Abstract
Tumor growth cascade is a complicated and multistep process with numerous obstacles. Until recently, evidences have shown the involvement of microRNAs (miRNAs) in tumorigenesis and tumor progression of various cancers, including colorectal cancer (CRC). In this study, we explored the role of miR-194 and its downstream pathway in CRC. We acquired data through miRNA microarray profiles, showing that the expression of miR-194 was significantly suppressed in CRC tissues compared with corresponding noncancerous tissues. Decreased miR-194 expression was obviously associated with tumor size and tumor differentiation, as well as TNM stage. Both Kaplan–Meier and multivariate survival analysis showed that downregulated miR-194 was associated with overall survival. Moreover, functional assays indicated that overexpression of miR-194 in CRC cell lines inhibited cell proliferation both in vitro and in vivo. In addition, using dual-luciferase reporter gene assay, we found MAP4K4 was the direct target of miR-194. Silencing of MAP4K4 resulted in similar biological behavior changes to that of overexpression of miR-194. We also observed through Human Gene Expression Array that MDM2 was one of the downstream targets of MAP4K4. Knockdown of MAP4K4 downregulated MDM2 expression through transcription factor c-Jun binding to the −1063 to -1057 bp of the promoter. These results suggest that miR-194, regulating the MAP4K4/c-Jun/MDM2 signaling pathway, might act as a tumor suppressor and serve as a novel target for CRC prevention and therapy.
Keywords: apoptosis, c-Jun, colorectal cancer, miR-194, MAP4K4, MDM2, proliferation
Introduction
Colorectal cancer (CRC) is the third most common form of cancer and the third leading cause of cancer death in men and women, with increased morbidity and mortality in recent years.1 High priority must be given to understanding the mechanisms of occurrence and development of CRC and identifying novel diagnostics and effective therapeutics.2,3 CRC develops through progressive accumulation of mutations in oncogenes and tumor-suppressor genes.4 Targeting driver pathways represent the best option to tailor cancer treatment and improve the overall survival in patients with advanced cancer.5
MicroRNAs (miRNAs) are small non-coding RNAs that bind to partially complementary recognition sequences in the 3′-untranslated region (3’UTR) of mRNA, causing degradation or inhibition of translation resulting in gene silencing.6,7 Recently, numerous miRNAs that were aberrantly expressed in cancers were identified, and their deregulation was linked to cancer progression and clinical outcome.8,9 Recent data have shown that miR-194 plays an important part in tumorigenesis. For example, miR-194 was found to act as a tumor suppressor in gastric cancer,10 endometrial cancer,11 renal cell carcinoma,12 lung cancer,13 and breast cancer,14 while upregulated as an oncogene in pancreatic cancer.15 In CRC, miR-194 has been reported to be downregulated in colon tissues 16 and can predict adenoma recurrence.17 Sundaram18 found that miR-194 could promote angiogenesis and facilitate tissue repair by targeting thrombospondin-1. However, the functions and mechanisms of miR-194 involved in CRC are not fully elucidated and the therapeutic application of miR-194 has not been reported.
Since a single miRNA can target hundreds of mRNAs, TargetScan, an online software for prediction of miRNA targets (http://www.targetscan.org/), was applied to predict the downstream target genes of miR-194. The result shows that MAP4K4 is a potential target of miR-194. MAPK kinase kinase kinase isoform 4 (MAP4K4) is a relatively novel gene belonging to the mammalian STE20/MAP4K family. It has been reported as an oncogene in multiple forms of cancer19 with poor prognosis, such as prostate cancer,20 lung cancer,21,22 hepatocellular cancer,21 and pancreatic cancer.23 However, little is known about the functions of MAP4K4 and not yet been fully clarified in CRC.
For further clarifying the mechanism of miR-194 and MAP4K4 in tumorigenesis of CRC, we used genome-wide expression microarray to explore the changed downstream target. Interestingly, MDM2 was found to be significantly downregulated after knockdown of MAP4K4. MDM2 oncogene promotes tumor formation by targeting tumor suppressor proteins such as p53.24,25 Accumulating evidence indicated that downregulation of MDM2 through various approaches resulted in repressing tumor growth and metastasis,26-28 including CRC.29,30
MAP4K4 is also involved in activation of c-Jun N-terminal kinase (JNK) pathway.31 C-Jun, the substrate of JNK, induced expression by phosphorylation of JNK.32 C-Jun was usually reported as a transcription factor to regulate multiple downstream genes that function as tumor suppressors33 or oncogenes34 in cancers. Therefore, the prediction software for transcription factors was applied to find whether c-Jun was one of the transcription factors regulating MDM2 expression.
In this study, we hypothesize that miR-194 suppresses tumorigenesis of CRC through regulating MAP4K4/c-Jun/MDM2 signaling pathway.
Results
Identification of ectopic miRNAs expression in CRC tissue samples
MiRNA microarray (Agilent) data (Fig. 1) analyzed between CRC and CRN tissues showed that a total of 9 downregulated miRNAs and 26 upregulated miRNAs in CRC tissue samples were screened out by a 2.5-fold change with a significant difference (Table 1, P < 0.05). Of these miRNAs, miR-135b showed the largest degree of upregulation whereas miR-139 was noted for the greatest changes of decreased expression (>2.5-fold).
Figure 1.
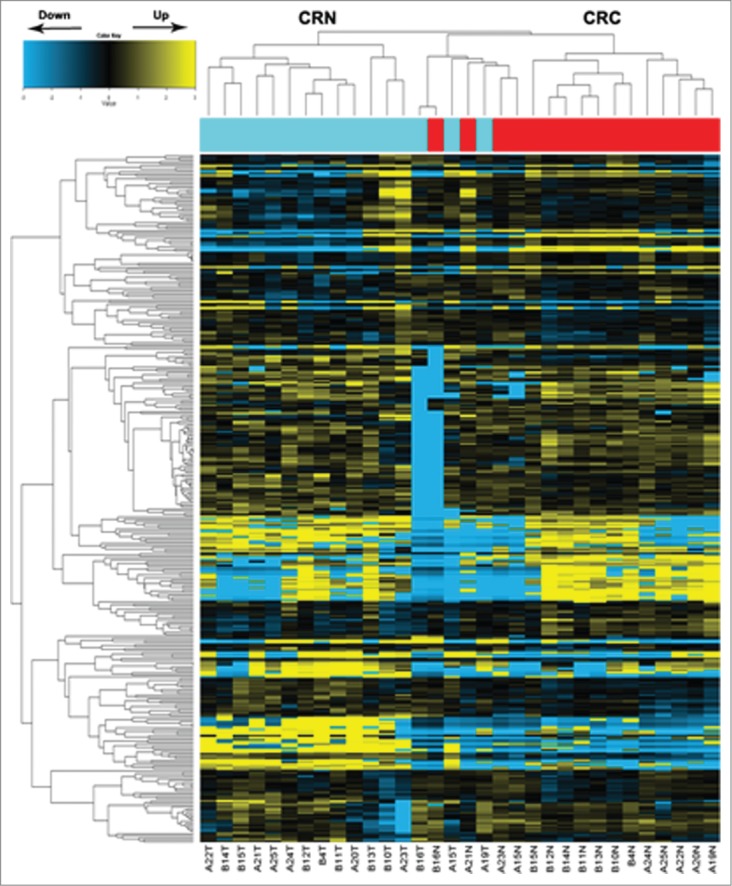
MicroRNA array analysis of miRNA expression in colorectal tissues. Hierarchical clustering of the expression values for mature miRNAs of CRN versus CRC tissues. Yellow indicates high relative expression, and blue indicates low relative expression. CRC: colorectal cancer tissues; CRN: matched adjacent noncancerous colorectal tissues.
Table 1.
MiRNAs expressed differentially in colorectal cancer tissues (CRC) compared with colorectal normal tissues (CRN)
| MiRNA | Fold change | Expressed in CRC | P value |
|---|---|---|---|
| hsa-miR-139–5p | 27.730995 | down | 0.000457488 |
| hsa-miR-133a | 14.123913 | down | 0.008218508 |
| hsa-miR-30a | 6.9308515 | down | 0.017357757 |
| hsa-miR-204 | 6.1801696 | down | 0.005467655 |
| hsa-miR-28–3p | 4.9092402 | down | 0.004293053 |
| hsa-miR-375 | 4.1634755 | down | 0.026016343 |
| hsa-miR-194 | 3.0145228 | down | 0.016694259 |
| hsa-miR-638 | 2.7310717 | down | 3.91053E-08 |
| hsa-miR-939 | 2.523975 | down | 0.011885638 |
| hsa-miR-135b | 233.2903 | up | 1.45697E-07 |
| hsa-miR-503 | 34.187862 | up | 1.58373E-06 |
| hsa-miR-183 | 20.887215 | up | 0.000161949 |
| hsa-miR-182 | 14.202393 | up | 0.000314092 |
| hsa-miR-31 | 13.799324 | up | 0.014758383 |
| hsa-miR-224 | 12.9758 | up | 0.000144943 |
| hsa-miR-205 | 11.577718 | up | 0.004554494 |
| hsa-miR-181d | 10.776051 | up | 0.000724382 |
| hsa-miR-135a | 10.168467 | up | 0.000661757 |
| hsa-miR-382 | 9.6167965 | up | 0.00095844 |
| hsa-miR-339–3p | 8.677848 | up | 0.000207839 |
| hsa-miR-501–3p | 8.025005 | up | 0.013724973 |
| hsa-miR-595 | 6.4784694 | up | 0.013636186 |
| hsa-miR-181c | 6.1030097 | up | 0.003669478 |
| hsa-miR-20a | 5.8202763 | up | 0.011896088 |
| hsa-miR-224 | 5.584402 | up | 0.000124752 |
| hsa-miR-424 | 4.872745 | up | 6.79045E-09 |
| hsa-miR-136 | 4.1466455 | up | 0.03151187 |
| hsa-miR-32 | 3.981137 | up | 0.03172399 |
| hsa-miR-29b-1 | 3.972951 | up | 0.019592708 |
| hsa-miR-34a | 3.3766873 | up | 0.014503202 |
| hsa-miR-21 | 3.0463343 | up | 2.75981E-10 |
| hsa-miR-18a | 2.6846995 | up | 0.008156499 |
| hsa-miR-95 | 2.664752 | up | 0.014796132 |
| hsa-miR-98 | 2.6145992 | up | 0.007363567 |
| hsa-miR-21 | 2.5509512 | up | 3.93848E-08 |
MiR-194 showed greatest inhibition effect on cell proliferation of the 9 decreased miRNAs
All significantly downregulated miRNAs showed in microarray profiles (>2.5-fold) were selected to transfect SW480 cell lines. CCK8 assays indicated that miR-194 could significantly inhibit the SW480 cell proliferation compared with the other 8 miRNAs (Fig. 2A). Hence, miR-194 arose our interest and was applied for a further study. qRT-PCR data indicated that miR-194 was significantly downregulated in CRC tissues compared with noncancerous tissues (Fig. 2B, P < 0.01), which exhibited a good consistency with the result of the microarray assay. In addition, compared with the NCM460 cells, miR-194 expression in 6 human CRC cell lines was significantly downregulated (Fig. 2C). Because SW480 and RKO were the 2 cell lines with the lowest miR-194 expression, these 2 cell lines were applied in the following study.
Figure 2.
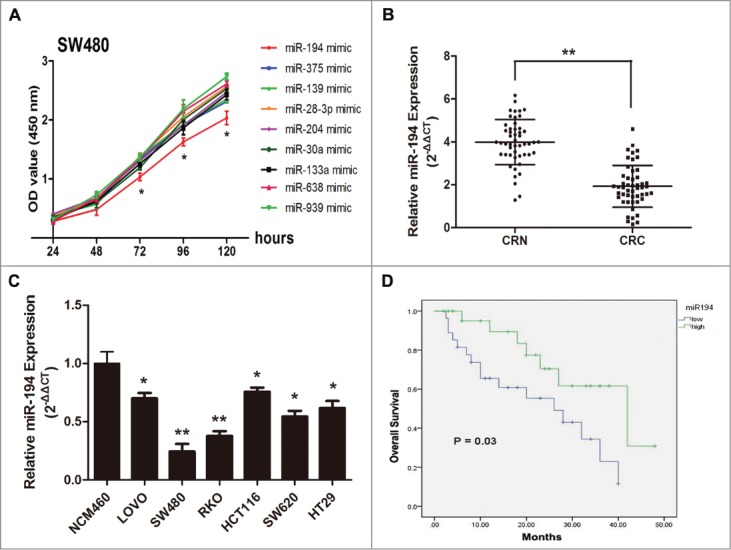
MiR-194 expression in colorectal tissues and cell lines and its relevance to overall survival. (A) MiR-194 exhibited greatest suppression effect on cell proliferation of 9 downregulated miRNAs. (B) The relative expression level of miR-194 in human CRC tissues (n = 50) and CRN tissues (n = 50), examined by qRT-PCR. (C) The relative miR-194 expression in the 6 CRC cell lines was significantly lower than that in normal colorectal cell line (NCM460). The average gene expression from NCM460 was appointed as 1. (D) Kaplan-Meier curves for overall survival analysis by miR-194 expression in CRC patients. P value was obtained by a log-rank test. *P<0.01.
Decreased miR-194 was associated with poorer prognosis in patients with CRC
The follow-up study showed that the low miR-194 expression group displayed a higher incidence of an increased tumor size (P = 0.025), poor tumor differentiation (P = 0.001) and late TNM stage (P = 0.027). However, no significant differences were observed with regard to age, gender, lymph node metastasis, distant metastasis, or vessel infiltration in CRC (Table 2). Furthermore, CRC patients with low levels of miR-194 expression had a significantly shorter median survival (26 ± 7.1 vs.42 ± 11.1 months, P = 0.03) than those with high levels of miR-194 expression (Fig. 2D). Meanwhile, Cox's multivariate analysis showed that miR-194 expression, tumor size, and tumor differentiation were significantly associated with overall survival of CRC patients as independent prognostic factors (Table 3). These results indicate that decreased miR-194 expression predicts poorer prognosis in CRC patients.
Table 2.
Clinicopathologic correlations of miR-194 expression in colorectal cancer
| |
miR-194 expression |
|||
|---|---|---|---|---|
| Parameters | High (n = 23) | Low (n = 27) | Total (n = 50) | P value |
| Age(y) | ||||
| ≤60 | 6 | 8 | 14 | 0.781 |
| > 60 | 17 | 19 | 36 | |
| Gender | ||||
| Female | 9 | 9 | 18 | 0.670 |
| Male | 14 | 18 | 32 | |
| Tumor size(cm) | ||||
| ≤2 | 10 | 4 | 14 | 0.025* |
| >2 | 13 | 23 | 36 | |
| Tumor differentiation | ||||
| Well/moderate | 19 | 10 | 29 | 0.001* |
| Poor | 4 | 17 | 21 | |
| TNM stage | ||||
| I+II | 14 | 8 | 22 | 0.027* |
| III+IV | 9 | 19 | 28 | |
| Lymph node metastasis | ||||
| Positive | 11 | 10 | 21 | 0.441 |
| Negative | 12 | 17 | 29 | |
| Distant metastasis | ||||
| Positive | 4 | 5 | 9 | 0.918 |
| Negative | 19 | 22 | 41 | |
| Vascular infiltration | ||||
| Positive | 10 | 13 | 23 | 0.741 |
| Negative | 13 | 14 | 27 | |
Statistically significant (P < 0.05).
Table 3.
Multivariate analysis of factors associated with overall survival in CRC patients
| |
Multivariate analysis |
|
|---|---|---|
| Variable | HR (95%CI) | P value |
| Age | 0.987 (0.949-1.027) | 0.530 |
| Gender | 1.452 (0.571-3.687) | 0.433 |
| Tumor size | 4.416 (1.296-15.042) | 0.018* |
| Tumor differentiation | 4.232 (1.235-14.505) | 0.022* |
| TNM stage | 0.218 (0.045-1.064) | 0.060 |
| Lymph node metastasis | 3.454 (0.695-17.177) | 0.130 |
| Distant metastasis | 1.273 (0.278-5.827) | 0.756 |
| miR-194 | 4.112 (1.380-12.251) | 0.011* |
HR, hazard ratio; CI, confidence interval;
Statistically significant (P < 0.05).
MiR-194 decreases CRC cell growth, colony formation, and induces G1 arrest and apoptosis
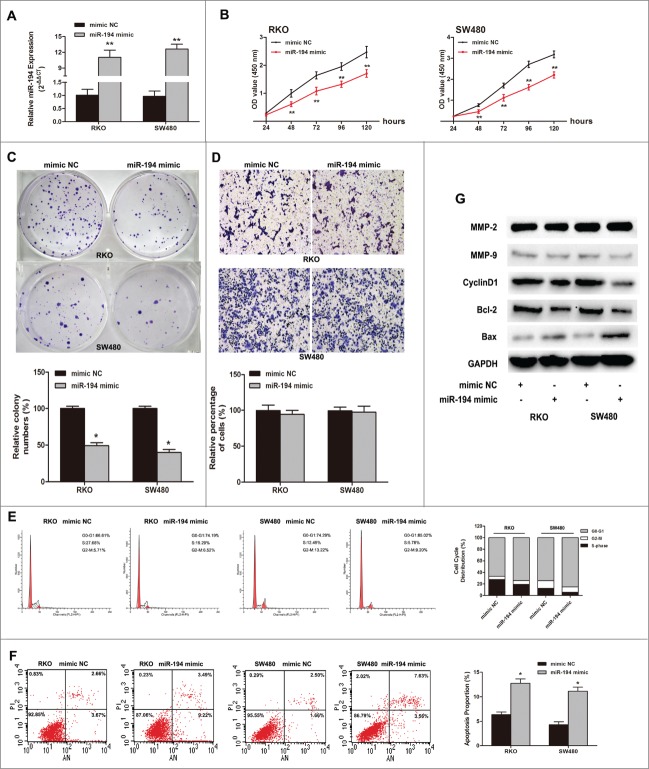
To explore the role of miR-194 in CRC cells, we transfected RKO and SW480 cells with miR-194 mimics for functional analysis. qRT-PCR was used to confirm the increased expression of miR-194 (Fig. 3A). CCK-8 assay revealed that the overexpression of miR194 significantly repressed the cell proliferation rate of these 2 cell lines (Fig. 3B). Furthermore, the colony numbers of RKO and SW480 cells transfected with miR-194 mimics were notably lower than those transfected with NC mimics (Fig. 3C). As shown in Figure 3E, upregulation of miR-194 resulted in an accumulation of both in RKO (66.83 ± 3.26 vs. 75.18 ± 3.52 %, P < 0.05) and SW480 (74.88 ± 4.82 vs. 85.84 ± 2.97 %, P < 0.05) cells in the G0/G1 phase of the cell cycle. Moreover, after transfected with miR-194 mimics, the cell apoptosis rates were significantly increased both in RKO (6.37 ± 0.53 vs. 12.74 ± 0.88 %, P < 0.01) and SW480 (4.32 ± 0.59 vs. 11.13 ± 0.84 %, P < 0.01) cells (Fig. 3F). However, upregulation of miR-194 had no significant effect on invasion of RKO and SW480 cells (Fig. 3D).
Figure 3.
Tumor suppressive effects of miR-194 in RKO and SW480 CRC cell lines. (A) Relative miR-194 expression after transfected with miR-194 mimics or mimic NC, detected by SYBR qRT-PCR. The average miRNA expression from mimic NC group was appointed as 1. (B) The effect of transient transfection of miR-194 mimics or mimic NC (50 nM) for 24–120 h was examined on the proliferation of RKO and SW480 cells by CCK8 assay. (C) Colony formation assay after upregulation of miR-194 expression. (D) Overexpression of miR-194 seemed to be little effect on cell invasion of RKO and SW480 cells. Quantification was performed by counting the stained cells that invaded to the lower chamber under a light microscopy. The two CRC cells were treated as mentioned above. Flow cytometry analysis showed miR-194 induced cell cycle arrest (E) and increase of apoptosis (F). Data are presented as mean ± SD of results from 3 independent experiments. (G) Western blot analysis showed the expression levels of invasion related protein MMP2 and MMP9, cell cycle related protein cyclinD1, and apoptosis associated protein Bax and Bcl-2 after overexpression of miR-194. *P<0.01.
Furthermore, we detected expression of the invasion, apoptosis, and cell cycle-associated proteins. CyclinD1 and Bcl-2 were significantly decreased in miR-194 mimic group, while the expression of Bax was obviously increased. The MMP2 and MMP9 proteins had no significant changes (Fig. 3G).
To further clarify the functions of miR-194 in CRC, we evaluated the SW480 cell growth ability treated with miR-194 inhibitor. The results showed that inhibition of miR-194 significantly promoted the cell proliferation and colony formation (Fig. S1).
MiR-194 suppresses tumor growth of CRC cells in vivo
SW480 cells infected with lentivirus/miR-194 (LV-miR-194) or lentivirus/miR-NC (LV-miR-NC), with better infected effect (Fig. S2A—a), were chosen to inoculate into the nude mice. After 32 days, the tumor volume of mice inoculated with LV-miR-NC cells was 787.52 ± 84.61 mm3, whereas that injected with LV-miR-194 cells was 84.53 ± 26.64 mm3 (Fig. S2B-a, C, P < 0.01). qRT-PCR showed that miR-194 expression levels were obviously increased in LV-miR-194-infected tumors compared with control tumors (Fig. S2E).
Association of MAP4K4 expression level with miR-194 expression in CRC tissue samples
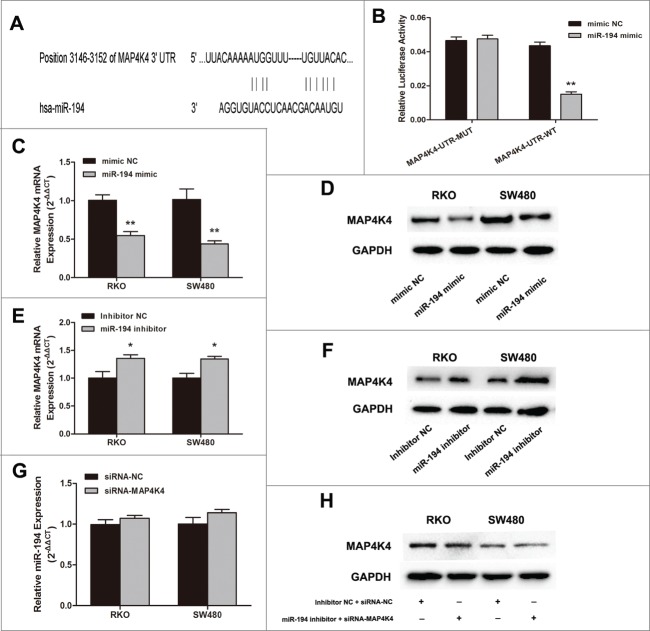
MAP4K4 was a putative target of miR-194 through bioinformatics analysis. To explore the actual association between MAP4K4 and miR-194, the relative expression of MAP4K4 and miR-194 was determined in CRC tissues and cell lines. As shown in Figure 4A–B, through immunohistochemical staining and qRT-PCR assay, we observed stronger MAP4K4 expression in CRC tissues than that in CRN tissues (Table S3). We also found that the stronger immunoreactivity of MAP4K4 was inversely correlated with lower miR-194 expression by Pearson analysis (Fig. 4C), further confirming that MAP4K4 was the target of miR-194.
Figure 4.
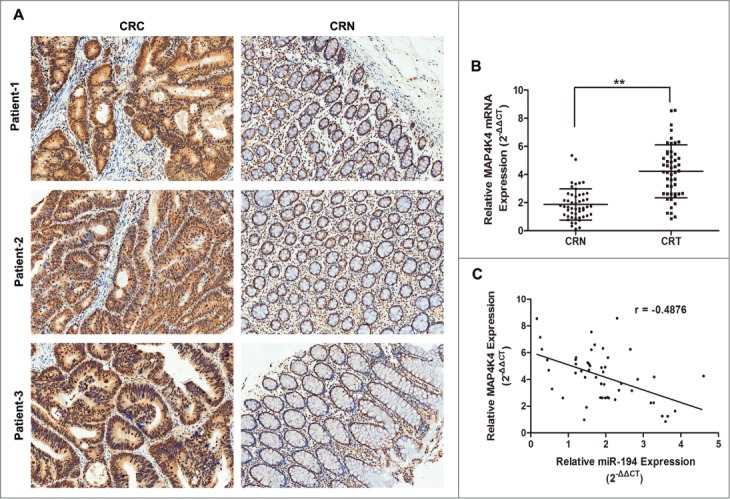
Relationship between expression of miR-194 and MAP4K4 in colorectal tissue samples. (A) Immunohistochemical analysis of MAP4K4 protein in representative colorectal tissue specimen is shown in ×100 magnification. Weak cytoplasmic and cytoblast staining for MAP4K4 was observed in CRN tissues while strong staining was observed in CRC tissues. (B) MAP4K4 was detected by qRT-PCR in 50 CRC tissues and matched CRN tissues. The MAP4K4 abundance was normalized against GAPDH. (C) The relationship between miR-194 and MAP4K4 expression was explored by Pearson's correlation in CRC tissues. **P<0.01.
MiR-194 directly targets MAP4K4 gene by interaction with the 3′UTR
To verify that MAP4K4 was one of the direct targets of miR-194, we cloned dual-luciferase reporters containing the 3′UTR of MAP4K4 with seed sequence recognizing sites of wild type or mutant one (Fig. 5A). Then miR-194 mimic or mimic NC was co-transfected with MAP4K4-UTR-WT or MAP4K4-UTR-MUT plasmid into SW480 cells. The relative luciferase activity of the reporter contained wild type 3’UTR was significantly decreased by 65.1% in the presence of miR-194 mimic (P < 0 .01). However, the luciferase activity of mutant reporter was unaffected by simultaneous transfection of miR-194 mimic (P > 0 .05), suggesting that miR-194 might directly target its predicted seed region of MAP4K4 (Fig. 5B). The effect of miR-194 on endogenous expression of MAP4K4 was subsequently observed by qRT-PCR and Western blot analysis. The results showed that overexpression of miR-194 significantly decreased the MAP4K4 expressions at mRNA level and protein level (P < 0.01, Fig. 5C–D). Furthermore, transfection of miR-194 inhibitor could upregulate the expression of MAP4K4 (P < 0.05, Fig. 5E-F). Our results also demonstrated that the expression level of MAP4K4 was dramatically downregulated in LV-miR-194 group tumors from nude mice (Fig. S2E).
Figure 5.
MAP4K4 is a direct target of miR-194. (A) Predicted binding site of miR-194 to the 3’UTR of human MAP4K4 by TargetScan. (B) Dual-luciferase reporter assay with cotransfection of wild-type or mutant 3’UTR vector (100 ng) and miR-194 mimics or mimic NC (50 nM) in SW480 cells. Firefly luciferase activity of each sample was normalized against Renilla luciferase activity. The effects of upregulation of miR-194 on MAP4K4 expression at mRNA level (C) and protein level (D). The effects of inhibition of miR-194 on MAP4K4 expression at mRNA level (E) and protein level (F). (G) Relative miR-194 expression after treated with siRNA-MAP4K4. (H) The expression of MAP4K4 protein after co-transfected with miR-194 inhibitor and siRNA-MAP4K4. All data from 3 separate experiments are presented as mean ± SD. *P<0.01.
However, the expression level of miR-194 had little changes after silencing of MAP4K4 (Fig. 5G). After treated with miR-194 inhibitor plus siRNA-MAP4K4, we observed no significant changes of MAP4K4 protein expression (Fig. 5H).
Knockdown of MAP4K4 repressed cell growth in vitro and in vivo
To investigate the role of MAP4K4 in CRC, we applied siRNA-mediated MAP4K4 inhibition method to analyze whether it could replicate the tumor suppressor effect of miR-194 in CRC cell lines. As shown in Figure S3, the MAP4K4 expression was greatly decreased in RKO and SW480 cells after transfection of siRNA-MAP4K4 compared with siRNA-NC. siRNA-mediated MAP4K4 knockdown also significantly inhibited cell proliferation and colony formation. At the same time, siRNA/MAP4K4 could promote G0/G1 arrest and induce apoptosis enhancement of RKO and SW480 cells. At 32 days, the average volume and weight of tumors formed from LV-MAP4K4-RNAi–infected SW480 cells was evidently lower than that formed from LV-RNAi-NC infected cells (Fig. S2Bb-D). These results indicate that MAP4K4 targeting might be a mechanism of the tumor suppressor function of miR-194 in CRC cells.
Silencing of MAP4K4 down regulates MDM2 expression in SW480 cell line
Having identified that MAP4K4 was the direct target of miR-194 and as a potential oncogene in CRC, we next sought to investigate its downstream signaling pathways using the Human Gene Expression Array (Fig. 6A). The most differentially expressed genes whose expression changed 1.5-fold or more in MAP4K4-siRNA–transfected SW480 cells compared with NC-transfected cells are summarized in Table S4. Among these changed genes excluding MAP4K4, 17 were downregulated and 37 were upregulated. In this study, we focused on the top 5 of the downregulated genes. To confirm the array data, we performed Western blot analysis in SW480 cells. We found that MDM2 protein expression level was significantly decreased while the other 4 proteins FAM172A, CAMLG, PLOD2 and MPEG1 had little obvious changes after knockdown of MAP4K4 (Fig. 6B). Therefore, we selected MDM2 as the candidate and hypothesized that MDM2 was one of the most important genes regulated by MAP4K4.
Figure 6.
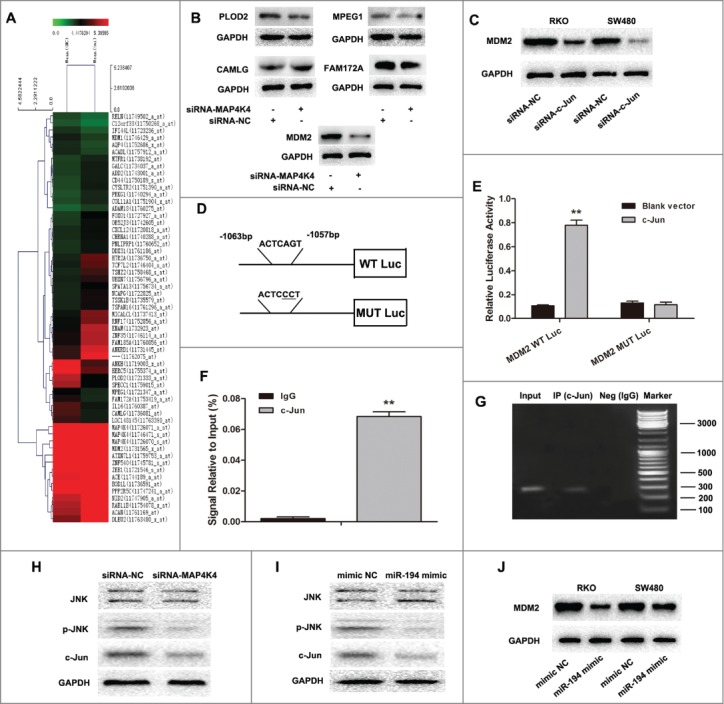
MAP4K4 influences MDM2 expression through regulating transcription factor c-Jun. (A) Gene expression profile array analysis in SW480 cells. Hierarchical clustering of the expression values for mRNA of siRNA-MAP4K4 cells vs. siRNA-NC cells. (B) Western blot analysis showed expression of MDM2, FAM172A, CAMLG, PLOD2 and MPEG1 protein after knockdown of MAP4K4. Silencing of transcription factor c-Jun resulted in downregulation of MAP4K4 expression in RKO and SW480 cells. (D) The human MDM2 promoter constructs containing a potential c-Jun binding motif (–1063 to –1057 bp). (E) The transcriptional activity of the MDM2 promoter. SW480 cells were co-transfected with the wild type or mutant type plasmids and c-Jun vector or blank vector for 48 h, and the luciferase activity was measured. (F) ChIP assays of c-Jun and its binding motif. Two antibodies (anti-IgG and c-Jun) were used in the ChIP assays in SW480 cells. QRT-PCR was performed to quantify the binding activity. (G) Products of qRT-PCR observed by electrophoresis method. (H) MAP4K4 knockdown influenced the expression of phosphorylated JNK and c-Jun, but had little effect on total JNK. MAP4K4-siRNA or siRNA-NC expressing SW480 transfectants were subjected to Western blot analysis. (I) Upregulation of miR-194 in SW480 cells had similar effects on expression of JNK, p-JNK and c-Jun to that of MAP4K4 knockdown. (J) The effects of overexpression of miR-194 on MDM2 expression. Representative images of 3 independent experiments with similar results are shown. **P<0.01.
MDM2 gene transcription was regulated by c-Jun through promoter occupancy
The mechanism of MAP4K4 regulating the expression of MDM2 was still unknown. JASPAR, TFSEARCH, and Gene-Regulation software was applied to predetermine the transcription factor of MDM2. Interestingly, all prediction software directed c-Jun, a downstream gene involved in MAPK signaling pathway, as one of the potential transcription factors binding to –1063 to –1057 bp of MDM2 promoter. Through Western Blot assay, we observed that MDM2 proteins were significantly downregulated when treated with siRNA-c-Jun (Fig. 6C).
Luciferase reporter results showed concurrent expression of c-Jun, with the MDM2 reporter construct showing increased MDM2 promoter activity, which was abrogated by mutation of the c-Jun DNA-binding site in the MDM2 promoter (Fig. 6D–E).
ChIP assay also verified that c-Jun could actually bind to the promoter of MDM2 on the –1063 to –1057 bp region (Fig. 6F–G). As shown in Figure 6H, we further confirmed that MAP4K4 could affect phosphorylated JNK and c-Jun expression whereas not having effect on total JNK in SW480 cells harboring MAP4K4-siRNA or NC. Additionally, upregulation of miR-194 presented the similar results to knockdown of MAP4K4 (Fig. 6I-J). These data suggested that MAP4K4 regulated MDM2 expression through the transcription factor c-Jun.
Discussion
MicroRNAs have emerged as important developmental regulators and control critical processes such as cell fate determination and cell death.6 Recently, increasing evidences have suggested that miRNAs are mutated or poorly expressed in human cancers and may act as tumor suppressors or oncogenes.35-37 Aberrant expressions of miRNAs, including upregulation or downregulation, are closely associated with tumorigenesis, and the identification of tumor-related miRNAs and their direct target genes as well as the involved signaling pathway is critical for understanding the biological significance of miRNAs in carcinogenesis. The microarray analysis of miRNA expression profiles provides us a key tool to screen tumor-associated miRNAs. In the present study, we focused on miR-194, which is significantly downregulated in CRC tissues and exhibit the most obviously inhibited effects on CRC cell proliferation. MiR-194 is a miRNA that play complicated role in various types of cancers, but the current knowledge about the effects of miR-194 on CRC and corresponding molecular mechanisms is still preliminary. Our study reveals, for the first time, that a decreased miR-194 expression is correlated with poor prognosis in patients with CRC and involved in tumorigenesis through regulation of the MAP4K4/c-Jun/MDM2 signaling pathway. These results strongly suggest that miR-194 might be a novel prognostic predictor in CRC.
Moreover, our clinical data showed that decreased miR-194 expression was significantly associated with large tumor size, poor tumor differentiation, and late TNM stage, whereas not related to lymphatic invasion and distant metastasis, suggesting that miR-194 might facilitate tumor growth of CRC rather than affect invasion and metastasis. In addition, multivariate analysis indicated that the low miR-194 expression, advanced TNM stage, and poor tumor differentiation were significant prognostic factors for a poor overall survival rate of CRC patients.
Previous studies have indicated that miR-194 was involved in CRC. Chiang et al. revealed that miR-194 was frequently downregulated in CRC tissues and cancer cell lines.16 However, Tan et al. indicated that miR-194 was overexpressed in ulcerative colitis-related CRC of mice.38 Sundaram et al. showed that miR-194 could promote angiogenesis and facilitate tissue repair by targeting thrombospondin-1 in colon cancer.18 Nevertheless, the effect of miR-194 in CRC was far from defined. For this reason, our study aimed to identify the function of miR-194 in CRC. We showed that overexpression of miR-194 significantly suppressed cell proliferation, colony formation, promoted G0/G1 arrest, and induced cell apoptosis. Furthermore, our in vivo study revealed that the growth of xenograft tumors in nude mice was significantly repressed after transfected with LV-miR-194. These results strongly implied that miR-194 might act as an inhibitor in the progress of CRC. Meanwhile, miR-194 played different role in different cancer types,39,40 suggesting that miR-194 might be tumor heterogeneous and highly dependent on its targets.
MAP4K4, involved in MAPK signaling pathway, has been reported to be frequently overexpressed in several types of human tumors.41,42 MAP4K4 participates in the initiation and development of cancers by promoting cell proliferation, inhibiting cell apoptosis, promoting angiogenesis, invasion, and metastasis.21,23 In this study, through bioinformatics analysis, we found that MAP4K4 might be one of the potential targets for miR-194. Thus, we applied dual-luciferase activity assay to identify the effect of miR-194 on MAP4K4 expression. The data showed that miR-194 can inhibit the expression of MAP4K4 by combining directly to the 3’UTR of MAP4K4, whereas inhibition of miR-194 expression resulted in upregulation of MAP4K4. However, in reverse, knockdown of MAP4K4 had little effects on miR-194 expression, suggesting that there is no loop-regulation between miR-194 and MAP4K4. Thereafter, we inhibited MAP4K4 expression by RNA interference and found that knockdown of MAP4K4 could significantly prohibit proliferation of CRC cells both in vitro and in vivo, which were similar to the effects of ectopic miR-194 expression. Furthermore, the inverse correlation between miR-194 and MAP4K4 expression in CRC and CRN tissues was further confirmed that miR-194 downregulation resulted in MAP4K4 overexpression. Thus, we concluded that downregulation of MAP4K4 might be a mechanism by which miR-194 exerts its tumor suppressor functions. This study is the first to show that MAP4K4 functions as an oncogene and might be a promoter in CRC. It is also the first to show that MAP4K4 is negatively regulated by miR-194.
To further clarify the mechanism of MAP4K4 involved in cancer cell growth, an array analysis was applied after knockdown of MAP4K4 in CRC cells. We found that MDM2 expression was significantly decreased after inhibition of MAP4K4. MDM2, played a major oncogenic role on blocking p53 transcriptional activity, and it was reported to be frequently overexpressed in various cancer types.28,43,44 Inhibition of MDM2 was supposed to be an effective therapeutic method to treat human cancer24,25 through the mechanism of apoptosis induction26 and invasion repression.27 There have been accumulating evidences that the antitumor effects of MAP4K4 depletion are likely mediated through the JNK pathway.21,31 Phosphorylation of JNK induced c-Jun expression.32 Interestingly, through bioinformatics assay we found that c-Jun was one of the potential transcription factors for MDM2 promoter. Thus, ChIP assay was performed to verify the role of c-Jun on MDM2 expression. The results revealed that c-Jun could directly bind to –1063 to –1057 bp of MDM2 promoter to regulate MDM2 expression. Moreover, inhibition of MAP4K4 resulted in downregulation of phosphorylated JNK and c-Jun, as well as MDM2, suggesting that downregulation of MDM2 through c-Jun might be a mechanism by which MAP4K4 depletion display anticancer effects.
In summary, our present study indicated that miR-194 was frequently downregulated in CRC and the level of miR-194 was closely associated with overall survival of CRC patients. This study also implied that miR-194 negatively regulated MAP4K4/c-Jun/MDM2 signaling pathway and inhibited CRC cell growth in vitro and in vivo. Therefore, our findings highlighted that miR-194 might act as an inhibitor in CRC by targeting MAP4K4/c-Jun/MDM2 signaling pathway and also support the development of effective therapeutic strategies that target miR-194/MAP4K4/c-Jun/MDM2 interactions as a novel therapeutic application for CRC patients.
Materials and Methods
Cell lines and patient tissue samples
Human CRC cell lines SW480, SW620, RKO, HT29, HCT116, and LoVo cells were purchased from American Type Culture Collection (Manassas, VA). NCM460 cell was purchased from American INCELL Corporation. They were all tested and authenticated for genotypes by DNA fingerprinting. These cell lines were passaged for less than 6 months after resuscitation, and no reauthorization was done; SW480 and SW620 cells were cultured in Leibovitz's L-15 medium and the others were cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS) (all from Gibco), 100 IU/mL penicillin, and 100 μg/mL streptomycin at 37°C with 5% CO2. Tissue samples were obtained from patients undergoing surgery. The patients were treated with coloproctectomy, depending on the National Comprehensive Cancer Network (NCCN) guideline for colon/rectal cancer (version 1. 2013). One part of the sample was embedded in paraffin and the other was immediately snap-frozen and stored at –80°C until RNA or protein extraction. All patients provided written informed consent before samples were collected. The study was approved by the local Research Ethics Committee of Peking University.
MiRNA microarray analysis
Total RNA, containing miRNA, was extracted from tissue samples using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The low-molecular-weight RNA was isolated using a polyethylene glycol (PEG) solution precipitation method45 and was labeled using the T4 RNA ligase labeling method.46 They were then hybridized to Agilent human miRNA 8 × 60 k microarray (V16.0). Images were scanned using the Agilent Scan Array Express array scanner (G2565CA). The resulting images were quantified with Agilent's Feature Extraction software (v10.7). Data were normalized and analyzed with Agilent Gene Spring software.
MAP4K4 knockdown, overexpression, and inhibition of miR-194
The MAP4K4 interference sequence was obtained as described previously.47 Cells were seeded onto plate wells, and transfected the next day with 50 nmol/L MAP4K4 siRNA, miR-194 mimics, miR-194 inhibitor, or their corresponding negative controls (NC; siRNA NC, mimics NC, or miRNA inhibitor NC; Ribobio Co., Guangzhou, China). Lipofectamine 2000 (Invitrogen) was used for cell transfection according to the manufacturer's instructions. Cells were collected after an additional 48–72 h.
Quantitative real-time reverse transcription polymerase chain reaction
The reverse transcription was performed using a reverse transcription kit (Takara, Japan). The expression of mature miRNAs and potential target genes were measured by Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) with SYBR Green PCR Kit (Takara) on CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Human U6 RNA or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA was amplified as an internal control respectively for miRNA or mRNA. The miRNA or mRNA levels were calculated according to 2−ΔΔCt. The reverse primers of U6 and miR-194 were the universal primer provided by Takara. The miR-194 expression was determined as high when the expression level was equal to or above the median of the cohort and low when it was below the median of the cohort.48 Primer sequences are shown in Table S1.
Genome-wide expression microarray analysis
The genomic expression level of total RNA from cells transfected with MAP4K4 siRNA or NC was detected by GeneChip® PrimeView™ Human Gene Expression Array (Affymetrix, USA). Fold-changes in gene expression were determined with the normalized signal intensities. Genes were identified to exhibit significant differential expression if they fit the criteria of P-values <0.05 and fold change > |1.5|.
Immunohistochemistry
Tissue samples were embedded in paraffin. The paraffin sections (4 μm) were deparaffinized with xylene, rehydrated, permeabilized in 0.1% Triton X-100, and incubated overnight at 4°C with MAP4K4 antibody (sc-100445; SantaCruz Biotechnology, Santa Cruz, CA; 1:100) overnight. The sections were incubated with biotinylated goat anti-mouse secondary antibody (Boster, Wuhan, China) for 1 h at room temperature. 3, 3-Diaminobenzidine was used to detect the antigen–antibody complex, and hematoxylin was used for counter staining. NC sections were incubated in phosphate buffered saline (PBS) instead of primary antibody.
The staining results were evaluated by 2 experienced pathologists who were blinded to patient information. For MAP4K4, using the median value of 10% as a cutoff, positive expression was defined as cytoplasmic staining of >10% of the cells.49
Western blot analysis
We used cell lysis buffer with proteinase inhibitors (Roche Diagnostics, Basel, Switzerland) to extract total protein. Lysates were denatured with sodium dodecyl sulfate (SDS) buffer at 100°C for 10 min and separated in 10% or 15% polyacrylamide gels and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Hertfordshire, UK). Membranes were blocked with 5% non-fat milk powder in Tris-buffered saline containing 0.1% Tween 20 (TBST) and probed with primary antibodies overnight at 4°C. Membranes were treated with secondary antibodies on the next day. Band signals were visualized using enhanced chemiluminescence (ECL; Pierce, Rockford, IL), exposed to a ChemiDoc™ XRS+ System (Bio-Rad), and the band density was evaluated by Bio-Rad Quantity One software. Primary antibodies are shown in Table S2.
Cell proliferation and colony formation assay
Cells were seeded onto 96-well plates and incubated at 37°C after transfection. After incubation for 1 to 5 days, each well was added 10 μl of CCK8 solution and the plates were continued to be incubated for another 4 h at 37°C. The absorbance was measured at 450 nm using a microplate reader (Bio-Rad). The CCK8 assay was repeated 3 times with 6 replicates.
The colony formation was performed as follows: after transfection, cells were seeded onto each well of a 6-well plate and then incubated for another 10 d The wells were then washed with PBS, fixed with 4% paraformaldehyde, and stained with 0.1% crystal violet. The colony assay was repeated 3 times in duplicate.
Cell apoptosis and cell cycle analysis
For cell apoptosis analysis, cells were collected 72 h after transfection, and the process was performed with Alexa Fluor\R488 annexin V/Dead cell apoptosis kit (Invitrogen). For cell cycle assay, cells were collected and stained using BD cycletestTM plus DNA reagent kit (BD Biosciences) according to the manufactures’ instructions. All cells were detected by flow cytometry (BD Biosciences). Data were analyzed with FlowJo V7 software (Tree Star, Ashland, OR).
Matrigel invasion assay
The Matrigel invasion chamber was used to assess cell invasion ability (24-well plates, 8-μm pore size, Corning). In brief, cells were seeded in the upper chamber with the media containing 0.1% bovine serum albumin, while the media containing 30% fetal bovine serum was placed in the lower well. After incubation for 48 h at 37°C, the non-invading cells were removed with cotton swabs. Invasive cells at the bottom of the membrane were stained with 0.1% crystal violet and were counted under microscopic observation. This invasion detection was repeated 3 times in duplicate.
Luciferase assay
SW480 cells were seeded in 96-well plates at 5000 cells per well and cotransfected pMIR-REPORT Luciferase vector (Ribobio Co.) containing MAP4K4 3’UTR (3441 bp) or mutated ones with miR-194 mimic or mimic NC; or cotransfected luciferase vector containing the −1063 to –1057 MDM2 promoter sequence with or without c-Jun luciferase reporter plasmid. After incubation for 48 h, luciferase activity was determined using the Dual-Luciferase reporter assay system (Promega, Madison, WI). The relative luciferase activities were detected by normalizing to Renilla Luciferase activities.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using SW480 cells and carried out as described previously.28,50 Briefly, cells were cross-linked by incubation in 1% formaldehyde-containing medium for 10 min at 37°C and then sonicated to make soluble chromatin with DNA fragments between 200 and 1000 bp. The chromatin fragments were immunoprecipitated with the following antibodies: c-Jun (9165#, Cell Signal Technology, 1:50), and normal rabbit IgG (2729#, Cell Signal Technology). The protein–DNA complex was collected with protein A Sepharose beads (Millipore), eluted, and reverse cross-linked. Following treatment with protease K (Sigma-Aldrich, St. Louis, MO), samples were extracted with phenol-chloroform, and precipitated with ethanol. The recovered DNA was resuspended in TE buffer and used for qPCR amplification.
In vivo tumor growth assay
We used Lentiviral vector (GeneChem Co. Ltd, Shanghai, China) containing miR-194 (LV-miR-194) or MAP4K4 (LV-MAP4K4-RNAi) RNA interference sequences to infected SW480 cells. The fluorescence intensity and range were observed to identify the efficiency of infection. BALB/c female nude mice (4 weeks old) were purchased from Beijing Vital River Laboratories (China). These mice were maintained in a specific pathogen-free environment in the Experimental Animal Center of Peking University People's Hospital. The animal experiments were approved and reviewed by the Animal Research Committee of the Peking University People's Hospital. Care and handling of the animals were in accordance with the guidelines for Institutional and Animal Care and Use Committees.
Mice were randomly allocated to 4 groups (NC, MAP4K4 RNAi, miR-194 and miR-NC), with 4 mice in each group. Infected SW480 cells (1 × 107 cells/mouse) were injected subcutaneously into mice. Tumor volumes were measured every 4 d and calculated according to the formula V = 0.5 × L (length) × W2 (width). At 40 d after the cells inoculation, mice were sacrificed and tumors were excised to measure the volume and weight. The expression of MAP4K4 in tumors was detected by qRT-PCR and Western blot analysis.
Statistical analysis
All results are expressed as mean ± SD. Differences between groups were assessed using Student's t-test and Fisher's exact test. The relationship between the expression of miR-194 and MAP4K4 was analyzed using Pearson's correlation. The relationship between miR-194 expression and clinicopathologic features of CRC was analyzed using the Pearson χ2 test. Kaplan-Meier method was used to calculate overall survival of 2 patients groups whose differences were analyzed by log-rank test. P < 0.05 was considered to be statistically significant. All data, unless otherwise explained specifically, were analyzed using the SPSS 20.0 software (SPSS, Chicago, IL).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81372290, 81372291), Peking University People's Hospital Funds (RDB 2013-15).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- 1. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin 2014; 64:104-17; PMID:24639052; http://dx.doi.org/ 10.3322/caac.21220 [DOI] [PubMed] [Google Scholar]
- 2. Chen DL, Wang ZQ, Zeng ZL, Wu WJ, Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, Ren C, et al. Identification of MicroRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology 2014; 60:598-609; PMID:24616020; http://dx.doi.org/ 10.1002/hep.27118 [DOI] [PubMed] [Google Scholar]
- 3. Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F, et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014; 25:469-83; PMID:24735923; http://dx.doi.org/ 10.1016/j.ccr.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science 2006; 314:268-74; PMID:16959974; http://dx.doi.org/ 10.1126/science.1133427 [DOI] [PubMed] [Google Scholar]
- 5. De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol 2011; 12:594-603; PMID:21163703; http://dx.doi.org/ 10.1016/S1470-2045(10)70209-6 [DOI] [PubMed] [Google Scholar]
- 6. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 7. Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009; 11:228-34; PMID:19255566; http://dx.doi.org/ 10.1038/ncb0309-228 [DOI] [PubMed] [Google Scholar]
- 8. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. nature 2005; 435:834-8; PMID:15944708; http://dx.doi.org/ 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- 9. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature Reviews Cancer 2006; 6:857-66; PMID:17060945; http://dx.doi.org/ 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- 10. Li Z, Ying X, Chen H, Ye P, Shen Y, Pan W, Zhang L. MicroRNA-194 Inhibits the Epithelial-Mesenchymal Transition in Gastric Cancer Cells by Targeting FoxM1. Dig Dis Sci 2014; 59:2145-52; PMID:24748184; http://dx.doi.org/ 10.1007/s10620-014-3159-6 [DOI] [PubMed] [Google Scholar]
- 11. Zhai H, Karaayvaz M, Dong P, Sakuragi N, Ju J. Prognostic significance of miR-194 in endometrial cancer. Biomark Res 2013; 1; PMID:24040515; http://dx.doi.org/ 10.1186/2050-7771-1-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khella HW, Bakhet M, Allo G, Jewett MA, Girgis AH, Latif A, Girgis H, Von Both I, Bjarnason GA, Yousef GM. miR-192, miR-194 and miR-215: a convergent microRNA network suppressing tumor progression in renal cell carcinoma. Carcinogenesis 2013; 34:2231-9; PMID:23715501; http://dx.doi.org/ 10.1093/carcin/bgt184 [DOI] [PubMed] [Google Scholar]
- 13. Wu X, Liu T, Fang O, Leach LJ, Hu X, Luo Z. miR-194 suppresses metastasis of non-small cell lung cancer through regulating expression of BMP1 and p27kip1. Oncogene 2014; 33:1506-14; PMID:23584484; http://dx.doi.org/ 10.1038/onc.2013.108 [DOI] [PubMed] [Google Scholar]
- 14. Le XF, Almeida MI, Mao W, Spizzo R, Rossi S, Nicoloso MS, Zhang S, Wu Y, Calin GA, Bast RC, Jr. Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer. PLoS One 2012; 7:e41170; PMID:22829924; http://dx.doi.org/ 10.1371/journal.pone.0041170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang J, Zhao CY, Zhang SH, Yu DH, Chen Y, Liu QH, Shi M, Ni CR, Zhu MH. Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma. Oncol Rep 2014; 31:1157-64; PMID:24398877 [DOI] [PubMed] [Google Scholar]
- 16. Chiang Y, Song Y, Wang Z, Liu Z, Gao P, Liang J, Zhu J, Xing C, Xu H. microRNA-192, -194 and -215 are frequently downregulated in colorectal cancer. Exp Ther Med 2012; 3:560-6; PMID:22969930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang ZH, Ren LL, Zheng P, Zheng HM, Yu YN, Wang JL, Lin YW, Chen YX, Ge ZZ, Chen XY, et al. MiR-194 as a predictor for adenoma recurrence in patients with advanced colorectal adenoma after polypectomy. Cancer Prev Res (Phila) 2014; 7:607-16; PMID:24691499; http://dx.doi.org/ 10.1158/1940-6207.CAPR-13-0426 [DOI] [PubMed] [Google Scholar]
- 18. Sundaram P, Hultine S, Smith LM, Dews M, Fox JL, Biyashev D, Schelter JM, Huang Q, Cleary MA, Volpert OV, et al. p53-responsive miR-194 inhibits thrombospondin-1 and promotes angiogenesis in colon cancers. Cancer Res 2011; 71:7490-501; PMID:22028325; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wright JH, Wang X, Manning G, LaMere BJ, Le P, Zhu S, Khatry D, Flanagan PM, Buckley SD, Whyte DB, et al. The STE20 kinase HGK is broadly expressed in human tumor cells and can modulate cellular transformation, invasion, and adhesion. Mol Cell Biol 2003; 23:2068-82; PMID:12612079; http://dx.doi.org/ 10.1128/MCB.23.6.2068-2082.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rizzardi AE, Rosener NK, Koopmeiners JS, Isaksson VR, Metzger GJ, Forster CL, Marston LO, Tiffany JR, McCarthy JB, Turley EA, et al. Evaluation of protein biomarkers of prostate cancer aggressiveness. BMC Cancer 2014; 14:244; PMID:24708576; http://dx.doi.org/ 10.1186/1471-2407-14-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu AW, Cai J, Zhao XL, Jiang TH, He TF, Fu HQ, Zhu MH, Zhang SH. ShRNA-targeted MAP4K4 inhibits hepatocellular carcinoma growth. Clin Cancer Res 2011; 17:710-20; PMID:21196414; http://dx.doi.org/ 10.1158/1078-0432.CCR-10-0331 [DOI] [PubMed] [Google Scholar]
- 22. Qiu MH, Qian YM, Zhao XL, Wang SM, Feng XJ, Chen XF, Zhang SH. Expression and prognostic significance of MAP4K4 in lung adenocarcinoma. Pathol Res Pract 2012; 208:541-8; PMID:22824148; http://dx.doi.org/ 10.1016/j.prp.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 23. Zhao G, Wang B, Liu Y, Zhang JG, Deng SC, Qin Q, Tian K, Li X, Zhu S, Niu Y, et al. miRNA-141, downregulated in pancreatic cancer, inhibits cell proliferation and invasion by directly targeting MAP4K4. Mol Cancer Ther 2013; 12:2569-80; PMID:24013097; http://dx.doi.org/ 10.1158/1535-7163.MCT-13-0296 [DOI] [PubMed] [Google Scholar]
- 24. Bartel F, Taubert H, Harris LC. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell 2002; 2:9-15; PMID:12150820; http://dx.doi.org/ 10.1016/S1535-6108(02)00091-0 [DOI] [PubMed] [Google Scholar]
- 25. Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer 2013; 13:83-96; PMID:23303139; http://dx.doi.org/ 10.1038/nrc3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang M, Zhang H, Liu T, Tian D, Gu L, Zhou M. Triptolide inhibits MDM2 and induces apoptosis in acute lymphoblastic leukemia cells through a p53-independent pathway. Mol Cancer Ther 2013; 12:184-94; PMID:23243057; http://dx.doi.org/ 10.1158/1535-7163.MCT-12-0425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jung CH, Kim J, Park JK, Hwang SG, Moon SK, Kim WJ, Um HD. Mdm2 increases cellular invasiveness by binding to and stabilizing the Slug mRNA. Cancer Lett 2013; 335:270-7; PMID:23438693; http://dx.doi.org/ 10.1016/j.canlet.2013.02.035 [DOI] [PubMed] [Google Scholar]
- 28. Kaindl U, Morak M, Portsmouth C, Mecklenbrauker A, Kauer M, Zeginigg M, Attarbaschi A, Haas OA, Panzer-Grümayer R. Blocking ETV6/RUNX1-induced MDM2 overexpression by Nutlin-3 reactivates p53 signaling in childhood leukemia. Leukemia 2014; 28:600-8; PMID:24240203; http://dx.doi.org/ 10.1038/leu.2013.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carvajal D, Tovar C, Yang H, Vu BT, Heimbrook DC, Vassilev LT. Activation of p53 by MDM2 antagonists can protect proliferating cells from mitotic inhibitors. Cancer Res 2005; 65:1918-24; PMID:15753391; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-3576 [DOI] [PubMed] [Google Scholar]
- 30. Chaar I, Amara S, Khiari M, Ounissi D, Dhraif M, Ben HAE, Gharbi L, Mzabi S, Bouraoui S. Relationship between MDM2 and p53 alterations in colorectal cancer and their involvement and prognostic value in the Tunisian population. Appl Immunohistochem Mol Morphol 2013; 21:228-36; PMID:22914606 [DOI] [PubMed] [Google Scholar]
- 31. Tesz GJ, Guilherme A, Guntur KV, Hubbard AC, Tang X, Chawla A, Czech MP. Tumor necrosis factor alpha (TNFalpha) stimulates Map4k4 expression through TNFalpha receptor 1 signaling to c-Jun and activating transcription factor 2. J Biol Chem 2007; 282:19302-12; PMID:17500068; http://dx.doi.org/ 10.1074/jbc.M700665200 [DOI] [PubMed] [Google Scholar]
- 32. Shimokawa N, Qiu CH, Seki T, Dikic I, Koibuchi N. Phosphorylation of JNK is involved in regulation of H(+)-induced c-Jun expression. Cell Signal 2004; 16:723-9; PMID:15093613; http://dx.doi.org/ 10.1016/j.cellsig.2003.11.005 [DOI] [PubMed] [Google Scholar]
- 33. Toh WH, Siddique MM, Boominathan L, Lin KW, Sabapathy K. c-Jun regulates the stability and activity of the p53 homologue, p73. J Biol Chem 2004; 279:44713-22; PMID:15302867; http://dx.doi.org/ 10.1074/jbc.M407672200 [DOI] [PubMed] [Google Scholar]
- 34. Xu YM, Zhu F, Cho YY, Carper A, Peng C, Zheng D, Yao K, Lau AT, Zykova TA, Kim HG, et al. Extracellular signal-regulated kinase 8-mediated c-Jun phosphorylation increases tumorigenesis of human colon cancer. Cancer Res 2010; 70:3218-27; PMID:20395206; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-4306 [DOI] [PubMed] [Google Scholar]
- 35. Chao CH, Chang CC, Wu MJ, Ko HW, Wang D, Hung MC, Yang JY, Chang CJ. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Invest 2014; 124:3093-106; PMID:24911147; http://dx.doi.org/ 10.1172/JCI73351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han TS, Hur K, AUID Oho, Xu G, Choi B, Okugawa Y, Toiyama Y, Oshima H, Oshima M, Lee HJ, et al. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut 2014; 64:203-14; PMID:24870620; http://dx.doi.org/ 10.1136/gutjnl-2013-306640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang Y, Liu S, Sun X, Wang M, Hou Y, et al. A microRNA 221- and 222-Mediated Feedback Loop, via PDLIM2, Maintains Constitutive Activation of NFkappaB and STAT3 in Colorectal Cancer Cells. Gastroenterology 2014; 147:847-859; PMID:24931456; http://dx.doi.org/ 10.1053/j.gastro.2014.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tan YG, Zhang YF, Guo CJ, Yang M, Chen MY. Screening of differentially expressed microRNA in ulcerative colitis related colorectal cancer. Asian Pac J Trop Med 2013; 6:972-6; PMID:24144030; http://dx.doi.org/ 10.1016/S1995-7645(13)60174-1 [DOI] [PubMed] [Google Scholar]
- 39. Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove R, Xu R, Huang W. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology 2010; 52:2148-57; PMID:20979124; http://dx.doi.org/ 10.1002/hep.23915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Selth LA, Townley SL, Bert AG, Stricker PD, Sutherland PD, Horvath LG, Goodall GJ, Butler LM, Tilley WD. Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br J Cancer 2013; 109:641-50; PMID:23846169; http://dx.doi.org/ 10.1038/bjc.2013.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wright JH, Wang X, Manning G, LaMere BJ, Le P, Zhu S, Khatry D, Flanagan PM, Buckley SD, Whyte DB, et al. The STE20 kinase HGK is broadly expressed in human tumor cells and can modulate cellular transformation, invasion, and adhesion. Mol Cell Biol 2003; 23:2068-82; PMID:12612079; http://dx.doi.org/ 10.1128/MCB.23.6.2068-2082.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Qiu MH, Qian YM, Zhao XL, Wang SM, Feng XJ, Chen XF, Zhang SH. Expression and prognostic significance of MAP4K4 in lung adenocarcinoma. Pathol Res Pract 2012; 208:541-8; PMID:22824148; http://dx.doi.org/ 10.1016/j.prp.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 43. Ye Y, Li X, Yang J, Miao S, Wang S, Chen Y, Xia X, Wu X, Zhang J, Zhou Y, et al. MDM2 is a useful prognostic biomarker for resectable gastric cancer. Cancer Sci 2013; 104:590-8; PMID:23347235; http://dx.doi.org/ 10.1111/cas.12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu Q, Li Y, Mu K, Li Z, Meng Q, Wu X, Wang Y, Li L. Amplification of Mdmx and overexpression of MDM2 contribute to mammary carcinogenesis by substituting for p53 mutations. Diagn Pathol 2014; 9:71; PMID:24667108; http://dx.doi.org/ 10.1186/1746-1596-9-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, Imai H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett 2005; 579:318-24; PMID:15642338; http://dx.doi.org/ 10.1016/j.febslet.2004.11.067 [DOI] [PubMed] [Google Scholar]
- 46. Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods 2004; 1:47-53; PMID:15782152; http://dx.doi.org/ 10.1038/nmeth704 [DOI] [PubMed] [Google Scholar]
- 47. Collins CS, Hong J, Sapinoso L, Zhou Y, Liu Z, Micklash K, Schultz PG, Hampton GM. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc Natl Acad Sci U S A 2006; 103:3775-80; PMID:16537454; http://dx.doi.org/ 10.1073/pnas.0600040103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hwang JH, Voortman J, Giovannetti E, Steinberg SM, Leon LG, Kim YT, Funel N, Park JK, Kim MA, Kang GH, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PLoS One 2010; 5:e10630; PMID:20498843; http://dx.doi.org/ 10.1371/journal.pone.0010630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee BL, Lee HS, Jung J, Cho SJ, Chung HY, Kim WH, Jin YW, Kim CS, Nam SY. Nuclear factor-κB activation correlates with better prognosis and Akt activation in human gastric cancer. Clinical cancer research 2005; 11:2518-25; PMID:15814628; http://dx.doi.org/ 10.1158/1078-0432.CCR-04-1282 [DOI] [PubMed] [Google Scholar]
- 50. Li L, Wang L, Song P, Geng X, Liang X, Zhou M, Wang Y, Chen C, Jia J, Zeng J. Critical role of histone demethylase RBP2 in human gastric cancer angiogenesis. Mol Cancer 2014; 13:81; PMID:24716659; http://dx.doi.org/ 10.1186/1476-4598-13-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.