Abstract
Purpose
Ocular hypertension is a major risk factor for glaucoma and the inner wall of Schlemm's canal (SC) endothelia participates in the regulation of aqueous humor outflow resistance. This study aimed to identify differentially expressed genes in primary cultures of SC cells from glaucoma patients.
Methods
This study examined SC samples from three glaucoma cases and four controls. Schlemm's canal cells were isolated from eight different postmortem human eyes. Total RNA was extracted, labeled, and hybridized to Illumina HumanWG-6 BeadChips containing probes for approximately 47,000 human transcripts. After extracting the data using Illumina GenomeStudio software, the data were normalized and analyzed using the R package limma in Bioconductor. Using Protein ANalysis THrough Evolutionary Relationships (PANTHER) software, gene ontology analysis of highly expressed genes was executed in controls and glaucoma groups separately. Pathway analysis was performed with differentially expressed genes using WebGestalt (WEB-based GEne SeT AnaLysis Toolkit). Selected genes were validated using droplet digital PCR (ddPCR).
Results
Gene ontology analysis indicated similar functional categories in cases and controls. Differential analysis identified a total of 113 genes with at least 2-fold expression changes in cases. Pathway analysis indicated significant enrichment of genes in cell adhesion, heparin binding, glycosaminoglycan binding, filopodium, and extracellular matrix remodeling. Eighteen selected genes with differential expression were successfully validated using ddPCR.
Conclusions
This study represents the first genome-wide expression study of human primary SC cells from glaucoma patients and provides a potential list of targets regulating SC cell stiffness and pore formation, eventually the outflow resistance in glaucoma individuals.
Keywords: glaucoma, gene expression, Schlemm's canal, aqueous flow
We have identified a number of differentially expressed genes in primary Schlemm's canal (SC) endothelial cells derived from individuals with and without glaucoma. The novel information will improve our understanding of how SC cells contribute to the pathological process of glaucoma.
Glaucoma is the leading cause of irreversible vision loss and blindness around the world, affecting over 60 million people.1–4 Primary open-angle glaucoma (POAG) is the most common type and is inherited as a complex trait. POAG is characterized with progressive loss of retinal ganglion cells, optic nerve head excavation, and visual field loss without an identifiable clinical cause.4 Major risk factors for POAG include increased age, elevated intraocular pressure (IOP), positive family history, and African ancestry.2 Genetics has been shown to play an important role in POAG. Family-based linkage analyses and case-control based genome-wide association studies (GWAS) have identified a number of genes contributing to pathogenesis of POAG5–18 (see review in Refs 2, 14, and 15).
A number of genetic factors are associated with elevated IOP, the only modifiable major risk factor in glaucoma, such as variants in or near TMOC1 (transmembrane and coiled-coil domains 1), growth arrest-specific 7 (GAS7), ATP-binding cassette, sub-family A, member 1 (ABCA1), and fibronectin type III domain containing 3B (FNDC3B).12,19,20 It remains unknown how these genes are involved in IOP regulation. Because IOP, whether elevated or not, contributes to disease progression, all available clinical treatments (medical, procedural, or surgical) attempt to lower IOP.21 Elevated IOP is caused by increased outflow resistance generated deep in the conventional outflow tract, where trabecular meshwork (TM) cells and Schlemm's canal (SC) cells interact.22 Expression pattern alterations and/or changes of TM cells and SC cells likely mediate increased resistance to outflow and elevated IOP, leading to the pathological retinal changes of glaucoma.
While the majority of resistance is generated by extracellular matrix in the juxtacanalicular region of the TM, SC cells appear to modulate resistance by forming pressure-dependent openings (pores) in its continuous monolayer.23,24 Recently, the pore formation process has been shown to be affected by an increased stiffness of SC cells in glaucomatous eyes.25,26 Elevated extracellular matrix stiffness has been observed to increase SC cell stiffness and decrease pore formation in vitro.27 For the first time, using eight of the same SC cell strains examined in these previous studies, we compared the expression profiles of normal and glaucomatous cell strains. The goal is to identify differentially expressed genes, which will improve our understanding of the mechanism(s) underlying stiffer glaucomatous cells and decreased pore formation.
Methods
SC Cell Isolation and Culture
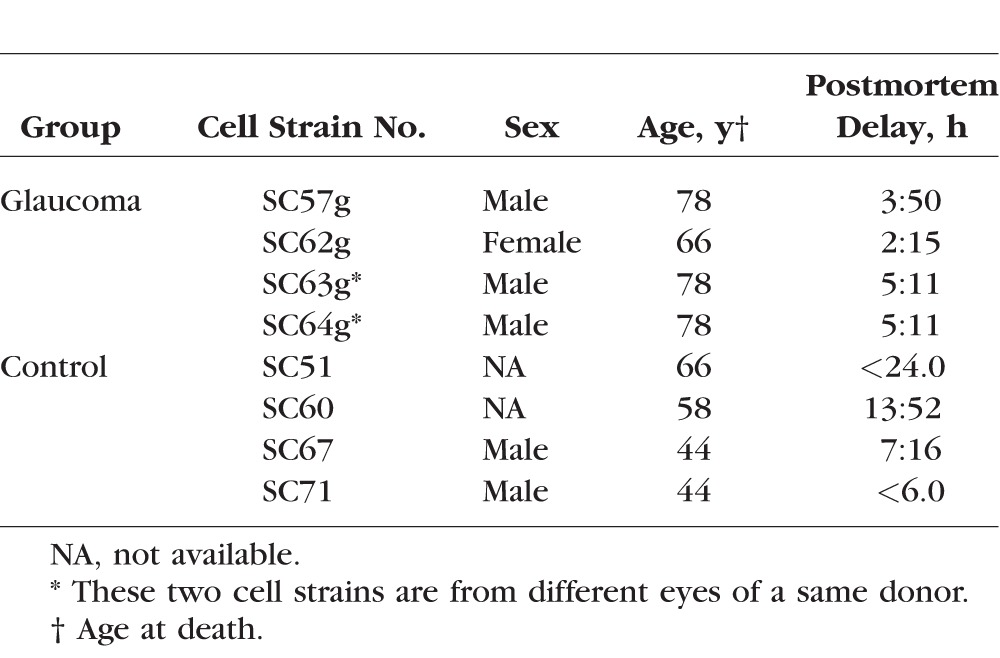
Primary cultures of human SC endothelial cells were isolated from donor eyes provided by Midwest Eye Bank (Ann Arbor, MI, USA), National Disease Research Interchange (Philadelphia, PA, USA), or Life Legacy (Tucson, AZ, USA) within 36 hours of death with enucleation occurring less than 6 hours after death (Table 1). Selective isolation and culture of the SC endothelial cells was processed according to the techniques previously established.28 All SC cell strains were characterized by (1) the expression of vascular endothelial cadherin, (2) a net transendothelial electrical resistance greater than or equal to 10 ohms·cm2, and (3) lack of myocilin induction upon treatment with dexamethasone. A total of eight strains of SC cells were isolated from four donors without glaucoma and three donors with glaucoma. All the SC cells were tested between the third and fifth passages. The determination of glaucoma was relied on documentation provided by eye banks with ocular hypertension/glaucoma history or presence of glaucoma treatment eye drops on patient medication list. All cells examined in the present study have been used in previous studies.29
Table 1.
Clinical Characteristics of Glaucoma and Control Donors
RNA Extraction
Total RNA was extracted using the Qiagen (Valencia, CA, USA) AllPrep Kit for both genomic DNA and total RNA isolation, following the manufacturer's protocol. Briefly, RNA was first harvested from cultured SC cells by adding the provided RLT Plus buffer and homogenizing using a QIAshredder spin column (Qiagen, Valencia, CA, USA). DNA was isolated and retained with AllPrep DNA Mini spin column (Qiagen). RNA was selectively isolated from the column flow through using an RNeasy mini spin column (Qiagen). Fifty microliters of RNase-free water was added to the spin column to recover RNA for each primary SC cell line. The quality and quantity of extracted total RNA was evaluated using the Agilent RNA 6000 Nano Kit with the Agilent Bioanalyzer 2100 system (Agilent, Santa Clara, CA, USA). Two hundred nanograms of total RNA from each SC cell line was used for Illumina BeadChip-based expression analysis (Illumina, San Diego, CA, USA) and the remaining RNA was used for droplet digital PCR (ddPCR) for expression validation.
Differential Expression Analysis
Illumina HumanWG-6 BeadChips were used to hybridize labeled RNA and scanned by Illumina iScan System following the manufacturer's protocol as previously described.30 The experiment was performed by the Molecular Genomics Core Facility at Duke Molecular Physiology Institute (Durham, NC, USA). Briefly, 200 ng of high quality RNA from all eight SC samples was labeled using the TotalPrep RNA Amplification kit (Illumina). Each sample was duplicated during the whole process. The labeled RNA was quantitated and 1.5 μg was hybridized to Illumina BeadChips overnight. The chips were scanned using the Illumina iScan system. Probe and intensity data were exported using Illumina's software GenomeStudio. The software used a detection P value to measure the probability of seeing a specific probe intensity level. Probes with detection P greater than or equal to 0.05 were excluded and probes were removed if they were present in fewer than half of samples of each group. The RNA expression data were imported into the R package limma in Bioconductor (http://www.bioconductor.org, in the public domain). All raw data were log2 transformed, and the quantile normalization function in limma was applied to normalize the intensity measurement among all eight samples as previously described. Data from duplicated samples were averaged after normalization.30 The glaucoma samples were compared with the control samples to identify differentially expressed genes (P ≤ 0.05) with at least 2-fold expression changes (up- or downregulated).
Pathway Analysis
First, gene ontology analysis was performed with all expressed genes in SC cells from cases or controls. The significantly expressed genes in each group were defined using detection P value. While a P value below 0.01 or 0.05 indicates the detection of an expressed transcript, a P value of less than 0.001 was used for those significantly expressed transcripts. Because the analysis focused on functional classification, unique gene IDs were used in the analysis, regardless of different probes for the same gene. These unique gene ID lists were imported into Protein Analysis THrough Evolutionary Relationships (PANTHER; in the public domain, http://www.pantherdb.org/) Classification System to examine the distribution of these genes in molecular function, biological process, cellular component, protein class, and pathway.31 The analysis was performed for cases and controls separately and the results were compared.
Second, the list of differentially expressed genes (P < 0.05 and fold change <0.5 or >2) was uploaded into WebGestalt (WEB-based GEne SeT AnaLysis Toolkit; in the public domain, http://bioinfo.vanderbilt.edu/webgestalt/), a gene ontology analysis website, to examine potentially enriched pathways or categories in molecular function, cellular component, and biological process.32 WebGestalt revealed a full picture of each class and where genes located, as well as how genes related in each class.
Validation of Differentially Expressed Genes
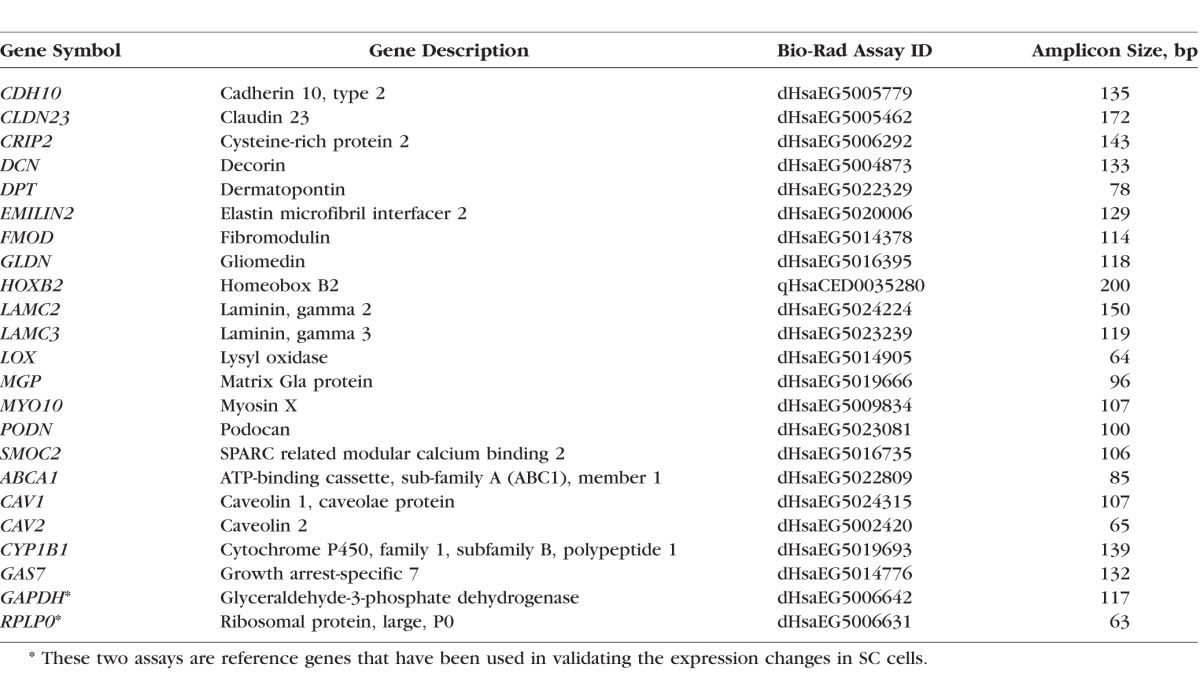
Approximately 300 to 500 ng total RNA extracted from each strain of SC cells was reverse transcribed to cDNA using SuperScript III first-strand synthesis kit from Life Technologies (Grand Island, NY, USA). The generated cDNA was diluted 8-fold and 1 μL cDNA was used in each ddPCR reaction. Droplet digital PCR is a precise digital PCR to measure absolute quantification of target cDNA sequences. QX200 droplet digital PCR system from Bio-Rad (Hercules, CA, USA) was used for the validation experiments. Predesigned and experimentally-tested Bio-Rad PrimerPCR/ddPCR Expression EvaGreen assays were selected to validate the differential expression identified from Illumina BeadChip analysis (Table 2). If possible, the selected assays were required to have amplicons spanning an intron to reduce artifact from potential DNA contamination. If two assays for the same gene were available, the one with relatively larger amplicon was chosen for its stable property.
Table 2.
Selected Droplet Digital PCR Gene Expression Assays From Bio-Rad
After the preparation of the reaction mix using QX200 ddPCR EvaGreen Supermix from Bio-Rad, a Bio-Rad QX200 droplet generator was used to partition each prepared sample into up to 20,000 nanoliter-sized droplets. Generated ddPCR droplets were transferred to a clean 96-well plate and sealed with the PX1 PCR plate sealer from Bio-Rad prior to PCR amplification using Bio-Rad C1000 touch thermal cycler with deep wells. The parameters for PCR were: 95°C for 10 minutes to activate the polymerase, followed by 40 cycles of 94°C for 30 seconds and 60°C for 1 minute, ended with 98°C for 10 minutes to deactivate the polymerase. After thermal cycling, ddPCR droplets from each reaction were read and quantified by the QX200 droplet reader using QuantaSoft software from Bio-Rad. Expression data was extracted and analyzed using QuantaSoft software. For quality control, any PCR reactions with less than 10,000 droplets were removed. Each assay with each sample was run in duplicate. Two reference assays were integrated in our experiment: glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ribosomal protein, large, P0 (RPLP0). The absolute number of cDNA copies for each selected gene was derived from ddPCR and normalized to the average number of copies of GAPDH and RPLP0 in each SC sample. The fold change of normalized gene expression in cases versus controls was analyzed for each selected gene using Student's t-test. A P value of less than 0.05 was considered significant.
Results
Clinical Phenotypes
This study included four cell strains isolated from four donor eyes without a history of eye disease and four cell strains from four eyes of three donors (one donor provided both eyes) with glaucoma. Although two glaucomatous eyes were from the same donor, the severity of glaucoma seems to be different as indicated by the different level of stiffness in previous study.33 Our results were consistent with this published finding, showing that expression-based correlation analysis among all eight SC cell strains did not indicate significantly higher similarity between the two SC cells from the same donor than other paired-sample comparisons (data not shown here). The average age at death from nonglaucoma individuals was 53 years while the average age at death from glaucoma-affected individuals was 75 years. The average postmortem delay in glaucoma group was approximately 4 hours, ranging from 2 to 5 hours. The average postmortem delay in nonglaucoma control was over 10 hours, ranging from 7 to 14 hours. Detailed information is listed in Table 1. The stiffness in glaucoma-affected SC lines was noted to be significantly elevated compared with nonglaucoma SC lines using multiple approaches to measure the stiffness.33
Gene Ontology Analysis
Based on the detection P value, our expression analysis using Illumina BeadChips identified 8193 highly expressed genes in all four glaucomatous SC cells and 8508 highly expressed genes in all four nonglaucomatous SC cells. These two lists of expressed genes were uploaded into the PANTHER classification system for molecular function-based gene ontology analysis (Fig.). Genes were categorized into multiple different functional groups, including binding, catalytic activity, enzyme regulator, nucleic acid binding transcription factor activity, protein binding transcription factor activity, receptor activity, structure molecule activity, transporter activity, and others. The percentage of genes in each main category and subcategory was similar between glaucoma and nonglaucoma SC cell lines, with less than 0.3% overall difference. This similarity indicates the lack of significant categorical changes in glaucomatous SC endothelial cells in terms of molecular function. We also performed gene ontology analyses with biological process, cellular component, and protein class and pathway and did not identify significant categorical expression shifts in glaucomatous SC cells in comparison to nonglaucoma SC cells (Supplementary Fig. S1).
This expression analysis identified a total of 113 genes that were differentially expressed in glaucomatous SC cells with at least two fold changes compared with control SC cells (Supplementary Table S1). Fifty-one genes were upregulated and 62 genes were downregulated in glaucomatous SC cells. The list of these 113 genes was further analyzed for significantly enriched pathways using the online toolkit WebGestalt in terms of biological process, molecular function, and cellular component. Pathway analysis indicated significant enrichment of genes in cell adhesion, heparin binding, glycosaminoglycan binding, oxidoreductase activity, filopodium, and extracellular matrix with P values from 2.3 × 10−6 to 1 × 10−2 (Supplementary Fig. S2, indicated with the red color). A number of genes contribute to more than one of these functional categories, such as cell adhesion, glycosaminoglycan binding, and extracellular matrix (Supplementary Material).
Expression Validation With ddPCR
To validate the differential expression from Illumina BeadChip, based on their potential involvement in extracellular matrix regulation, cell adhesion, glycosaminoglycan binding, and glaucoma, we selected and validated the differential expression of 16 genes using ddPCR successfully (Table 3). We considered a P value less than 0.05 to be significant. Results from ddPCR confirmed the differential expression of these 16 genes, providing confidence in the Illumina BeadChip data. We also examined the potential differential expression of known glaucoma-associated genes. These genes included CDKN2B-AS1, TMCO1, SIX1, SIX6, CAV1, CAV2, LRP12, ZFPM2, GAS7, ATOH7, FNDC3B, CYP1B1, MYOC, OPTN, WDR36, SRBD1, TBK1, GALC, ABCA1, PMM2, AFAP1, and GMDS (Supplementary Table S2). Based on suggestive results showing differential expression in glaucomatous SC cells in Illumina BeadChips experiments, we selected five genes for further validation with ddPCR assays, including ABCA1, CAV1, CAV2, CYP1B1, and GAS7. Droplet digital PCR experiments identified and validated the differential expression of two genes CYP1B1 and ABCA1 (Table 4). Although significant in microarray experiment, the expression changes of CAV1 and CAV2 were near, but did not reach statistical significance (P = 0.07 and 0.08, respectively). These data indicate the potential role of these four genes in the function of SC cells with aqueous outflow.
Table 3.
List of Selected Differentially Expressed Genes That Were Validated Using Droplet Digital PCR

Table 4.
List of Expression of Glaucoma-Associated Genes in Glaucomatous SC Endothelial Cells in Comparison With Nonglaucoma SC Cells

Discussion
This study represents the first comprehensive differential expression analysis in primary human SC endothelial cells derived from glaucoma-affected and nonglaucoma individuals. A unique feature of this study is the biomechanical characterization of these established SC cells, elevated cell stiffness, reduced cell strain, and decreased propensity to form pores in glaucomatous SC cells.24 Using microarray and ddPCR, a number of differentially expressed genes have been identified in glaucomatous SC cells that may be related with the elevated stiffness, thus potentially increased outflow resistance and elevated IOP. This foundational information provides candidates to explore and advances the understanding on how SC endothelial cells contribute to the regulation of aqueous outflow plus potential targets that may regulate IOP by affecting the stiffness of SC cells.
The differentially expressed genes are enriched in several pathways, including cell adhesion, heparin binding, glycosaminoglycan binding, and extracellular matrix, all of which may contribute to cell stiffness status and decreased pore-forming ability in glaucomatous SC cells.33–36 In comparison to trabecular meshwork tissues,30 glaucomatous SC cells shared similar expression changes in genes involved with extracellular matrix remodeling and cell adhesion. Schlemm's canal cells have more changes in genes involved in glycosaminoglycan binding, oxidoreductase activity, and heparin binding while TM cells have more changes in genes involved with signal peptides, glycoproteins, and exosome-related components. We confirmed a previously reported lower expression of DCN (decorin) in glaucomatous SC cells with over 2-fold reduction.33 Interestingly, the expression of DCN may be further regulated by environmental factors, such as the elevated substrate stiffness of juxtacanalicular tissue, only in glaucomatous but not normal SC cells. It will be important to study the underlying regulatory mechanisms related with the expression of DCN, especially in glaucomatous SC cells. Another gene, lysyl oxidase (LOX), a cross-linking enzyme, results in resistance to matrix degradation and increased tissue stiffness.37 The significantly elevated expression of LOX in glaucomatous SC cells indicates its potential role in regulating SC cell stiffness. The observed expression alterations may represent a glaucoma trigger, or a secondary consequence of the medical treatment of glaucoma. Further study is needed to clarify the sequence of alteration.
Our study also examined the potential for differential expression of glaucoma, or IOP-associated genes. Our ddPCR experiment validated the significantly reduced expression of two genes, CYP1B1 and ABCA1 in glaucomatous SC cells. Mutations in CYP1B1 have been known to cause primary congenital glaucoma, juvenile open-angle glaucoma, and POAG.2 Variants in and near ABCA1 have been associated with elevated risk of POAG and IOP.8–10,19,20 Both CYP1B1 and ABCA1 are involved in synthesis and transportation of lipids.38,39 Reduced expression of these two genes in glaucomatous SC cells might lead to the alteration of phospholipids involved in signaling pathways regulating the stiffness of SC cells, including lipid raft domains. In fact, the expression change of residents of lipid rafts such as CAV1/CAV2 was significant, but was marginal in ddPCR validation. Variants in the CAV1/CAV2 locus are associated with elevated IOP.40,41 Knockdown of caveolin expression elevated IOP in mice and altered conventional outflow in organ-cultured human eyes or perfused mouse eyes42 (Elliott MH, et al. IOVS 2014;55:ARVO E-Abstract 2888). It will be necessary to further study the protein level of caveolin in these SC cells. It will be interesting to know whether the expression of these four genes are caused by glaucoma or their reduced expression may contribute to the elevated IOP and glaucoma. Mouse models of these glaucoma-related genes may provide more helpful information about their specific roles in glaucoma. The knowledge of how the expression of these genes is regulated will significantly improve our understanding of their involvement in regulating SC cell stiffness and IOP potentially.
An interesting aspect of our study is the successful integration of ddPCR to validate gene expression. Droplet digital PCR is a relatively novel technology that measures the absolute copy number of DNA/RNA templates by dividing a 20 μL PCR reaction mixture into 20,000 droplets followed by end-point detection of the amplification for each individual droplet. Compared with traditional realtime PCR, ddPCR significantly increases the accuracy of quantification at low-target concentration.43 Although ddPCR does not need to use reference gene assays, two widely used reference assays (GAPDH and RPLP0) were integrated to normalize the varying amounts of starting RNA materials from different SC samples. The absolute quantification of target genes using ddPCR made the experimental validation much easier and more accurate. More application of this novel technology is expected in other expression profiling studies.
This study was limited by several factors. First, this study used primary cultures of SC cells instead of the intact tissue. While cells were a confluent monolayer for approximately 7 weeks before isolation of RNA, cell culture will certainly alter the expression of some genes; however, both cases and controls were handled identically. Second, all the donated glaucoma eyes were under medical treatment, which may have resulted in expression changes in the isolated primary SC cells. Third, while the most comprehensive study to date, the sample size is relatively small. This study included only eight SC strains of cells derived from three individuals with glaucoma and four individuals without glaucoma. This small sample size is due to the difficulty of obtaining viable postmortem eyes affected with glaucoma and deriving primary SC cells from these rare specimens. Although it is not easy, it will be critical to collect more postmortem eye tissues from individuals with glaucoma. It will be necessary to replicate the identified expression alterations in other independent studies with larger sample size. If possible, it will be ideal to match glaucoma cases and controls in terms of sex, age, and ethnicity.
In summary, for the first time, this study has successfully characterized the functional classification of all expressed genes in SC cells from nonglaucoma and glaucoma-affected subjects. The identification and validation of underlying related gene expression changes in primary glaucomatous SC cells will provide novel information for investigating the possible mechanisms underlying the biomechanical property changes in these SC cells and generate a potential list of targets regulating the SC cell stiffness and pore formation, eventually the outflow resistance in glaucoma individuals.
Figure.
Gene ontology analysis using PANTHER with molecular function for all highly expressed genes (detection P < 0.012) in nonglaucoma control or glaucoma cases, using Illumina BeadChip data.
Supplementary Material
Acknowledgments
The authors thank all the individuals who donated their eye tissues for this project. Without their support, this study would not be feasible. The authors also thank the genomic service by the Molecular Genomics Core facility at Duke Molecular Physiology Institute for performing the Illumina BeadChip experiment.
Supported by grants from The Glaucoma Research Foundation, The Glaucoma Foundation, BrightFocus Foundation, Research to Prevent Blindness (New York, NY, USA), National Institutes of Health/National Eye Institute P30 EY005722, EY019696 (Bethesda, MD, USA).
Disclosure: J. Cai, None; K.M. Perkumas, None; X. Qin, None; M.A. Hauser, None; W.D. Stamer, None; Y. Liu, None
References
- 1. Liu Y,, Allingham RR. Genetics of glaucoma. : Encyclopeida of Life Sciences (ELS). Chichelster: John Wiley & Sons Ltd.; 2010: doi:http://dx.doi.org/10.1002/9780470015902.a0021429. [Google Scholar]
- 2. Liu Y,, Allingham RR. Molecular genetics in glaucoma. Exp Eye Res. 2011; 93: 331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tham YC,, Li X,, Wong TY,, Quigley HA,, Aung T,, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014; 121: 2081–2090. [DOI] [PubMed] [Google Scholar]
- 4. Kwon YH,, Fingert JH,, Kuehn MH,, Alward WL. Primary open-angle glaucoma. N Engl J Med. 2009; 360: 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stone EM,, Fingert JH,, Alward WL,, et al. Identification of a gene that causes primary open angle glaucoma. Science. 1997; 275: 668–670. [DOI] [PubMed] [Google Scholar]
- 6. Stoilov I,, Akarsu AN,, Sarfarazi M. Identification of three different truncating mutations in cytochrome P4501B1 (CYP1B1) as the principal cause of primary congenital glaucoma (Buphthalmos) in families linked to the GLC3A locus on chromosome 2p21. Hum Mol Genet. 1997; 6: 641–647. [DOI] [PubMed] [Google Scholar]
- 7. Thorleifsson G,, Walters GB,, Hewitt AW,, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010; 42: 906–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gharahkhani P,, Burdon KP,, Fogarty R,, et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014; 46: 1120–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hysi PG,, Cheng CY,, Springelkamp H,, et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014; 46: 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y,, Lin Y,, Vithana EN,, et al. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat Genet. 2014; 46: 1115–1119. [DOI] [PubMed] [Google Scholar]
- 11. Burdon KP,, Macgregor S,, Hewitt AW,, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011; 43: 574–578. [DOI] [PubMed] [Google Scholar]
- 12. van Koolwijk LM,, Ramdas WD,, Ikram MK,, et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012; 8: e1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wiggs JL,, Yaspan BL,, Hauser MA,, et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012; 8: e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cooke Bailey JN,, Sobrin L,, Pericak-Vance MA,, Haines JL,, Hammond CJ,, Wiggs JL. Advances in the genomics of common eye diseases. Hum Mol Genet. 2013; 22: R59–R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Allingham RR,, Liu Y,, Rhee DJ. The genetics of primary open-angle glaucoma: a review. Exp Eye Res. 2009; 88: 837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Y,, Garrett ME,, Yaspan BL,, et al. DNA copy number variants of known glaucoma genes in relation to primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2014; 55: 8251–8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fingert JH. Primary open-angle glaucoma genes. Eye (Lond). 2011; 25: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Y,, Gibson J,, Wheeler J,, et al. GALC deletions increase the risk of primary open-angle glaucoma: the role of Mendelian variants in complex disease. PLoS One. 2011; 6: e27134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hysi PG,, Cheng CY,, Springelkamp H,, et al. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014; 46: 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Springelkamp H,, Iglesias AI,, Cuellar-Partida G,, et al. ARHGEF12 influences the risk of glaucoma by increasing intraocular pressure. Hum Mol Genet. 2015; 24: 2689–2699. [DOI] [PubMed] [Google Scholar]
- 21.Braunger BM,, Fuchshofer R,, Tamm ER.The aqueous humor outflow pathways in glaucoma: a unifying concept of disease mechanisms and causative treatment [ Eur J Pharm Biopharm. 2015]. published online ahead of print May 7, . doi: http://dx.doi.org/10.1016/j.ejpb.2015.04.029. [DOI] [PubMed]
- 22. Allingham RR,, Shields MB. Shields' textbook of glaucoma. 6th ed. Philadelphia: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2011. [Google Scholar]
- 23. Allingham RR,, de Kater AW,, Ethier CR. Schlemm's canal and primary open angle glaucoma: correlation between Schlemm's canal dimensions and outflow facility. Exp Eye Res. 1996; 62: 101–109. [DOI] [PubMed] [Google Scholar]
- 24. Hann CR,, Vercnocke AJ,, Bentley MD,, Jorgensen SM,, Fautsch MP. Anatomic changes in Schlemm's canal and collector channels in normal and primary open-angle glaucoma eyes using low and high perfusion pressures. Invest Ophthalmol Vis Sci. 2014; 55: 5834–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson M,, Chan D,, Read AT,, Christensen C,, Sit A,, Ethier CR. The pore density in the inner wall endothelium of Schlemm's canal of glaucomatous eyes. Invest Ophthalmol Vis Sci. 2002; 43: 2950–2955. [PubMed] [Google Scholar]
- 26. Allingham RR,, de Kater AW,, Ethier CR,, Anderson PJ,, Hertzmark E,, Epstein DL. The relationship between pore density and outflow facility in human eyes. Invest Ophthalmol Vis Sci. 1992; 33: 1661–1669. [PubMed] [Google Scholar]
- 27. Russell P,, Johnson M. Elastic modulus determination of normal and glaucomatous human trabecular meshwork. Invest Ophthalmol Vis Sci. 2012; 53: 117. [DOI] [PubMed] [Google Scholar]
- 28. Stamer WD,, Roberts BC,, Howell DN,, Epstein DL. Isolation, culture, and characterization of endothelial cells from Schlemm's canal. Invest Ophthalmol Vis Sci. 1998; 39: 1804–1812. [PubMed] [Google Scholar]
- 29. Perkumas KM,, Stamer WD. Protein markers and differentiation in culture for Schlemm's canal endothelial cells. Exp Eye Res. 2012; 96: 82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y,, Allingham RR,, Qin X,, et al. Gene expression profile in human trabecular meshwork from patients with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2013; 54: 6382–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mi H,, Muruganujan A,, Casagrande JT,, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013; 8: 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang J,, Duncan D,, Shi Z,, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013; 41: W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Overby DR,, Zhou EH,, Vargas-Pinto R,, et al. Altered mechanobiology of Schlemm's canal endothelial cells in glaucoma. Proc Natl Acad Sci U S A. 2014; 111: 13876–13881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pattabiraman PP,, Maddala R,, Rao PV. Regulation of plasticity and fibrogenic activity of trabecular meshwork cells by Rho GTPase signaling. J Cell Physiol. 2014; 229: 927–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang M,, Maddala R,, Rao PV. Novel molecular insights into RhoA GTPase-induced resistance to aqueous humor outflow through the trabecular meshwork. Am J Physiol Cell Physiol. 2008; 295: C1057–C1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stamer WD,, Braakman ST,, Zhou EH,, et al. Biomechanics of Schlemm's canal endothelium and intraocular pressure reduction. Prog Retin Eye Res. 2015; 44: 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wordinger RJ,, Sharma T,, Clark AF. The role of TGF-beta2 and bone morphogenetic proteins in the trabecular meshwork and glaucoma. J Ocul Pharmacol Ther. 2014; 30: 154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mookherjee S,, Acharya M,, Banerjee D,, Bhattacharjee A,, Ray K. Molecular basis for involvement of CYP1B1 in MYOC upregulation and its potential implication in glaucoma pathogenesis. PLoS One. 2012; 7: e45077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmitz G,, Langmann T. Structure, function and regulation of the ABC1 gene product. Curr Opin Lipidol. 2001; 12: 129–140. [DOI] [PubMed] [Google Scholar]
- 40. Kim S,, Kim K,, Heo DW,, et al. Expression-associated polymorphisms of CAV1-CAV2 affect intraocular pressure and high-tension glaucoma risk. Mol Vis. 2015; 21: 548–554. [PMC free article] [PubMed] [Google Scholar]
- 41. Chen F,, Klein AP,, Klein BE,, et al. Exome array analysis identifies CAV1/CAV2 as a susceptibility locus for intraocular pressure. Invest Ophthalmol Vis Sci. 2015; 56: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aga M,, Bradley JM,, Wanchu R,, Yang YF,, Acott TS,, Keller KE. Differential effects of caveolin-1 and -2 knockdown on aqueous outflow and altered extracellular matrix turnover in caveolin-silenced trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2014; 55: 5497–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gutierrez-Aguirre I,, Racki N,, Dreo T,, Ravnikar M. Droplet digital PCR for absolute quantification of pathogens. Methods Mol Biol. 2015; 1302: 331–347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



