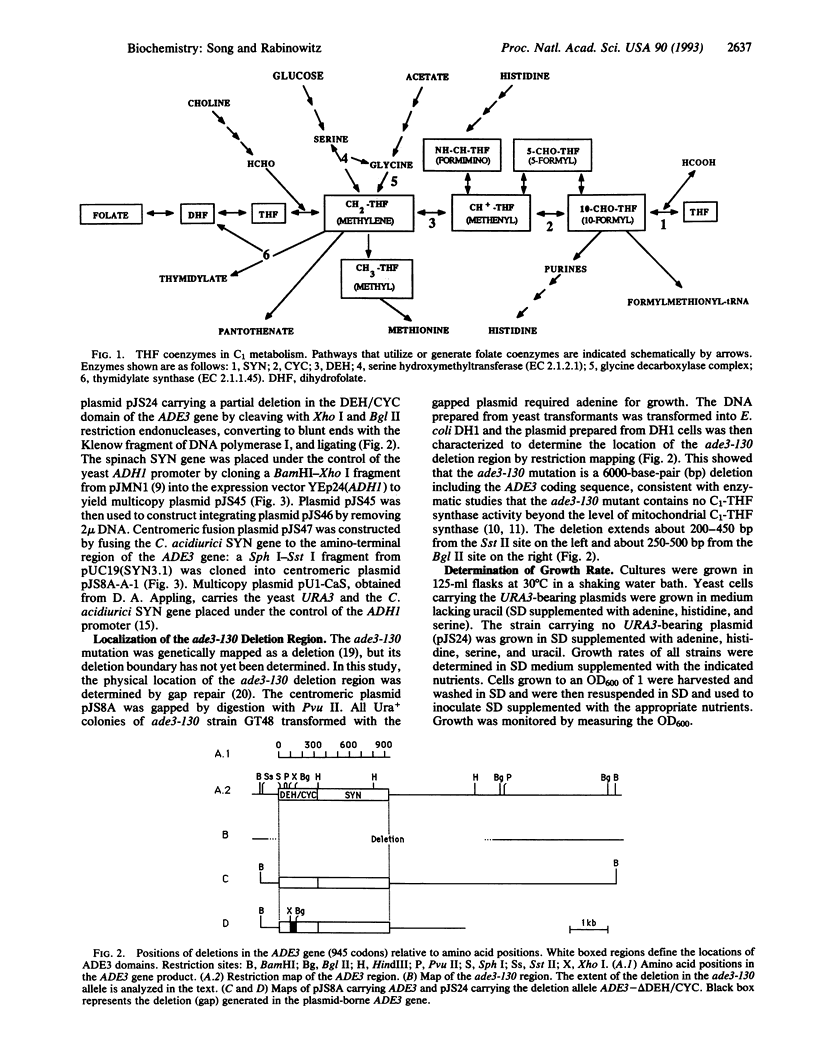
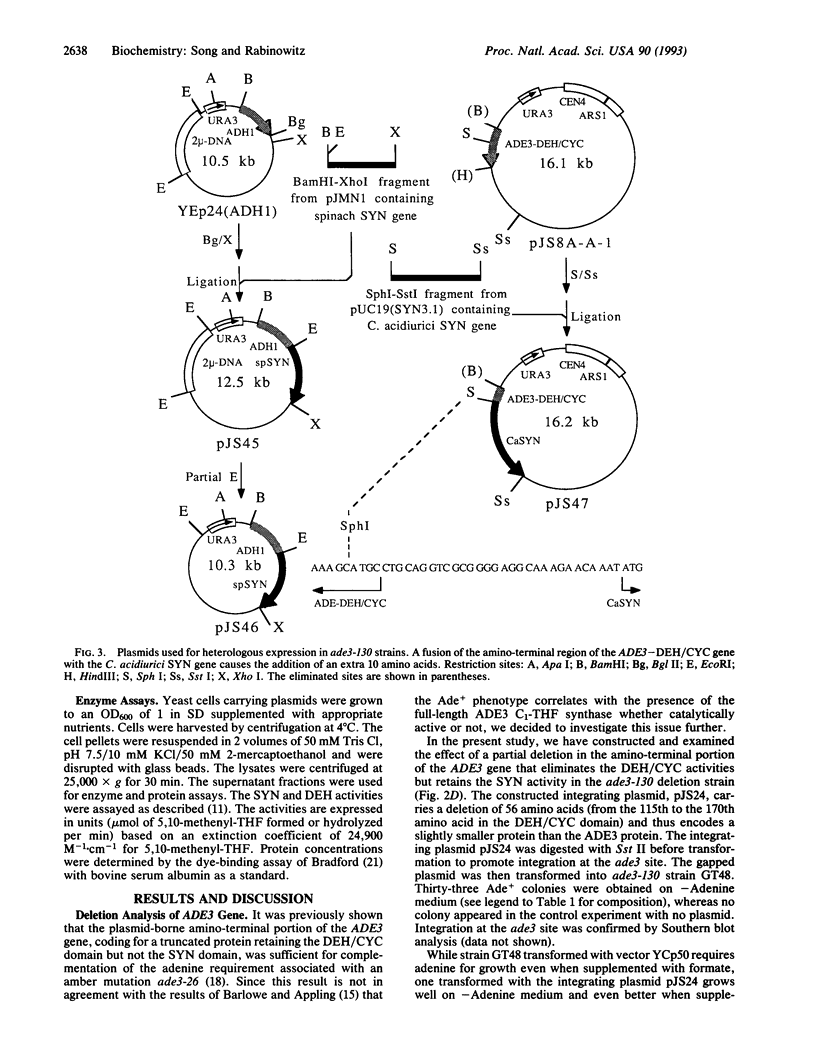
Abstract
The protein product of the ADE3 gene of the yeast Saccharomyces cerevisiae has been identified as the cytoplasmic trifunctional C1-tetrahydrofolate (THF) synthase, which possesses 10-formyl-THF synthetase (EC 6.3.4.3), 5,10-methenyl-THF cyclohydrolase (EC 3.5.4.9), and 5,10-methylene-THF dehydrogenase (EC 1.5.1.5) activities. However, it has been suggested that the ADE3-encoded C1-THF synthase does not play a role in providing the enzymes involved in the generation of one-carbon intermediates in the biosynthesis of the purine bases but functions in maintaining the structural integrity of the enzyme complex involved in purine biosynthesis [Barlowe, C. K. & Appling, D. A. (1990) Mol. Cell. Biol. 10, 5679-5687]. This hypothesis is based on their finding that the presence of the full-length ADE3 C1-THF synthase, whether catalytically active or not, is correlated with the Ade+ phenotype. In contrast to their results, our deletion analysis of the ADE3 gene indicates that the presence of either the synthetase or dehydrogenase/cyclohydrolase domains of C1-THF synthase is enough to complement the adenine requirement in ade3 strains. These results are also consistent with those obtained in heterologous expression of spinach and Clostridium acidiurici monofunctional synthetases in ade3 strains. Heterologous expression studies show that the high synthetase activity may be correlated with the increased growth in medium lacking adenine. These results suggest that the catalytic activity of the C1-THF synthase is involved in purine biosynthesis.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barlowe C. K., Appling D. R. Molecular genetic analysis of Saccharomyces cerevisiae C1-tetrahydrofolate synthase mutants reveals a noncatalytic function of the ADE3 gene product and an additional folate-dependent enzyme. Mol Cell Biol. 1990 Nov;10(11):5679–5687. doi: 10.1128/mcb.10.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Curthoys N. P., Rabinowitz J. C. Formyltetrahydrofolate synthetase. Binding of folate substrates and kinetics of the reverse reaction. J Biol Chem. 1972 Apr 10;247(7):1965–1971. [PubMed] [Google Scholar]
- Curthoys N. P., Scott J. M., Rabinowitz J. C. Folate coenzymes of Clostridium acidi-urici. The isolation of (l)-5,10-methenyltetrahydropteroyltriglutamate, its conversion to (l)-tetrahydropteroyltriglutamate and (l)-10-( 14 C)formyltetrahydropteroyltriglutamate, and the synthesis of (l)-10-formyl-(6,7- 3 H 2 )tetrahydropteroyltriglutamate and (l)-(6,7- 3 H 2 )tetrahydropteroyltriglutamate. J Biol Chem. 1972 Apr 10;247(7):1959–1964. [PubMed] [Google Scholar]
- Dev I. K., Harvey R. J. A complex of N5,N10-methylenetetrahydrofolate dehydrogenase and N5,N10-methenyltetrahydrofolate cyclohydrolase in Escherichia coli. Purification, subunit structure, and allosteric inhibition by N10-formyltetrahydrofolate. J Biol Chem. 1978 Jun 25;253(12):4245–4253. [PubMed] [Google Scholar]
- Green J. M., MacKenzie R. E., Matthews R. G. Substrate flux through methylenetetrahydrofolate dehydrogenase: predicted effects of the concentration of methylenetetrahydrofolate on its partitioning into pathways leading to nucleotide biosynthesis or methionine regeneration. Biochemistry. 1988 Oct 18;27(21):8014–8022. doi: 10.1021/bi00421a007. [DOI] [PubMed] [Google Scholar]
- Hum D. W., Bell A. W., Rozen R., MacKenzie R. E. Primary structure of a human trifunctional enzyme. Isolation of a cDNA encoding methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase. J Biol Chem. 1988 Nov 5;263(31):15946–15950. [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. W. Bipartite structure of the ade3 locus of Saccharomyces cerevisiae. Genetics. 1977 Feb;85(2):209–223. doi: 10.1093/genetics/85.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzbach C., Stokstad E. L. Partial purification of a 10-formyl-tetrahydrofolate: NADP oxidoreductase from mammalian liver. Biochem Biophys Res Commun. 1968 Jan 25;30(2):111–117. doi: 10.1016/0006-291x(68)90456-7. [DOI] [PubMed] [Google Scholar]
- Lovell C. R., Przybyla A., Ljungdahl L. G. Primary structure of the thermostable formyltetrahydrofolate synthetase from Clostridium thermoaceticum. Biochemistry. 1990 Jun 19;29(24):5687–5694. doi: 10.1021/bi00476a007. [DOI] [PubMed] [Google Scholar]
- McKenzie K. Q., Jones E. W. Mutants of formyltetrahydrofolate interconversion pathway of Saccharomyces cerevisiae. Genetics. 1977 May;86(1):85–102. doi: 10.1093/genetics/86.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour J. M., Rabinowitz J. C. Isolation, characterization, and structural organization of 10-formyltetrahydrofolate synthetase from spinach leaves. J Biol Chem. 1991 Sep 25;266(27):18363–18369. [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Paukert J. L., Williams G. R., Rabinowitz J. C. Formyl-methenyl-methylenetetrahydrofolate synthetase (combined); correlation of enzymic activities with limited proteolytic degradation of the protein from yeast. Biochem Biophys Res Commun. 1977 Jul 11;77(1):147–154. doi: 10.1016/s0006-291x(77)80176-9. [DOI] [PubMed] [Google Scholar]
- Rios-Orlandi E. M., Zarkadas C. G., MacKenzie R. E. Formyltetrahydrofolate dehydrogenase-hydrolase from pig liver: simultaneous assay of the activities. Biochim Biophys Acta. 1986 May 12;871(1):24–35. doi: 10.1016/0167-4838(86)90129-9. [DOI] [PubMed] [Google Scholar]
- Schirch V., Strong W. B. Interaction of folylpolyglutamates with enzymes in one-carbon metabolism. Arch Biochem Biophys. 1989 Mar;269(2):371–380. doi: 10.1016/0003-9861(89)90120-3. [DOI] [PubMed] [Google Scholar]
- Shannon K. W., Rabinowitz J. C. Isolation and characterization of the Saccharomyces cerevisiae MIS1 gene encoding mitochondrial C1-tetrahydrofolate synthase. J Biol Chem. 1988 Jun 5;263(16):7717–7725. [PubMed] [Google Scholar]
- Song J. M., Liebman S. W. Mutations in ADE3 reduce the efficiency of the omnipotent suppressor sup45-2. Curr Genet. 1989 Dec;16(5-6):315–321. doi: 10.1007/BF00340709. [DOI] [PubMed] [Google Scholar]
- Staben C., Rabinowitz J. C. Nucleotide sequence of the Saccharomyces cerevisiae ADE3 gene encoding C1-tetrahydrofolate synthase. J Biol Chem. 1986 Apr 5;261(10):4629–4637. [PubMed] [Google Scholar]
- Taylor R. T., Hanna M. L. Folate-dependent enzymes in cultured Chinese hamster cells: folypolyglutamate synthetase and its absence in mutants auxotrophic for glycine + adenosine + thymidine. Arch Biochem Biophys. 1977 May;181(1):331–334. doi: 10.1016/0003-9861(77)90512-4. [DOI] [PubMed] [Google Scholar]
- Thigpen A. E., West M. G., Appling D. R. Rat C1-tetrahydrofolate synthase. cDNA isolation, tissue-specific levels of the mRNA, and expression of the protein in yeast. J Biol Chem. 1990 May 15;265(14):7907–7913. [PubMed] [Google Scholar]
- Whitehead T. R., Park M., Rabinowitz J. C. Distribution of 10-formyltetrahydrofolate synthetase in eubacteria. J Bacteriol. 1988 Feb;170(2):995–997. doi: 10.1128/jb.170.2.995-997.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead T. R., Rabinowitz J. C. Nucleotide sequence of the Clostridium acidiurici ("Clostridium acidi-urici") gene for 10-formyltetrahydrofolate synthetase shows extensive amino acid homology with the trifunctional enzyme C1-tetrahydrofolate synthase from Saccharomyces cerevisiae. J Bacteriol. 1988 Jul;170(7):3255–3261. doi: 10.1128/jb.170.7.3255-3261.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]


