Abstract
The developing embryo is a paradigmatic model to study molecular mechanisms of time control in Biology. Hox genes are key players in the specification of tissue identity during embryo development and their expression is under strict temporal regulation. However, the molecular mechanisms underlying timely Hox activation in the early embryo remain unknown. This is hindered by the lack of a rigorous temporal framework of sequential Hox expression within a single cluster. Herein, a thorough characterization of HoxB cluster gene expression was performed over time and space in the early chick embryo. Clear temporal collinearity of HoxB cluster gene expression activation was observed. Spatial collinearity of HoxB expression was evidenced in different stages of development and in multiple tissues. Using embryo explant cultures we showed that HoxB2 is cyclically expressed in the rostral presomitic mesoderm with the same periodicity as somite formation, suggesting a link between timely tissue specification and somite formation. We foresee that the molecular framework herein provided will facilitate experimental approaches aimed at identifying the regulatory mechanisms underlying Hox expression in Time and Space.
Keywords: chick embryo, Hox genes, Hox code, spatial collinearity, temporal collinearity, temporal control
Abbreviations
- A-P
anterior-posterior
- PSM
presomitic mesoderm
- HH
Hamburger and Hamilton
- ps
primitive streak
- nt
neural tube
- nc
notochord
Introduction
Vertebrate embryo development proceeds from a single cell to an autonomous individual through highly complex, yet coordinated mechanisms functioning together in time and space. This is achieved through strictly regulated gene expression within specific tissues and at precise times.1 Time and space are closely intertwined in the gastrulating embryo, where epiblast cells undergoing epithelial-to-mesenchymal transitions earlier in time, are specified to anterior body structures and those remaining longer in the stem cell-rich epiblast will give rise to more posterior structures. This process occurs under the control of Hox transcription factors and temporal control of Hox gene expression during gastrulation ultimately defines cell fate specification along the entire body axis.2-4
Hox genes are expressed in defined, often overlapping domains along the embryo anterior-posterior (A-P) body axis and their joint activity distinctly patterns the various body segments.5 In the genome, Hox genes are positioned in a 3′ to 5′ direction along chromosomal clusters, in a sequence that reflects their anterior limit of expression along the embryo A-P axis, defined as “spatial collinearity.”4 Their chromosomal alignment is also critical for the timing of expression initiation, as 3′ Hox genes in each cluster are expressed first, whereas more 5′ Hox genes are expressed later in time, a phenomenon coined “temporal collinearity.”2,4 Initial activation of Hox genes occurs during gastrulation and different signaling pathways such as Wnt, Fgf or Retinoic acid have been proposed to regulate this event (reviewed in ref.6). However, the molecular mechanisms underlying temporal collinearity of Hox gene activation are far from being understood.
The best known molecular mechanism of temporal control operating during embryo development is the molecular clock underlying somitogenesis,7,8 characterized by cyclic gene expression of Notch, FGF and Wnt signaling pathway effectors and/or downstream targets.9 Several experimental evidences have suggested that temporal regulation of Hox gene expression may be linked to the somitogenesis clock. Notch-dependent dynamic transcriptional bursts of HoxD1 gene expression were described in the rostral presomitic mesoderm (PSM)10 and loss of notch-signaling gene expression cycles in transgenic mice altered Hox gene expression and produced anterior shifts in vertebrae axial identity.11 Further evidence come from Xenopus embryos, where a feedback loop between Hox genes and X-Delta-2 was identified as early as gastrulation, when Hox gene expression is first initiated.12 In fact, a timer mechanism in the early non-organizer mesoderm driving Hox gene expression initiation and the existence of coordination between somite formation and antero-posterior patterning have been independently postulated.12,13
A significant impediment to experimentally interrogate the molecular mechanisms underlying Hox temporal collinearity has been the lack of knowledge on the initial momentum of gene expression induction of sequential Hox genes within a unique cluster. Also, it is not clear if different combinations of Hox gene expression-Hox codes – are present along the early embryo A-P axis or if they are only established later in development.14 Herein, we present a detailed study of HoxB cluster gene expression in the chick embryo by in situ hybridization, identifying the developmental stage of activation of each HoxB gene (Time), as well as its domains of expression (Space). We experimentally show temporal collinearity of HoxB activation during gastrulation, as well as spatial collinearity in different embryonic tissues. Cyclic expression of one of the HoxB genes is described and a summary of our results is presented in the form of tissue-specific “Hox codes” of gene expression along the embryo A-P axis.
Results
HoxB1
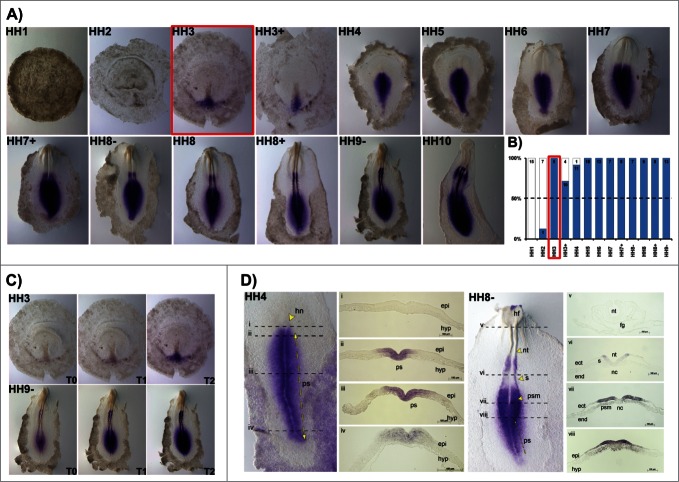
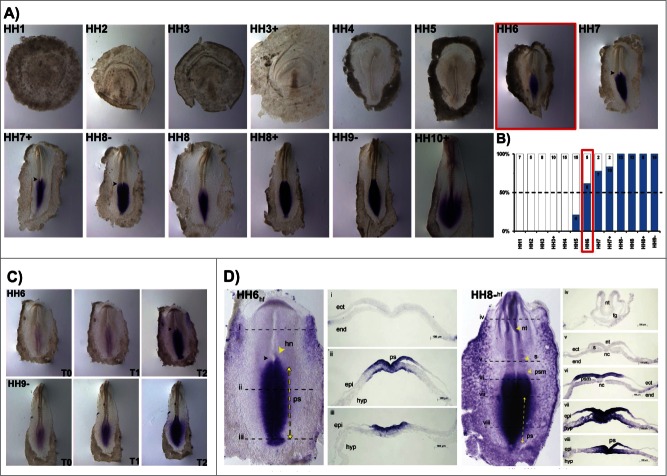
A detailed characterization of HoxB cluster gene expression domains during early stages of chick embryonic development (from late blastula to 6-somite stage) was performed by whole mount in situ hybridization analysis and cross section histology. HoxB1 expression was never detected in late blastula stage embryos (HH1, Fig. 1A, B) and from stage HH2 to HH4, HoxB1 was detected in some, but not all embryos analyzed for each of these stages. From HH3 onwards, however, HoxB1 expression was consistently observed in more than 50% of the studied embryos (Fig. 1A, B) and we have considered this condition (over 50%) as the activation stage of HoxB gene expression. From HH5 onwards HoxB1 was expressed in 100% of the tested embryos (Fig. 1B).
Figure 1.
HoxB1 expression patterns. (A) HoxB1 gene expression evaluated by in situ hybridization during early chick development. (B) Representation of the percentage of embryos that display HoxB1 expression. Numbers indicate the experimental N. Red boxes highlight the developmental stage where over 50% of the tested embryos present HoxB1 staining. (C) In situ hybridization images obtained with increasing times of staining, evidencing graded nature of HoxB1 expression. (D) Transverse section analysis of HoxB1 expression patterns in different developmental stages. hn – Hensen's node; ps – primitive streak; epi – epiblast; hyp – hypoblast; hf – head fold; nt – neural tube; s – somite; psm – presomitic mesoderm; fg – foregut; nc – notochord; ect – ectoderm; end – endoderm.
HoxB1 is expressed along the primitive streak in gastrulation stages, excluded from the Hensen's node. During early somitogenesis, HoxB1 is present in the caudal tissues and in the neural tube, caudal to the otic placode, where it is also strongly expressed (Fig. 1A). When allowed to stain for increasing periods of time, HoxB1 expression was detected in the heart tube while absent from the anterior intestinal portal (HH10, Fig. 1A). The spatial patterns obtained were highly suggestive of graded HoxB1 expression in the caudal embryo, which was confirmed by photographing in situ hybridization staining reactions of the same embryos over time (Fig. 1C). We found that HoxB1 presents a clear gradient of expression: HoxB1 expression initially appeared along the primitive-streak and progressively extended radially along the adjacent tissues. As a result, we can conclude that HoxB1 exhibits higher levels of expression near the primitive streak while in the surrounding cells the expression is progressively lower.
We further analyzed cross-sections of HH4 (gastrulation) and HH8- (somitogenesis) stained embryos. HoxB1 transcripts were excluded from the Hensen's node (Fig. 1Di) and detected along the primitive streak (ps) and in the nascent mesoderm (Fig. 1D ii-iv). Graded expression is observed in the epiblast cells adjacent to the ps in a medial-lateral direction. At stage HH8-, HoxB1 continues to be expressed in the caudal streak, in the ingressing epiblast and nascent mesoderm (Fig. 1D viii). Rostrally, HoxB1 expression is detected in the neural plate and presomitic mesoderm, while there is no expression in both endoderm and notochord (Fig. 1D vii). Finally, HoxB1 expression is detected in the dorsal neural tube (Fig. 1D vi).
HoxB2
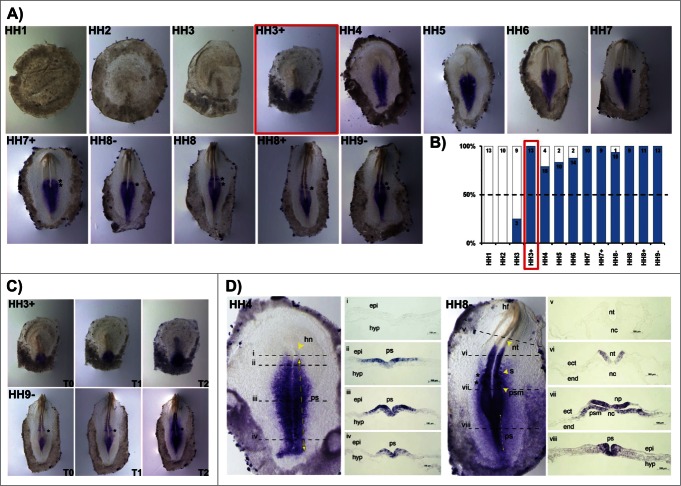
HoxB2 transcripts were detected at the beginning of gastrulation (HH3) but only consistently after HH3+ (Fig. 2A, B). HoxB2 gene expression initially appears in the posterior primitive streak (HH3+) and extends rostrally along the primitive streak progressively in time. As soon as somitogenesis takes place, it is possible to observe 1–2 bands of HoxB2 expression in the rostral PSM, located in the prospective somite region (Fig. 2A, asterisks). The anterior boundary of HoxB2 expression is situated at the last somite formed. HoxB2 is also expressed in a gradient, with highest levels near the streak (Fig. 2C). Regarding the cross-section analysis, at stage HH4 the entire primitive streak excluding the Hensen's node, displays HoxB2 transcripts (Fig. 2D i-iv). A striking feature of HoxB2 expression is its salt-and-pepper pattern in both the epiblast and nascent mesoderm. The analysis of HH8- embryos reveals that the expression of HoxB2 is present along the primitive streak, in ingressing epiblast and prospective mesoderm cells (Fig. 2D viii); HoxB2 is also localized in the neural plate and in the neural tube (nt) (Fig. 2D, vi, vii); no expression is detected in the notochord, endoderm or non-neural ectoderm (Fig. 2D v, vi).
Figure 2.
HoxB2 expression patterns. (A) HoxB2 gene expression by in situ hybridization during early chick development. (B) Representation of the percentage of embryos that display HoxB2 expression. Numbers indicate the experimental N. Red boxes highlight the developmental stage where over 50% of the tested embryos present HoxB2 staining. (C) In situ hybridization images obtained with increasing times of staining, evidencing graded nature of HoxB2 expression. (D) Transverse section analysis of HoxB2 expression patterns in different developmental stages. hn – Hensen's node; ps – primitive streak; epi – epiblast; hyp – hypoblast; hf – head fold; nt – neural tube; s – somite; psm – presomitic mesoderm; nc – notochord; ect – ectoderm; end – endoderm.
HoxB3
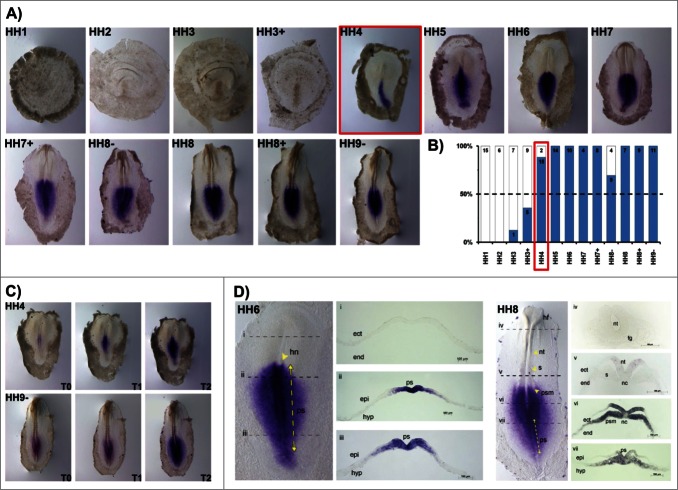
HoxB3 expression was consistently observed from stage HH4 onwards (Fig. 3A, B). At this stage HoxB3 was detected only in the posterior half primitive-streak, contrarily to HoxB1 and HoxB2 which were present along the entire A-P axis. At somitogenesis stages, neither head, somites nor neural tube are stained by this probe and the expression is restricted to the caudal tissues. HoxB3 displayed a graded expression with maximum levels near the streak, consistent with previous genes (Fig. 3C). Cross-sections from HH6 embryos showed graded medial-lateral HoxB3 expression in the primitive streak and nascent mesoderm (Fig. 3D ii, iii), while absent from the Hensen's node (Fig. 3D i). Embryos at stage HH8 exhibited HoxB3 expression in the primitive streak, PSM and neural plate (Fig. 3D vi-vii). No expression was detected in the endoderm, notochord or foregut (Fig. 3D iv, v).
Figure 3.
HoxB3 expression patterns. (A) Evaluation of HoxB3 gene expression by in situ hybridization. (B) Representation of the percentage of embryos that display HoxB3 expression. Numbers indicate the experimental N. Red boxes highlight the developmental stage where over 50% of the tested embryos present HoxB3 staining. (C) In situ hybridization images obtained with increasing times of staining, evidencing graded HoxB3 expression. (D) Transverse section analysis of HoxB3 expression patterns in different developmental stages. hn – Hensen's node; ps – primitive streak; ect – ectoderm; end – endoderm; epi – epiblast; hyp – hypoblast; hf – head fold; nt – neural tube; s – somite; psm – presomitic mesoderm; fg – foregut; nc – notochord.
HoxB5
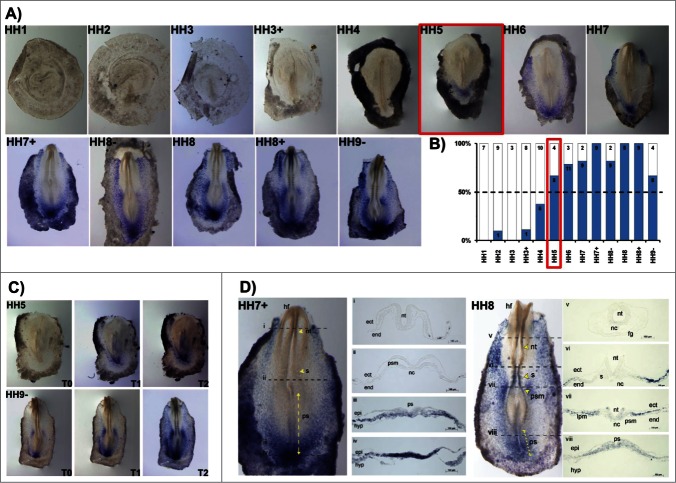
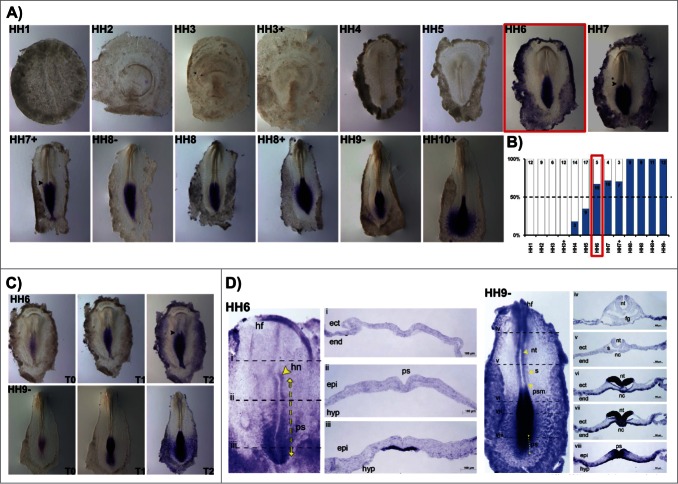
HoxB5 gene expression activation (in situ staining in more than 50% of the studied embryos) occurred at stage HH5 (Fig. 4A, B). HoxB5 expression patterns differed significantly from the other HoxB cluster genes. While most HoxB genes initially appeared at primitive-streak level and quickly extended to the mesodermal tissues, HoxB5 expression was initiated in cells of the posterior primitive streak that will contribute to extraembryonic mesoderm and only expanded anteriorly along the streak later in time. In fact, HoxB5 was mostly expressed in the reminiscent Kohler´s Sickle and in the lateral plate mesoderm, including extraembryonic mesoderm (Fig. 4A, C). At HH7+, HoxB5 transcripts are clearly localized in the caudal-most primitive streak region, where the nascent mesoderm is strongly stained with a “salt-and-pepper” type pattern (Fig. 4D iii-iv). At HH8, HoxB5 is further expressed as “salt-and-pepper” in the lateral plate, extraembryonic and presomitic mesoderm (Fig. 4D vi-viii).
Figure 4.
HoxB5 expression patterns. (A) Evaluation of HoxB5 gene expression by in situ hybridization. (B) Representation of the percentage of embryos that display HoxB5 expression. Numbers indicate the experimental N. Red boxes highlight the developmental stage where over 50% of the tested embryos present HoxB5 staining. (C) In situ hybridization images obtained with increasing times of staining, evidencing graded HoxB5 expression. (D) Transverse section analysis of HoxB5 expression patterns. nt – neural tube; s – somite; ps – primitive streak; ect – ectoderm; end – endoderm; psm – presomitic mesoderm; nc – notochord; epi – epiblast; hyp – hypoblast; hf – head fold; fg – foregut; lpm – lateral plate mesoderm.
HoxB8
HoxB8 expression was active from stage HH6 onwards (Fig. 5A, B). HoxB8 was expressed along the primitive streak and Hensen´s node, occasionally in an asymmetrical manner (black arrowheads in Fig. 5, see also Fig. 7B). At later stages (HH10+) HoxB8 was also detected in the lateral plate mesoderm. HoxB8 was expressed as a radial gradient in the caudal tissues surrounding the primitive streak and neural plate (Fig. 5A,C). Cross-section analysis at stage HH6 evidenced that HoxB8 transcripts are strongly expressed in the caudal-most epiblast, while absent from the hypoblast (Fig. 5D iii). At primitive streak-level, Hoxb8 was expressed in the epiblast adjacent to the streak and in the endoderm and nascent mesoderm, although with lower intensity (Fig. 5D i-ii). Graded expression was observed in the epiblast cells adjacent to the ps in a medial-lateral direction. HoxB8 expression at HH8- was spread as a medial-lateral gradient at the streak level, in the PSM and neural plate (Fig. 5D vi-viii). More anteriorly, HoxB8 was expressed in the neural plate and was absent from mesodermal and endodermal tissues (Fig. 5D vi). No expression was detected at the level of formed somites, neural tube and foregut (Fig. 5D iv-v).
Figure 5.
HoxB8 expression patterns. (A) Evaluation of HoxB8 gene expression by in situ hybridization. Black arrowheads highlight asymmetrical expression around the Henson's node. (B) Representation of the percentage of embryos that display HoxB8 expression. Numbers indicate the experimental N. Red boxes highlight the developmental stage where over 50% of the tested embryos present HoxB8 staining. (C) In situ hybridization images obtained with increasing times of staining, evidencing graded HoxB8 expression. (D) Transverse section analysis of HoxB8 expression patterns in different developmental stages. hf – head fold; hn – Hensen's node; ps – primitive streak; ect – ectoderm; end – endoderm; epi – epiblast; hyp – hypoblast; nt – neural tube; s – somite; psm – presomitic mesoderm; fg – foregut; nc – notochord.
Figure 7.
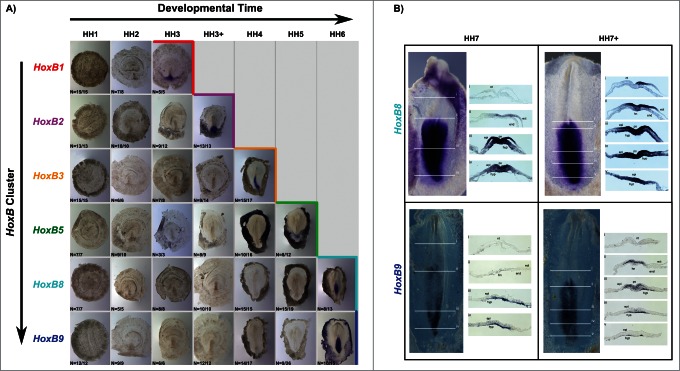
Temporal collinearity of HoxB cluster activation in the early chick embryo. (A) HoxB cluster expression by in situ hybridization evidences temporal collinearity of gene expression activation. The developmental stages where over 50% of the tested embryos were stained (as indicated by N in each figure) was considered the first stage of gene activation. (B) Transverse section analysis of HoxB8 and HoxB9 expression patterns in sequential developmental stages evidences that HoxB8 is expressed in the embryo proper before HoxB9. hn – Hensen's node; ps – primitive streak; ect – ectoderm; end – endoderm; epi – epiblast; hyp – hypoblast; nt – neural tube.
HoxB9
HoxB9 expression was very similar to HoxB8. HoxB9 was consistently detected after stage HH6 (Fig. 6A, B), present along the primitive streak and neural plate as a radial gradient (Fig. 6C), frequently with asymmetry around the Hensen's node (black arrowheads in Fig. 6). At later stages (HH10+) HoxB9 was also strongly expressed in the lateral plate mesoderm (Fig. 6A). Cross-section analysis of HH6 embryo revealed that maximum HoxB9 expression is present in the caudal-most hypoblast while excluded from the epiblast (Fig. 6D iii). Furthermore, HoxB9 transcripts cannot be detected in the endoderm, ectoderm and nascent mesoderm (Fig. 6D i). In HH9-, the caudal-most endoderm is strongly stained along the entire medial-lateral extension, while it is only present in the ectoderm and mesoderm nearby the streak (Fig. 6D viii). More rostrally HoxB9 is expressed in the endoderm and in the neural plate and early notochord (Fig. 6D vii), which loses its expression at more anterior levels (Fig. 6D vi). HoxB9 is also not expressed in the rostral presomitic mesoderm (Fig. 6D vi). No expression was detected in somites, neural tube and foregut (Fig. 6D iv-v).
Figure 6.
HoxB9 expression patterns. (A) Evaluation of HoxB9 gene expression by in situ hybridization. Black arrowheads highlight asymmetrical expression around the Henson's node. (B) Representation of the percentage of embryos that display HoxB9 expression. Numbers indicate the experimental N. Red boxes highlight the developmental stage where over 50% of the tested embryos present HoxB9 staining. (C) In situ hybridization images obtained with increasing times of staining reaction, evidencing graded HoxB9 expression. (D) Transverse section analysis of HoxB9 expression patterns in different developmental stages. hf – head fold; hn – Hensen's node; ps – primitive streak; ect – ectoderm; end – endoderm; epi – epiblast; hyp – hypoblast; nt – neural tube; s – somite; psm – presomitic mesoderm; fg – foregut; nc – notochord.
Temporal collinearity of HoxB cluster gene expression
A characteristic feature of Hox gene expression is temporal collinearity of Hox activation along the genomic cluster. We show this property for the first time along a single cluster in the early vertebrate embryo. The activation of HoxB cluster genes over time, defined as the HH stage where more than 50% of the tested embryos presented in situ hybridization staining, is summarized in Figure 7. HoxB genes are sequentially activated during developmental time progression, reflecting their relative positions in the genomic cluster. Although HoxB8 and HoxB9 expression was detected within the same developmental stage, cross section analysis revealed that only HoxB8 is expressed in the embryo proper at HH6 (Figs. 5D and 6D). Cross section analysis of HH7 and HH7+ embryos stained for HoxB8 and HoxB9 (Fig. 7B) confirmed that HoxB8 expression is initiated in the embryonic tissue earlier than HoxB9, thus confirming the temporally collinear nature of HoxB cluster gene expression initiation.
Spatial collinearity of HoxB cluster gene expression
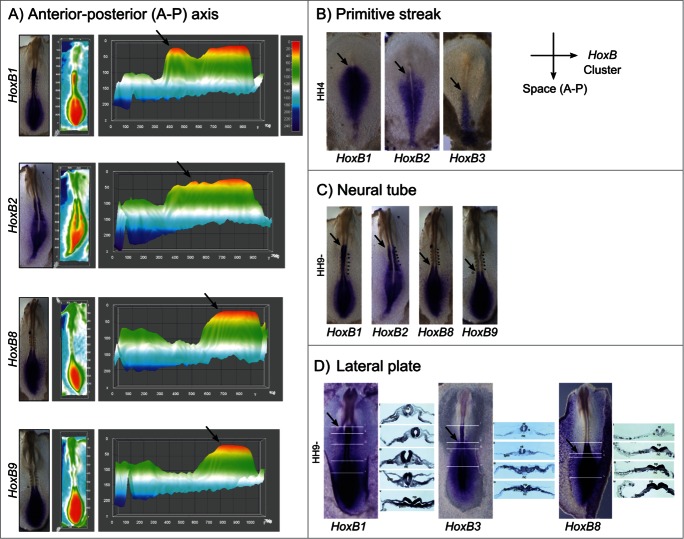
Spatial collinearity of HoxB cluster gene expression was further visualized by in situ hybridization in multiple embryonic tissues. This was clearly observable along the embryo anterior-posterior axis (Fig. 8A). Interactive surface plots representing the HoxB expression patterns obtained experimentally highlight HoxB cluster spatial collinearity (Fig. 8A): the anterior limit of maximum expression of 3′ Hox genes is significantly rostralized relatively to the 5′ Hox genes. Spatial collinearity was further observed in specific tissues, namely along the primitive streak at HH4, where the anterior limit of expression of HoxB1, HoxB2 and HoxB3 was progressively caudalized (Fig. 8B), in the neural tube (Fig. 8C) and in the lateral plate mesoderm (Fig. 8D).
Figure 8.
Spatial collinearity of HoxB cluster gene expression evidenced by in situ hybridization. (A) Heatmap surface plots of HH9- embryos evidencing progressively posterior peaks of expression (arrows) of HoxB genes positioned along the genomic cluster. (B) Spatial collinearity of early HoxB gene expression along the primitive streak of gastrulating embryos (HH4). Spatial collinearity is also observed in specific tissues of somitogenesis-staged embryos (HH9-), namely along the neural tube (C) and in the lateral plate mesoderm (D). Arrows indicate rostral limit of expression. lpm – lateral plate mesoderm; nc – notochord; np – neural plate; nt – neural tube; s – somite; psm – presomitic mesoderm.
Cyclic HoxB2 gene expression
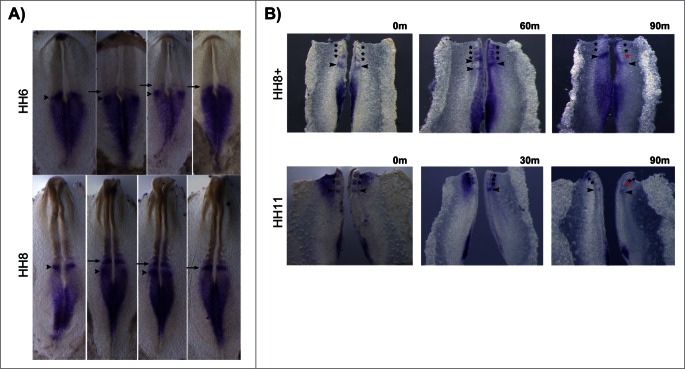
HoxB2 expression patterns varied considerably in the rostral PSM, even among embryos of the same HH stage (Fig. 9A). HoxB2 was consistently expressed along the primitive streak and neural plate, however some embryos displayed an expression band overlapping the prospective somite (Fig. 9A, arrows) while others had an additional band in the PSM (Fig. 9A, arrowheads). In order to clarify the dynamic nature of this expression profile, the caudal portion of chick embryos was divided into 2 halves by sectioning along the midline; one half was fixed immediately, while the other half was cultured in vitro for varying amounts of time. When the experimental half was cultured for 30 or 60 minutes, different profiles of HoxB2 expression were observed in the 2 halves (n = 9/9) (Fig. 9B). We concluded that HoxB2 displays dynamic stripes of expression in the chick PSM. When the experimental half was incubated for 90 minutes a new somite was generated in the incubated explant and both halves showed identical HoxB2 expression patterns (n = 7/7), indicating that HoxB2 has a cyclic expression pattern in the rostral PSM, with the same periodicity as somite formation.
Figure 9.
Cyclic HoxB2 expression in the rostral PSM. (A) In situ hybridization of the same-staged embryos reveals dynamic stripes of HoxB2 expression in the rostral PSM (arrows and arrowheads). (B) Explant culture experiments evidencing cyclic expression of HoxB2 stripes (arrowheads) with a 90 min periodicity. The left explants were immediately fixed and the right explants were cultured for the amount of time indicated above the figure. Both explants were processed for in situ hybridization simultaneously.
Discussion
Ingression of epiblast cells into the primitive streak in a temporally controlled manner during early embryogenesis is fundamental for proper specification and spatial distribution of adult body tissues, skeletal structures and organs. Timely activation of Hox gene expression sets the tempo for cell ingression during gastrulation,3 coupling temporal collinearity with spatial distribution of Hox expression and consequent tissue specification. Thus, it is critical to identify the tissues that express each HoxB gene over development as well as the developmental time when each Hox gene is first expressed in the early embryo.
Temporal collinear expression of Hox genes is known to occur in chick,3,14 mouse15 and xenopus.13 However, previous attempts to observe temporal collinearity of gene expression along one single Hox cluster by in situ hybridization have produced inconsistent results. Gaunt and Strachan14 described this property only in the rostral primitive streak, while they couldn't observe sequential gene activation in the caudal streak. More recently, Barak et al.16 challenged the idea that temporal collinearity takes place in early gastrulation stages. In the present study a large number of embryos per gene and stage were analyzed and we observed that HoxB cluster genes are not turned on from one HH stage to another, but rather along a period of time, from HH3 to HH6 (Fig. 7). Temporal collinearity was clearly observed and an activation-stage for each HoxB gene was established by considering the stage when over 50% of the embryos presented staining. Although expression of HoxB8 and HoxB9 was activated in the same stage (HH6), we found that these genes are differentially required for tissue patterning, since HoxB8 is expressed in the epiblast/ectoderm while HoxB9 is present only in the hypoblast/endoderm.
Spatial collinearity is another characteristic feature of Hox gene expression that was readily observed in the present study: we found that the rostral-most boundary of expression of 3’HoxB cluster genes is more anterior than that of 5’ genes (Fig. 8A). This is also visible along the primitive streak during gastrulation (Fig. 8B), in the neural tube (Figs. 8C and D) and lateral plate mesoderm (Fig. 8D) during early somitogenesis. This observation suggests that the molecular mechanisms underlying A-P patterning and specification are common to mesodermal-derived tissues.
We describe a singular expression pattern for HoxB5, since it is restricted to the caudal-most gastrulating region of the embryo and lateral plate mesoderm, including extraembryonic mesoderm. This observation suggests that HoxB5 may be primarily involved in patterning tissues and organs derived from the lateral plate mesoderm, while the axial and paraxial mesodermal tissues are patterned by the other HoxB genes. A very interesting observation was the asymmetric expression of both HoxB8 and HoxB9 around the Hensen's node in early stages (see Figs. 5 and 6). This is a characteristic feature of genes participating in the establishment of embryonic left-right asymmetry.17 A Hox gene was recently implicated in left-right asymmetry establishment in Drosophila18 and our work provides the first indication that Hox genes may also play a similar role in vertebrate embryos. Our results are in overall agreement with previous data,3,14,16 while providing greater detail on the expression domains of each HoxB gene.
An intriguing observation was made regarding the dynamics of HoxB2 expression. HoxB2 in situ hybridization of embryos belonging to the same HH stage produced varied expression patterns and cyclic pulses of HoxB2 expression were shown to underlie this observation (Fig. 9). This is a characteristic feature of somitogenesis clock genes7 and is reminiscent of what was previously reported for mouse HoxD1,10 further supporting the hypothesis that temporal regulation of Hox gene expression initiation may be linked to the somitogenesis clock.10,11,13
Materials and Methods
Eggs and embryos
Fertilized chick (Gallus gallus) eggs obtained from commercial sources were incubated at 37.8°C in a 49% humidified atmosphere and were staged according to the Hamburger and Hamilton (HH) classification.19 A somite pair was considered to be formed when definite clefts separating somites from both sides of the PSM were observed.
RNA probes
Antisense digoxigenin-labeled RNA probes were produced as previously described: HoxB1,20 HoxB3,20 HoxB821 and HoxB9.21 HoxB2 probe was generated by amplifying a 383 bp fragment of chick HoxB2 (XM_003642792) using sense 5′-GCAGCGAAGCCACGGATGAGG-3′ and antisense 5′-TCGGGGTGGGACAGAAAGGGATAA-3′ oligos. HoxB5 probe was generated by amplifying a 580 bp fragment of chick HoxB5 (NM_001025355) using sense 5′-CACCCGCAGGAGGAGAATAGAGAT-3′ and antisense 5′-CAGGAAGGGGAAAAAGCAACAAAT-3′ oligos. PCR products were cloned into the vector pGEM-T Easy (Promega) and orientation confirmed upon sequencing.
In situ hybridization
In situ hybridization was performed as previously described.22 Embryos were photographed over time during the staining reaction using an Olympus SZX16 stereomicroscope. Interactive surface plots were obtained using ImageJ software. The luminance of an image is interpreted as height for the plot. The pixels were counted for each gray value in 8-bit or 16-bit image in the defined interval and then transformed the picture in heatmap with calibration bar.
Cross-section preparation
Cross-section analysis of stained embryos was performed as previously described.23 Briefly, selected embryos were embedded in 15% sucrose, 7.5% gelatin, frozen, and sectioned (14 μm) using a Leica CM3050 S cryostat. Sections were examined and photographed using an Axio Imager Z2 Fluorescence microscope with a Zeiss Axio Cam IC.
Explant cultures
Explant culture experiments were performed as previously described.24 Briefly, ventrally positioned chick embryos in stages HH7–HH9− or HH11–14 were divided into 2 halves by cutting across the 3 germ layers at the midline level, bisecting both neural tube (nt) and notochord (nc), producing 2 equivalent nt/nc+ explants. All explants were delimited rostrally above the second or third formed somites. Tissues thus isolated, originated control and experimental explants, which were cultured individually in a dorsal position for varying amounts of time.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Aknowledgments
The authors thank R. Magno for helpful discussions, C. Tabin and G. Sheng for HoxB8/9 and HoxB1/3 probe plasmids, respectively, A. Moreira for technical assistance with interactive surface plot analyses and the staff at Light microscopy Facility, DCBM/UAlg.
Funding
AG, HMM and LG were supported by Fundação para a Ciência e a Tecnologia (FCT), Portugal (grants PTDC/SAU-OBD/105111/2008, UMINHO/BI/7/2014 and SFRH/BPD/65652/2009, respectively). RPA was funded by a Ciencia2007 Program Contract and Programa Operacional Regional do Norte (ON.2) – NORTE-07–0124-FEDER-000017. This work was supported by national Portuguese funding through FCT (National and FEDER COMPETE Program funds: PTDC/SAU-BID/121459/2010 and PTDC/SAU-OBD/099758/2008 to RPA and IP, respectively), and by PEst-OE/EQB/LA0023/2011.
References
- 1. Furlong EEM. A Topographical Map of Spatiotemporal Patterns of Gene Expression. Dev Cell 2008; 14:639-40; PMID:18477445; http://dx.doi.org/ 10.1016/j.devcel.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 2. Deschamps J, van Nes J. Developmental regulation of the Hox genes during axial morphogenesis in the mouse. Development 2005; 132:2931-42; PMID:15944185; http://dx.doi.org/ 10.1242/dev.01897. [DOI] [PubMed] [Google Scholar]
- 3. Iimura T, Pourquie O. Collinear activation of Hoxb genes during gastrulation is linked to mesoderm cell ingression. Nature 2006; 442:568-71; PMID:16760928; http://dx.doi.org/ 10.1038/nature04838. [DOI] [PubMed] [Google Scholar]
- 4. Tschopp P, Tarchini B, Spitz F, Zakany J, Duboule D. Uncoupling time and space in the collinear regulation of Hox genes. PLoS genetics 2009; 5:e1000398; PMID:19266017; http://dx.doi.org/ 10.1371/journal.pgen.1000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wellik DM. Hox genes and vertebrate axial pattern. Curr Top Dev Biol 2009; 88:257-78; PMID:19651308; http://dx.doi.org/ 10.1016/S0070-2153(09)88009-5. [DOI] [PubMed] [Google Scholar]
- 6. Iimura T, Denans N, Pourquie O. Establishment of Hox vertebral identities in the embryonic spine precursors. Curr Top Dev Biol 2009; 88:201-34; PMID:19651306; http://dx.doi.org/ 10.1016/S0070-2153(09)88007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 1997; 91:639-48; PMID:9393857; http://dx.doi.org/ 10.1016/S0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- 8. Andrade RP, Palmeirim I, Bajanca F. Molecular clocks underlying vertebrate embryo segmentation: A 10-year-old hairy-go-round. Birth Defects Res C Embryo Today 2007; 81:65-83; PMID:17600780; http://dx.doi.org/ 10.1002/bdrc.20094. [DOI] [PubMed] [Google Scholar]
- 9. Krol AJ, Roellig D, Dequeant ML, Tassy O, Glynn E, Hattem G, Mushegian A, Oates AC, Pourquie O. Evolutionary plasticity of segmentation clock networks. Development 2011; 138:2783-92; PMID:21652651; http://dx.doi.org/ 10.1242/dev.063834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zakany J, Kmita M, Alarcon P, de la Pompa JL, Duboule D. Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell 2001; 106:207-17; PMID:11511348; http://dx.doi.org/ 10.1016/S0092-8674(01)00436-6. [DOI] [PubMed] [Google Scholar]
- 11. Cordes R, Schuster-Gossler K, Serth K, Gossler A. Specification of vertebral identity is coupled to Notch signalling and the segmentation clock. Development 2004; 131:1221-33; PMID:14960495; http://dx.doi.org/ 10.1242/dev.01030. [DOI] [PubMed] [Google Scholar]
- 12. Peres JN, McNulty CL, Durston AJ. Interaction between X-Delta-2 and Hox genes regulates segmentation and patterning of the anteroposterior axis. Mech Dev 2006; 123:321-33; PMID:16644189; http://dx.doi.org/ 10.1016/j.mod.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 13. Wacker SA, Jansen HJ, McNulty CL, Houtzager E, Durston AJ. Timed interactions between the Hox expressing non-organiser mesoderm and the Spemann organiser generate positional information during vertebrate gastrulation. Dev Biol 2004; 268:207-19; PMID:15031117; http://dx.doi.org/ 10.1016/j.ydbio.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 14. Gaunt SJ, Strachan L. Temporal colinearity in expression of anterior Hox genes in developing chick embryos. Dev Dyn 1996; 207:270-80; PMID:8922526; http://dx.doi.org/ 10.1002/(SICI)1097-0177(199611)207:3%3c270::AID-AJA4%3e3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 15. Forlani S, Lawson KA, Deschamps J. Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development 2003; 130:3807-19; PMID:12835396; http://dx.doi.org/ 10.1242/dev.00573. [DOI] [PubMed] [Google Scholar]
- 16. Barak H, Preger-Ben Noon E, Reshef R. Comparative spatiotemporal analysis of Hox gene expression in early stages of intermediate mesoderm formation. Dev Dyn 2012; 241:1637-49; PMID:22930565; http://dx.doi.org/ 10.1002/dvdy.23853. [DOI] [PubMed] [Google Scholar]
- 17. Raya A, Izpisua Belmonte JC. Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nat Rev Genet 2006; 7:283-93; PMID:16543932; http://dx.doi.org/ 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- 18. Coutelis JB, Geminard C, Speder P, Suzanne M, Petzoldt AG, Noselli S. Drosophila left/right asymmetry establishment is controlled by the Hox gene abdominal-B. Dev Cell 2013; 24:89-97; PMID:23328400; http://dx.doi.org/ 10.1016/j.devcel.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 19. Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn 1992; 195:231-72; PMID:1304821; http://dx.doi.org/ 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- 20. Alev C, Wu Y, Kasukawa T, Jakt LM, Ueda HR, Sheng G. Transcriptomic landscape of the primitive streak. Development 2010; 137:2863-74; PMID:20667916; http://dx.doi.org/ 10.1242/dev.053462. [DOI] [PubMed] [Google Scholar]
- 21. Burke AC, Nelson CE, Morgan BA, Tabin C. Hox genes and the evolution of vertebrate axial morphology. Development 1995; 121:333-46; PMID:7768176. [DOI] [PubMed] [Google Scholar]
- 22. Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature 1995; 375:787-90; PMID:7596411; http://dx.doi.org/ 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- 23. Goncalves L, Vinhas M, Pereira R, Pais De Azevedo T, Bajanca F, Palmeirim I. Circadian clock genes Bmal1 and Clock during early chick development. Dev Dyn 2012; 241:1365-73; PMID:22700438; http://dx.doi.org/ 10.1002/dvdy.23821. [DOI] [PubMed] [Google Scholar]
- 24. Resende TP, Ferreira M, Teillet MA, Tavares AT, Andrade RP, Palmeirim I. Sonic hedgehog in temporal control of somite formation. Proc Natl Acad Sci U S A 2010; 107:12907-12; PMID:20615943; http://dx.doi.org/ 10.1073/pnas.1000979107. [DOI] [PMC free article] [PubMed] [Google Scholar]