was chosen because this represents a period when recruitment of cod in west Greenland was driven by the local West Greenlandic offshore population.
We used the exponential decay function to search for the equilibrium fishing mortality (Feq) for the age groups 5 to 12 years that would lead to an average spawning biomass of 5.56 kg and 1.06 kg. Age group specific fishing mortality and natural mortality, weight-at-age and maturity-at-age for the West Greenlandic offshore population was also obtained from30 whereas these data were obtained from38 for the Icelandic offshore population. We used the exponential decay function (1): Nt+1 = Nt * exp−(F+M), where Nt is the number of fish alive at time t, F is the instantaneous fishing mortality and M is the instantaneous natural mortality, to calculate the numbers of fish alive at in each age group (3 to 12 years) given the F and M. These numbers were then multiplied by the weight-at-age and maturity-at-age to yield the spawning stock biomass, and, subsequently scaled to one initial recruit, i.e. the stock size at age 3 was set to one fish (Supplementary Table S3 and Supplementary Table S4). The calculated average Feq was scaled to the age-specific fishing pattern obtained from a virtual population analysis (VPA) for the period 1961–1963. The low productivity of Greenlandic cod only allowed for an equilibrium F(5–12) of 0.14 (Supplementary Table S3). A similar calculation for the Icelandic cod stock using SSB/R of 1.06 kg corresponded to an equilibrium F(5–12) of 0.82 (Supplementary Table S4). A fishing mortality that exceeds Feq will lead to reduced SSB and subsequently reduced R until the collapse of the stock. Consequently, a fishing mortality between 0.14 and 0.82 that would sustain the Icelandic offshore cod population would lead to the collapse of the West Greenland offshore cod population. It should be noted that the equilibrium fishing mortality for the Icelandic offshore cod is calculated for the total cod population, i.e. including the individuals residing in Icelandic waters. Hence, the actual fishing pressure of this population will be an abundance weighted mean of the fishing pressure in Icelandic and West-Greenlandic waters. To test the impact of changes in growth and maturity trajectories, and errors in the estimation of recruitment and spawning stock biomass on the equilibrium fishing mortalities we performed a sensitivity analyses outline in Supplementary Information.
Habitat suitability modeling
The Greenland Institute of Natural Resources at Nuuk conducts scientific trawling primarily for shrimp and halibut each year, and also collects information on all other fish species (approximately 200 species, total, including cod). The surveys (with R/V Paamiut) are designed and stratified to provide core information relevant for stock assessment purposes. We modeled habitat suitability by combining the data from all four cod spawning populations, and at single population level (i.e. only including records identified as belonging to the Iceland offshore population) and projected/predicted these models to annual hydrodynamical scenarios (i.e. one from each year). For the mixed stock (all spawning populations combined), we modeled presence and absence of cod occurrences in the Paamiut scientific trawls. These data included n = 1263 presence and n = 1708 absence records. To compute the habitat suitability model at the population level, we modeled presences of Iceland offshore fish identified from genotyping of fish caught in the Paamiut trawls from 2000 to 2011, and absences were drawn from the absences recorded from the Paamiut surveys during the same period (n = 357 presence and 1708 absence records).
We used an ensemble forecasting approach that combined predictions of four modeling techniques including generalized linear models (GLM), generalized additive model (GAM), generalized boosting methods (GBM) and random forest (RF) as applied in39. We used a binomial linear model and down-weighted the absence records to match the number of presences40. For each modeling technique, 10 repetitions were performed using random sets of 80% of the initial occurrences to calibrate the model. The remaining 20% were used for the evaluation of the model using the Area Under the Curve41. The final model was then projected to past and future hydrographical scenarios (1948–2011), at 1 year-intervals averaging the four modeling techniques. Calculations were performed in R42.
A regime shift detection analysis (Fig. 3c) based on logarithm-transformed average habitat suitability (based on the random forest algorithm) for Icelandic offshore cod was performed to reveal periods of differing habitat suitability43. Further details about hydrodynamical modeling data and evaluation of population level suitability with dispersal are reported in Supplementary Information.
Additional Information
How to cite this article: Bonanomi, S. et al. Archived DNA reveals fisheries and climate induced collapse of a major fishery. Sci. Rep. 5, 15395; doi: 10.1038/srep15395 (2015).
Supplementary Material
Acknowledgments
We are grateful to the Greenland Institute of Natural Resources for contributing historical samples. The study was carried out with financial support from the Danish Agency for Science, Technology and Innovation as part of the Greenland Climate Research Centre and from the Nordic Centre for Research on Marine Ecosystem and Resources under Climate Change, ‘NorMER’.
Footnotes
Author Contributions E.E.N. planned and oversaw the project. S.B. conducted the SNP genotyping together with D.M. The genetic data analysis was conducted by S.B. with input from N.O.T., R.B.H. and A.R. were in charge of the contemporary and historical sample collection. A.N. generated the probability distribution of the genetic catch composition over time. The habitat suitability modelling and hindcast simulation were carried out by L.P. and M.S.W. Hydrodynamical data were provided by S.M.O. P.G. investigated spawning stock productivity and equilibrium fishing mortality. J.H.H. and C.P. discussed the results and critically commented on the manuscript at all stages. S.B., L.P., M.S.W., P.G. and E.E.N. wrote the manuscript with input from the other authors.
References
- Worm B. et al. Impacts of biodiversity loss on ocean ecosystem services. Science 314, 787–90 (2006). [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O. & Bruno J. F. The Impact of Climate Change on the World’s Marine Ecosystems. Science 328, 1523–1528 (2010). [DOI] [PubMed] [Google Scholar]
- Halpern B. S. et al. An index to assess the health and benefits of the global ocean. Nature 488, 615–620 (2012). [DOI] [PubMed] [Google Scholar]
- Jackson J. B. C. et al. Historical overfishing and the recent collapse of coastal ecosystems. Science 293, 629–637 (2001). [DOI] [PubMed] [Google Scholar]
- Brander K. M. Global fish production and climate change. Proceedings of the National Academy of Sciences of the United States of America 104, 19709–19714 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W. W. L., Watson R. & Pauly D. Signature of ocean warming in global fisheries catch. Nature 497, 365–368 (2013). [DOI] [PubMed] [Google Scholar]
- Pinsky M. L., Worm B., Fogarty M. J., Sarmiento J. L. & Levin S. A. Marine Taxa Track Local Climate Velocities. Science 341, 1239–1242 (2013). [DOI] [PubMed] [Google Scholar]
- Perry A., Low P., Ellis J. & Reynolds J. D. Climate change and distribution shifts in marine fishes. Science 308, 1912–1915 (2005). [DOI] [PubMed] [Google Scholar]
- Payne M. R. Fisheries: Climate change at the dinner table. Nature 497, 320–321 (2013). [DOI] [PubMed] [Google Scholar]
- Ruzzante et al. D. E. Biocomplexity in a highly migratory pelagic marine fish, Atlantic herring. Proceedings of the Royal Society B: Biological Sciences 273, 1459–1464 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss H., Hoarau G., Dickey-Collas M. & Wolff W. J. Genetic population structure of marine fish: mismatch between biological and fisheries management units. Fish and Fisheries 10, 361–395 (2009). [Google Scholar]
- Heath M. R. et al. Combination of genetics and spatial modelling highlights the sensitivity of cod (Gadus morhua) population diversity in the North Sea to distributions of fishing. ICES Journal of Marine Science: Journal du Conseil 71, 794–807 (2014). [Google Scholar]
- Schindler D. E. et al. Population diversity and the portfolio effect in an exploited species. Nature 465, 609–612 (2010). [DOI] [PubMed] [Google Scholar]
- Hilborn R., Quinn T. P., Schindler D. E. & Rogers D. E. Biocomplexity and fisheries sustainability. Proceedings of the National Academy of Sciences of the United States of America 100, 6564–6568 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozier L. G. & Hutchings J. A. Plastic and evolutionary responses to climate change in fish. Evolutionary Applications 7, 68–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen E. M. et al. Maturation trends indicative of rapid evolution preceded the collapse of northern cod. Nature 428, 932–935 (2004). [DOI] [PubMed] [Google Scholar]
- Hilborn R. & Litzinger E. Causes of decline and potential for recovery of Atlantic Cod Populations. The Open Fish Science Journal 2, 32–48 (2009). [Google Scholar]
- Swain D. P. Life-history evolution and elevated natural mortality in a population of Atlantic cod (Gadus morhua). Evolutionary Applications 4, 18–29 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. Lament For An Ocean: The Collapse Of The Atlantic Cod Fishery. Random House LLC (2013). [Google Scholar]
- Horsted S. A. A Review of Cod Fisheries at Greenland, 1910–1995. Journal of Northwest Atlantic Fishery Science 28, 1–112 (2000). [Google Scholar]
- ICES. Report of the North Western Working Group (NWWG), 25 April – 02 May 2013. ICES Headquarters, Copenhagen. ICES CM 2013/ACOM: 07. 1538 pp. Scientific report (2013).
- Jensen A. S. Concerning a change of climate during recent decades in the Arctic and Subartic regions from Greenland in the west to Eurasia in the east, and contemporary biological and geophysical changes. Det Kongelige Danske Videnskabelige Selskab, Biologiske Meddelelser 14, 1–77 (1939). [Google Scholar]
- Wieland K. & Hovgård H. Distribution and drift of Atlantic cod (Gadus morhua) eggs and larvae in Greenland waters. Journal of Northwest Atlantic Fishery Science 30, 61–76 (2002). [Google Scholar]
- Therkildsen N. O. et al. Spatiotemporal SNP analysis reveals pronounced biocomplexity at the northern range margin of Atlantic cod Gadus morhua. Evolutionary Applications 6, 690–705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therkildsen N. O., Nielsen E. E., Hüssy K., Meldrup D. & Geffen A. J. Does DNA extraction affect the physical and chemical composition of historical cod (Gadus morhua) otoliths? ICES Journal of Marine Science 67, 1251–1259 (2010). [Google Scholar]
- Nielsen E. E. & Bekkevold D. The memory remains: application of historical DNA for scaling biodiversity loss. Molecular Ecology 21, 1539–1541 (2012). [DOI] [PubMed] [Google Scholar]
- Nielsen E. E. et al. Gene-associated markers provide tools for tackling illegal fishing and false eco-certification. Nature Communications 3, 851 (2012). [DOI] [PubMed] [Google Scholar]
- ICES. Report of the North Western Working Group (NWWG), 24 April – 01 May 2014. ICES Headquarters, Copenhagen. ICES CM 2014/ACOM: 07. 902 pp. Scientific report (2014).
- Guisan A. et al. Predicting species distributions for conservation decisions. Ecology letters 16, 1424–1435 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovgård H. & Wieland K. Fishery and environmental aspects relevant for the emergence and decline of Atlantic cod (Gadus morhua) in West Greenland waters. Resiliency of gadid stocks to fishing and climate change. Alaska Sea Grant, University of Alaska Fairbanks (2008). [Google Scholar]
- ICES. Offshore cod in ICES Subarea XIV and NAFO Subarea 1 (Greenland cod). In Report of the ICES Advisory Committee, 2012. ICES Advice, 2012. Book 2, Section 2.4.1a. Scientific report (2012).
- Seeb J. E., Pascal C. E., Ramakrishnan R. & Seeb L. W. SNP genotyping by the 5′-nuclease reaction: advances in high-throughput genotyping with non-model organisms. Single Nucleotide Polymorphisms, Methods in Molecular Biology 578, 277–292 (2009). [DOI] [PubMed] [Google Scholar]
- Piry S. et al. GENECLASS2: a software for genetic assignment and first-generation migrant detection. Journal of Heredity 95, 536–539 (2004). [DOI] [PubMed] [Google Scholar]
- Rannala B. & Mountain J. L. Detecting immigration by using multilocus genotypes. Proceedings of the National Academy of Sciences 94, 9197–9201 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paetkau D., Slade R., Burden M. & Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Molecular ecology 13, 55–65 (2004). [DOI] [PubMed] [Google Scholar]
- Jombart T., Devillard S. & Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11, 94 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. A. & Mertz G. The limits of exploitation: a precautionary approach. Ecological Applications 8, S165–S169 (1998). [Google Scholar]
- ICES. Report of the North-Western Working Group (NWWG), 25 April – 4 May 2006. ICES Document CM 2006/ACFM: 26. Scientific report (2006).
- Pellissier L. et al. Combining food web and species distribution models for improved community projections. Ecology and Evolution 3, 4572–4583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisz M. S. & Guisan A. Do pseudo-absence selection strategies influence species distribution models and their predictions? An information-theoretic approach based on simulated data. BMC Ecology 9, 8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swets J. A. Measuring the accuracy of diagnostic systems. Science 240, 1285–1293 (1988). [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2011). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: http://cran.r-project.org/. (Accessed: 8th January 2014).
- Rodionov S. N. A sequential algorithm for testing climate regime shifts. Geophysical Research Letters 31, (2004). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.