Abstract
Protein synthesis is one of the most energy consuming processes in the cell. The mammalian/mechanistic target of rapamycin (mTOR) is a serine/threonine kinase that integrates a multitude of extracellular signals and intracellular cues to drive growth and proliferation. mTOR activity is altered in numerous pathological conditions, including metabolic syndrome and cancer. In addition to its well-established role in regulating mRNA translation, emerging studies indicate that mTOR modulates mitochondrial functions. In mammals, mTOR coordinates energy consumption by the mRNA translation machinery and mitochondrial energy production by stimulating synthesis of nucleus-encoded mitochondria-related proteins including TFAM, mitochondrial ribosomal proteins and components of complexes I and V. In this review, we highlight findings that link mTOR, mRNA translation and mitochondrial functions.
Keywords: 4E-BP1, eIF4E, mitochondria, metabolism, mRNA translation, mTOR, oxidative phosphorylation, TCA cycle
Background
Protein synthesis positively correlates with cell proliferation rates.1 It is therefore not surprising that upregulated mRNA translation is a common feature of pathological states that are characterized by aberrant proliferation including malignancies.2-5 In addition to playing an integral role in gene expression pathway, protein synthesis is one of the most energy consuming processes in the cell, and thus must be closely coordinated with cellular energy production.4,6,7 Similarly to protein synthesis, perturbations in energy metabolism are a frequent feature of cancer.8 These alterations in metabolic programs of cancer cells accommodate their elevated energy demand and provide building blocks (e.g. lipids, nucleotides) for continuous proliferation.9 Nonetheless, the mechanisms that coordinate mRNA translation and ATP production in mammals are still largely unknown.
The mechanistic/mammalian target of rapamycin (mTOR) is an evolutionarily conserved serine/threonine kinase that responds to a number of stimuli including hormones (insulin), growth factors (e.g., insulin-like growth factors [IGFs]), nutrients (amino acids), energy status, and oxygen levels to regulate cellular proliferation and growth rates.10,11 To bolster cellular proliferation and growth, mTOR stimulates anabolic processes including protein synthesis, and, as recent data show, acts as a major regulator of energy production in mitochondria.10-12 In turn, mTOR inhibits autophagy, which is a process that can eliminate mitochondria.13-16 As a consequence of inactivating mutations in tumor suppressor genes (e.g. PTEN, TSC1/2, NF1, LKB1) or hyperactivation of oncogenes (e.g., AKT, PI3K) mTOR signaling is upregulated in a plethora of malignancies.17 Recently, numerous mTOR mutations that lead to its hyperactivation have been described in cancer.18 Upregulated mTOR activity is thought to play a central role in tumorigenesis and progression of a wide variety of cancers.17 Taken together, these findings suggest that mTOR is well positioned to act as a central node of cellular networks that coordinate mRNA translation and cellular energy production. Indeed, we have recently uncovered that mTOR coordinates protein synthesis and mitochondrial functions by selectively modulating synthesis of nuclear-encoded mitochondrial proteins.12
mTOR signaling pathway
mTOR forms 2 distinct complexes, mTOR complex 1 (mTORC1) and 2 (mTORC2), which differ in their composition, downstream targets, and sensitivity to naturally occurring allosteric mTOR inhibitor rapamycin.19-21 mTORC1 stimulates protein synthesis and other anabolic processes to fuel cellular growth and proliferation, and is sensitive to acute rapamycin treatment.11,22 In most cell types, mTORC2 is insensitive to acute rapamycin treatment, regulates cytoskeletal organization, phosphorylates AGC kinases such as SGK1 and AKT, and has been implicated in the degradation of newly synthesized polypeptides.23
A multitude of extracellular signals and intracellular cues have been implicated in the modulation of mTORC1 signaling. In response to growth factors, mTORC1 is activated via the PI3K/AKT pathway.11 AKT phosphorylates and inactivates tuberous sclerosis complex (TSC1/2), which acts as a GTPase-activating protein (GAP) toward the small GTPase RAS homolog enriched in brain (RHEB).24,25 Inhibition of TSC complex increases levels of GTP-bound RHEB leading to activation of mTORC1. In addition, amino acids (in particular those with branched chains) signal to mTORC1 through the RAS-related GTP binding proteins (RAG) family of GTPases.26 RAG GTPases recruit mTORC1 to lysosomes where mTORC1 is stimulated by RHEB.27,28 A decrease in ATP/AMP ratio activates AMP kinase (AMPK) to suppress mTORC1 in a TSC complex-dependent or independent manner.29,30 Furthermore, hypoxia induces the expression of REDD1 that binds to TSC2 to stabilize the TSC complex, leading to inhibition of mTORC1.31-33 mTORC1 activation leads to phosphorylation of a number of substrates including eukaryotic translation initiation factor 4E (eIF4E)-binding proteins (4E-BPs), ribosomal protein S6 kinases (S6Ks), LARP1, Atg13, ULK1/2, which results in the upregulation of anabolic processes such as protein and lipid synthesis and inhibition of autophagy (reviewed in11).
Compared to mTORC1, upstream regulation of mTORC2 is less well understood.23 mTORC2 kinase activity is stimulated by growth factors, seemingly through a PI3K-dependent association of mTORC2 with ribosomes.34 Growth factors (e.g. insulin) also induce mTORC2 localization to the mitochondria-associated ER membrane (MAM), a sub-compartment of the ER.35 There, mTORC2 controls MAM integrity and mitochondrial function in an AKT-dependent manner (see below).
mTOR: a master regulator of mRNA translation
mTORC1 stimulates protein synthesis by phosphorylating a plethora of substrates.11,22 The 2 best established effectors of mTORC1 signaling on protein synthesis are the eukaryotic translation initiation factor 4E (eIF4E)-binding proteins (4E-BPs) and ribosomal protein S6 kinases (S6Ks) (Fig. 1).36 eIF4E is a cap binding subunit of the eIF4F translation initiation complex that also comprises large scaffolding protein eIF4G and DEAD box helicase eIF4A and facilitates recruitment of mRNA to the ribosome.37 Phosphorylation of 4E-BPs by mTORC1 stimulates their release from eIF4E, which allows eIF4E:eIF4G association and the assembly of the eIF4F complex, thereby increasing translation initiation rates.36,38-42 S6Ks phosphorylate a number of components of the translational machinery and related regulators such as ribosomal protein S6,43 eIF4B,44 and PDCD4.45 mTORC1 has also been shown to phosphorylate additional components of the translation initiation machinery (e.g., eIF4G46) and the eukaryotic translation elongation factor 2 kinase (eEF2K).47,48 Finally, mTORC1 is thought to increase translation by stimulating rRNA and tRNA synthesis, via activation of TIF-IA49 and inhibition of Maf1,50 respectively.
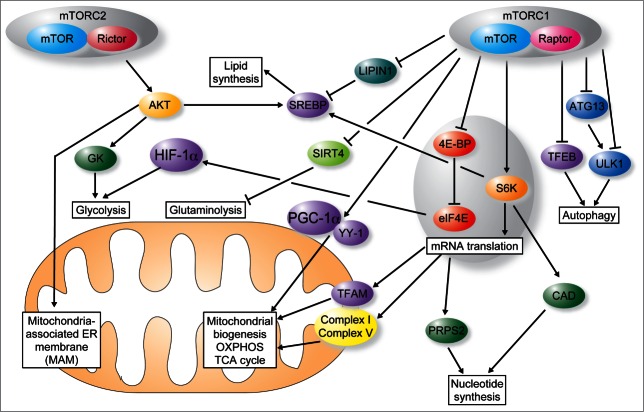
Figure 1.
mTOR coordinates protein synthesis, mitochondrial energy production, lipid and nucleotide synthesis and autophagy to fuel cell growth and proliferation. Proliferating cells have heightened requirement of building blocks (nucleotides, lipids and proteins) and energy as compared to quiescent cells. mTOR complex 1 (mTORC1) stimulates mitochondrial functions and biogenesis through the 4E-BP-mediated control of translation of nuclear-encoded mitochondrial mRNAs such as TFAM, mitochondrial ribosomal proteins and components of complex I and V. In addition, mTORC1 regulates mitochondrial function by modulating transcription of mitochondrial nuclear-encoded genes via Yin Yang 1 (YY1) and peroxisome proliferator-activated receptor-gamma coactivator 1 α (PGC-1α). Finally, mTORC1 modulates glycolysis through the 4E-BP1-dependent translational activation of the hypoxia-inducible factor 1 α (HIF1α) and glutaminolysis by inhibiting SIRT4. mTORC1 increases nucleotide synthesis through S6K dependent phosphorylation of carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, dihydroorotase (CAD) and induction of translation of phosphoribosyl-pyrophosphate synthetase 2 (PRPS2) mRNA mTORC1 also stimulates lipid synthesis by activating sterol regulatory element-binding proteins (SREBPs) via LIPIN1 and suppresses autophagy by inhibiting ULK1 (directly or via ATG13), and impeding nuclear translocation of transcription factor EB (TFEB). In contrast to mTORC1, following recruitment to the mitochondria-associated ER membrane (MAM) mTOR complex 2 (mTORC2) suppresses mitochondrial ATP production, membrane potential, and calcium uptake by phosphorylating MAM resident proteins. mTORC2 also regulates lipid metabolism by activating Akt and SREBP1c and stimulates glycolysis though the activity of Akt-stimulated glucokinase (GK). Arrows depict activation, and T-bars inhibition.
In addition to stimulating global protein synthesis, a large body of data indicate that mTOR selectively stimulates translation of a subset of transcripts including TOP mRNAs, which harbour a stretch of 4-14 pyrimidines following the cap structure, referred to as 5′ terminal oligopyrimidine (5′ TOP) motif.51 The vast majority of TOP mRNAs encode ribosomal proteins and other components of the protein synthesis machinery (e.g. eEF2). Albeit it has been initially thought that S6Ks mediate the effects of mTOR signaling on the synthesis of proteins encoded by TOP mRNAs, subsequent studies revealed that neither S6Ks nor phosphorylation of ribosomal protein S6 play a major role in regulating TOP mRNA translation.52,53 Two recent studies deployed high resolution translational profiling based on deep sequencing of ribosome-protected fragments to show that the effects of mTOR inhibitors on TOP mRNA translation are mediated by 4E-BPs.54,55 However, these results were challenged by a study showing that 4E-BPs are dispensable for the regulation of TOP mRNA translation under a variety of physiological stimuli including oxygen availability, amino acids and growth factors.56 These findings are consistent with a previous observation that eIF4E activity does not have a major influence on translation of TOP mRNAs.57 Therefore, the effectors of mTOR signaling on TOP mRNA translation appear to be context dependent. To this end, additional factors including TIA/TIAR-156,58 and LARP159 have been proposed to act as modulators of TOP mRNA translation.
In addition to regulating TOP mRNA translation, mTOR has been implicated in the regulation of synthesis of a number of cancer promoting proteins via inactivation of 4E-BPs and consequent increase in eIF4E activity. eIF4E exhibits oncogenic properties in vitro and in vivo and is overexpressed in the vast majority of cancers.3,60-66 These tumorigenic functions of eIF4E are a consequence of the selective upregulation of translation of mRNAs encoding cell cycle regulators (e.g., cyclins, ODC), survival promoting proteins (Bcl-xL, survivin, osteopontin), pro-angiogenic factors (e.g. VEGF) and oncogenes (e.g., Myc, Pim1).60,67 These mRNAs are thought to be “eIF4E-sensitive,” as a majority of them bear long and highly structured 5′UTRs,3,60,68 rendering them more dependent on the unwinding activity of eIF4A helicase69 than those mRNAs that are characterized by short, unstructured 5′UTRs such as those encoding housekeeping proteins. eIF4A is recruited to mRNA as a part of the eIF4F complex, and its activity is significantly higher when it is part of the eIF4F complex than as a single protein.70 Therefore, increase in eIF4E availability is thought to selectively stimulate translation of those mRNAs that critically depend on the dissolution of 5′UTR secondary structures by eIF4A.70-73
Using transcriptome-wide polysome profiling in conjunction with DNA microarrays, we demonstrated that, in addition to components of the translational machinery encoded by TOP mRNAs, mTOR inhibitors suppress the translation of transcripts encoding cell cycle and survival regulating proteins.74 Intriguingly, the most enriched mRNAs were those encoding for proteins implicated in the regulation of mitochondrial functions (Fig. 1).74
Notwithstanding the fact that the precise mechanisms by which mTOR regulates protein synthesis are still being debated, these findings demonstrate that changes in mTOR activity are paralleled not only by quantitative, but also by qualitative changes in the pools of mRNAs that are being translated. Moreover, the effects of mTOR on selective changes in pools of translating mRNAs are likely to be dependent on the nature of the stimulus and mediated by different downstream effectors.
mTOR regulates mitochondrial mass and functions by coordinating multiple levels of gene expression
Emerging data indicate that, in eukaryotes, coordinated expression of genes that are involved in the same biochemical processes is achieved via orchestration of different layers of gene expression machinery.75-77 Short-term (i.e. 12 h) mTOR inhibition does not induce major changes in the transcription of nuclear-encoded mitochondria-related genes.12 However, prolonged treatment with rapamycin downregulates the expression of pivotal transcriptional regulators of mitochondrial functions including PGC-1α, and ERR-α.78 This is paralleled by a decrease in mitochondrial respiration in skeletal muscle tissue and cell lines. The effect of mTOR on PGC-1α is mediated by ying-yang 1 (YY1), which belongs to the GLI-Kruppel class of zinc finger proteins and acts as a multifunctional transcriptional regulator.78 Depletion of YY1 results in downregulated expression of a number of nuclear encoded mitochondrial genes and decreased oxygen consumption.78 These findings suggest that, in addition to regulating the translation of nuclear-encoded mitochondria-related mRNAs, mTOR also regulates the transcription of nuclear encoded mitochondrial genes. Therefore, it appears that mTOR regulates the expression of nuclear-encoded mitochondrial genes by orchestrating their transcriptional and translational programs (Fig. 1). Moreover, it has been shown that mTOR directly governs the transcription of ERRα-target genes involved in energy metabolism including citric acid cycle and lipogenesis,79 further illustrating the coordination of transcriptional and translational energy homeostasis programs via mTOR.
mTOR links protein synthesis, mitochondrial function, and proliferation
Protein synthesis rates positively correlate with proliferation rates.1 In turn, mitochondrial ATP production is required to fuel protein synthesis and proliferation.6,7 These findings suggest that mitochondrial energy production, protein synthesis and proliferation are co-regulated, but the factors that orchestrate coordination of these processes are still largely unknown.
Experiments carried out in the model organism D. Melanogaster revealed that regulation of the expression of nuclear-encoded mitochondrial regulators at the level of translation plays a major role in lifespan extension by caloric restriction.80 Caloric restriction induces expression of d4E-BP (in contrast to mammals, flies express only one 4E-BP), which is paralleled by decreased phosphorylation of d4E-BP via downregulation of TOR signaling. Increased levels and decreased phosphorylation of d4E-BP resulted in suppression of global protein synthesis, but increased the translational efficacy of mRNAs encoding factors implicated in mitochondrial respiration.80 Although the precise mechanism underpinning the upregulation of the translation of mitochondria-regulating mRNAs under dietary restriction is unknown, these findings put forward a model whereby nutrient deprivation selectively induces the synthesis of mitochondrial regulators that impact the function of the electron transport chain via downregultion of TOR and activation of d4E-BP.
In mammalian cells, however, mTORC1 stimulates the synthesis of a number of nuclear-encoded mitochondrial regulators such as TFAM, mitochondrial ribosomal proteins and components of complex I and V by upregulating the translation of corresponding mRNAs12 (Fig. 1). Inhibition of mTOR signaling strongly decreases mitochondrial biogenesis and respiration in 4E-BP proficient cells, but not in those lacking 4E-BPs.12 The elevated mitochondrial respiratory capacity observed in cells where mTORC1 is hyperactivated by PTEN loss also appears to be mediated by the selective upregulation of expression of components of the electron transport chain via inactivation of 4E-BPs.81 In wild-type mice, mTOR inhibitors cause reduced systemic oxygen consumption and decreased locomotor activity and heat production. Mice lacking 4E-BPs were resistant to the systemic effects of mTOR inhibitors.12 Notwithstanding the apparent differences in the effects of the TOR/4E-BP pathway on translation of mRNAs encoding mitochondrial regulators in flies and mammals that likely stem from their different metabolic requirements, these findings indicate that TOR and 4E-BPs play a major role in coupling mitochondrial functions and translation. Moreover, it seems that translational control may play an even broader role in the regulation of mitochondrial functions, in as much as Largen, which is an important regulator of cells size, impacts mitochondrial activity by inducing selective perturbations in the translation of nuclear-encoded mitochondria-related mRNAs in a mTOR-independent manner.82 Collectively, these studies show that translational activity in the cell influences mitochondrial functions.
In addition to regulating synthesis of nuclear-encoded mitochondrial regulators, the mTORC1/4E-BP pathway regulates translation of mRNAs that encode proteins that promote proliferation such as cyclins, ODC, and Myc.74 Accordingly, 4E-BP status in the cell is a major determinant of the effects of mTOR on proliferation, inasmuch as cells lacking 4E-BPs maintain their proliferation under circumstances where mTORC1 signaling is inhibited by pharmacological (e.g. active-site mTOR inhibitors) or genetic (depletion of raptor) means or by nutrient deprivation (e.g., serum or amino-acid depletion).83 These results suggest that mTOR coordinates mitochondrial functions and proliferation, at least in part by modulating translation programs (Fig. 1).
In addition to mTORC1, mTORC2 appears to be an important regulator of mitochondrial functions (Fig. 1). Upon growth factor stimulation mTORC2 is recruited to the mitochondria-associated ER membrane (MAM), where it maintains MAM integrity via AKT dependent phosphorylation of MAM resident proteins including IP3 receptor and hexokinase 2.35 Loss of mTORC2 activity leads to disruption of MAM, paralleled by increase in mitochondrial membrane potential, and calcium uptake.35 This suggests that mTORC1 and mTORC2 play distinct, non-overlapping roles in regulating mitochondrial functions.
mTOR coordinates mRNA translation, availability of cellular building blocks and autophagy
mTORC1 promotes synthesis of building blocks that are required for cell proliferation (Fig. 1). To this end, mTORC1 stimulates lipid and sterol synthesis by activating sterol regulatory element-binding proteins (SREBPs),84 whereas it negatively regulates β-oxidation of free fatty acids.85 In vivo, depletion of raptor in adipose tissue of mice induces a lean phenotype, paralleled by increased energy expenditure and increased levels of mitochondrial uncoupling proteins.86 In addition, mTORC1 stimulates nucleotide synthesis via induction of the pentose phosphate pathway and S6K-mediated activation of carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, dihydroorotase (CAD).87,88 Analogous to enhancing mitochondrial functions by selectively increasing mRNA translation, mTORC1 appears to link protein synthesis and nucleotide availability inasmuch as eIF4E stimulates nucleotide synthesis via increased translation of phosphoribosyl-pyrophosphate synthetase 2 (PRPS2) mRNA.89 Increased aerobic glycolysis and glutaminolysis are hallmarks of energy metabolism reprogramming in cancer.9 mTORC1 increases glucose uptake and glycolysis through induction of the hypoxia-inducible factor 1 α (HIF1α).84,90,91 In addition, mTORC1 pathway stimulates glutamine anaplerosis and cell proliferation by repressing SIRT4 and thus promoting the activity of glutamate dehydrogenase.92 mTORC2 has also been shown to play a major role in the regulation of energy metabolism. For instance, liver-specific inhibition of mTORC2 signaling in conditional rictor knock-out mice revealed that mTORC2 regulates glycolysis and lipid metabolism through the Akt-dependent activation of glucokinase and SREBP1c, respectively93 (Fig. 1).
In parallel to stimulating anabolic processes, mTOR also inhibits autophagy (Fig. 1), which is a major catabolic process in the cell.94 mTORC1 inhibits autophagosome formation by phosphorylating the pro-autophagic kinase ULK1 and thus preventing its activation by AMPK,16 and by phosphorylating and inhibiting ATG13, a positive regulator of ULK1.13-15 mTORC1 also inhibits autophagy indirectly by blocking lysosome biogenesis through the phosphorylation and inhibition of the nuclear translocation of transcription factor EB (TFEB).95-97 mTORC1 inhibition by asTORi reduces mitochondrial mass by induction of autophagy, and these effects were alleviated by suppressing autophagy via depletion of ATG5.12 Therefore, mTORC1 induces mitochondrial biogenesis and functions by orchestrating synthesis of nuclear-encoded mitochondrial regulators and inhibiting autophagy.
Conclusions
mTOR activity is central to energy homeostasis, inasmuch as it coordinates protein synthesis, cell growth and proliferation, generation of metabolic intermediates, and mitochondrial biogenesis and functions (Fig. 1). Accordingly, dysregulation of mTOR signaling and mitochondrial dysfunction underpin aging and diseases such as cancer, diabetes, and neurodegeneration (reviewed in98,99). For instance, increased life span of female transgenic mice expressing non-steroidal anti-inflammatory drug-activated gene (NAG-1)/GDF15 is paralleled by downregulation of mTOR activity.100 Interestingly, many components of the electron transport chain (ETC) complexes that show alteration in expression in the process of aging,101 such as NDUFS6, ATP5D, ATP5L and ATP5O, were shown to be translationally controlled by mTOR.12 Cancer is characterized by aberrant proliferation, increased protein synthesis and perturbations in cellular energy metabolism.8 In turn, mTOR signaling is dysergulated in a vast majority of cancers, while low expression and high phosphorylation status of 4E-BPs, as well as upregulation in eIF4E levels, are also common in neoplasia.102 Notably, increase in eIF4E/4E-BP ratio appears to be a major mechanism of resistance to PI3K and mTOR-targeted therapies.103-107 This raises an intriguing possibility that cancer cells highjack mechanisms by which the hyperactivation of the mTOR/4E-BP/eIF4E pathway coordinates mitochondrial functions, nucleotide and lipid synthesis and translational programs to fuel neoplastic growth. A recent study established a mTOR-independent link between protein synthesis and mitochondrial functions, whereby Largen appears to regulate cell growth at least in part by modulating translation of nuclear-encoded mRNAs that code mitochondrial proteins in rapamycin-insensitive manner.82 Although the underlining mechanisms of this phenomenon remain to be established, these findings suggest that multiple pathways evolved to maintain cellular energy balance by coordinating mitochondrial functions, translational activity, cellular growth and proliferation. Therefore, the identification of specific translational programs mediated by mTOR-dependent and -independent pathways, their effectors, and the mechanisms by which they modify mitochondrial functions represent a promising avenue to improve the understanding of the molecular mechanisms of cellular energy homeostasis that contrast normal and malignant cells.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are thankful to Shannon McLaughlan for technical support and Valentina Gandin for invaluable comments.
Funding
This perspective is based on research projects in St-Pierre and Topisirovic labs that are supported by a Terry Fox Research Institute team grant (TFF-116128) to I.T., M.P. and J.S-P. and grants from the Canadian Institutes of Health Research (CIHR MOP-115195 to I.T; MOP-106603 to J.S-P.). I.T. is a recipient of CIHR New Investigator Salary Award. J.S-P. is an FRQS scholar. M.M. is a recipient of a CIHR-funded Chemical Biology Postdoctoral fellowship and Canadian Diabetes Association Postdoctoral fellowship.
References
- 1. Larsson O, Zetterberg A, Engstrom W. Cell-cycle-specific induction of quiescence achieved by limited inhibition of protein synthesis: counteractive effect of addition of purified growth factors. J Cell Sci 1985; 73:375-87; PMID:3894388 [DOI] [PubMed] [Google Scholar]
- 2. Ruggero D. Translational control in cancer etiology. Cold Spring Harb Perspect Biol 2013; 5:pii: a012336.; PMID:22767671; http://dx.doi.org/ 10.1101/cshperspect.a012336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer 2010; 10:254-66; PMID:20332778; http://dx.doi.org/ 10.1038/nrc2824 [DOI] [PubMed] [Google Scholar]
- 4. Topisirovic I, Sonenberg N. mRNA translation and energy metabolism in cancer: the role of the MAPK and mTORC1 pathways. Cold Spring Harb Symp Quant Biol 2011; 76:355-67; PMID:22123850 [DOI] [PubMed] [Google Scholar]
- 5. Larsson O, Li S, Issaenko OA, Avdulov S, Peterson M, Smith K, Bitterman PB, Polunovsky VA. Eukaryotic translation initiation factor 4E induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Res 2007; 67:6814-24; PMID:17638893; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-0752 [DOI] [PubMed] [Google Scholar]
- 6. Rolfe DF, Brown GC. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol Rev 1997; 77:731-58; PMID:9234964 [DOI] [PubMed] [Google Scholar]
- 7. Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J 1995; 312 (Pt 1):163-7; PMID:7492307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 9. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 2012; 21:297-308; PMID:22439925; http://dx.doi.org/ 10.1016/j.ccr.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med (Berl) 2011; 89:221-8; PMID:21301797; http://dx.doi.org/ 10.1007/s00109-011-0726-6 [DOI] [PubMed] [Google Scholar]
- 11. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274-93; PMID:22500797; http://dx.doi.org/ 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 2013; 18:698-711; PMID:24206664; http://dx.doi.org/ 10.1016/j.cmet.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 13. Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 2009; 284:12297-305; PMID:19258318; http://dx.doi.org/ 10.1074/jbc.M900573200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009; 20:1981-91; PMID:19211835; http://dx.doi.org/ 10.1091/mbc.E08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009; 20:1992-2003; PMID:19225151; http://dx.doi.org/ 10.1091/mbc.E08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; PMID:21258367; http://dx.doi.org/ 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011; 12:21-35; PMID:21157483; http://dx.doi.org/ 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, Jordan A, Beck AH, Sabatini DM. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer discov 2014; 4:554-63; PMID:24631838; http://dx.doi.org/ 10.1158/2159-8290.CD-13-0929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002; 110:177-89; PMID:12150926; http://dx.doi.org/ 10.1016/S0092-8674(02)00833-4 [DOI] [PubMed] [Google Scholar]
- 20. Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002; 110:163-75; PMID:12150925; http://dx.doi.org/ 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- 21. Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 2004; 14:1296-302; PMID:15268862; http://dx.doi.org/ 10.1016/j.cub.2004.06.054 [DOI] [PubMed] [Google Scholar]
- 22. Caron E, Ghosh S, Matsuoka Y, Ashton-Beaucage D, Therrien M, Lemieux S, Perreault C, Roux PP, Kitano H. A comprehensive map of the mTOR signaling network. Mol Syst Biol 2010; 6:453; PMID:21179025; http://dx.doi.org/ 10.1038/msb.2010.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oh WJ, Jacinto E. mTOR complex 2 signaling and functions. Cell Cycle 2011; 10:2305-16; PMID:21670596; http://dx.doi.org/ 10.4161/cc.10.14.16586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003; 17:1829-34; PMID:12869586; http://dx.doi.org/ 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol 2003; 13:1259-68; PMID:12906785; http://dx.doi.org/ 10.1016/S0960-9822(03)00506-2 [DOI] [PubMed] [Google Scholar]
- 26. Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol 2013; 15:555-64; PMID:23728461; http://dx.doi.org/ 10.1038/ncb2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol 2008; 10:935-45; PMID:18604198; http://dx.doi.org/ 10.1038/ncb1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320:1496-501; PMID:18497260; http://dx.doi.org/ 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003; 115:577-90; PMID:14651849; http://dx.doi.org/ 10.1016/S0092-8674(03)00929-2 [DOI] [PubMed] [Google Scholar]
- 30. Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 2008; 30:214-26; PMID:18439900; http://dx.doi.org/ 10.1016/j.molcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG, Jr,. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev 2004; 18:2893-904; PMID:15545625; http://dx.doi.org/ 10.1101/gad.1256804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reiling JH, Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev 2004; 18:2879-92; PMID:15545626; http://dx.doi.org/ 10.1101/gad.322704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev 2008; 22:239-51; PMID:18198340; http://dx.doi.org/ 10.1101/gad.1617608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell 2011; 144:757-68; PMID:21376236; http://dx.doi.org/ 10.1016/j.cell.2011.02.014 [DOI] [PubMed] [Google Scholar]
- 35. Betz C, Stracka D, Prescianotto-Baschong C, Frieden M, Demaurex N, Hall MN. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci U S A 2013; 110:12526-34; PMID:23852728; http://dx.doi.org/ 10.1073/pnas.1302455110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roux PP, Topisirovic I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol 2012; 4:pii: a012252; PMID:22888049; http://dx.doi.org/ 10.1101/cshperspect.a012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 1999; 68:913-63; PMID:10872469; http://dx.doi.org/ 10.1146/annurev.biochem.68.1.913 [DOI] [PubMed] [Google Scholar]
- 38. Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr., Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 1994; 371:762-7; PMID:7935836; http://dx.doi.org/ 10.1038/371762a0 [DOI] [PubMed] [Google Scholar]
- 39. Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J 1996; 15:658-64; PMID:8599949 [PMC free article] [PubMed] [Google Scholar]
- 40. Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr., Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 1997; 277:99-101; PMID:9204908; http://dx.doi.org/ 10.1126/science.277.5322.99 [DOI] [PubMed] [Google Scholar]
- 41. Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem 1997; 272:26457-63; PMID:9334222; http://dx.doi.org/ 10.1074/jbc.272.42.26457 [DOI] [PubMed] [Google Scholar]
- 42. Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev 1999; 13:1422-37; PMID:10364159; http://dx.doi.org/ 10.1101/gad.13.11.1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Banerjee P, Ahmad MF, Grove JR, Kozlosky C, Price DJ, Avruch J. Molecular structure of a major insulin/mitogen-activated 70-kDa S6 protein kinase. Proc Natl Acad Sci U S A 1990; 87:8550-4; PMID:2236064; http://dx.doi.org/ 10.1073/pnas.87.21.8550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J 2004; 23:1761-9; PMID:15071500; http://dx.doi.org/ 10.1038/sj.emboj.7600193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 2006; 314:467-71; PMID:17053147; http://dx.doi.org/ 10.1126/science.1130276 [DOI] [PubMed] [Google Scholar]
- 46. Raught B, Gingras AC, Gygi SP, Imataka H, Morino S, Gradi A, Aebersold R, Sonenberg N. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. EMBO J 2000; 19:434-44; PMID:10654941; http://dx.doi.org/ 10.1093/emboj/19.3.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J 2001; 20:4370-9; PMID:11500364; http://dx.doi.org/ 10.1093/emboj/20.16.4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang X, Regufe da Mota S, Liu R, Moore CE, Xie J, Lanucara F, Agarwala U, Pyr Dit Ruys S, Vertommen D, Rider MH, et al. Eukaryotic elongation factor 2 kinase activity is controlled by multiple regulatory inputs from oncogenic and anabolic pathways. Mol Cell Biol 2014; 34(22):4088-103; PMID:25182533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev 2004; 18:423-34; PMID:15004009; http://dx.doi.org/ 10.1101/gad.285504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Michels AA, Robitaille AM, Buczynski-Ruchonnet D, Hodroj W, Reina JH, Hall MN, Hernandez N. mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol 2010; 30:3749-57; PMID:20516213; http://dx.doi.org/ 10.1128/MCB.00319-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Meyuhas O, Dreazen A. Ribosomal protein S6 kinase from TOP mRNAs to cell size. Prog Mol Biol Transl Sci 2009; 90:109-53; PMID:20374740; http://dx.doi.org/ 10.1016/S1877-1173(09)90003-5 [DOI] [PubMed] [Google Scholar]
- 52. Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol 2004; 24:3112-24; PMID:15060135; http://dx.doi.org/ 10.1128/MCB.24.8.3112-3124.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 2005; 19:2199-211; PMID:16166381; http://dx.doi.org/ 10.1101/gad.351605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012; 485:109-13; PMID:22552098; http://dx.doi.org/ 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012; 485:55-61; PMID:22367541; http://dx.doi.org/ 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Miloslavski R, Cohen E, Avraham A, Iluz Y, Hayouka Z, Kasir J, Mudhasani R, Jones SN, Cybulski N, Ruegg MA, et al. Oxygen sufficiency controls TOP mRNA translation via the TSC-Rheb-mTOR pathway in a 4E-BP-independent manner. J Mol Cell Biol 2014; 6:255-66; PMID:24627160; http://dx.doi.org/ 10.1093/jmcb/mju008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shama S, Avni D, Frederickson RM, Sonenberg N, Meyuhas O. Overexpression of initiation factor eIF-4E does not relieve the translational repression of ribosomal protein mRNAs in quiescent cells. Gene Exp 1995; 4:241-52; PMID:7787416 [PMC free article] [PubMed] [Google Scholar]
- 58. Damgaard CK, Lykke-Andersen J. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev 2011; 25:2057-68; PMID:21979918; http://dx.doi.org/ 10.1101/gad.17355911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev 2014; 28:357-71; PMID:24532714; http://dx.doi.org/ 10.1101/gad.231407.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. De Benedetti A, Graff JR. eIF-4E expression and its role in malignancies and metastases. Oncogene 2004; 23:3189-99; PMID:15094768; http://dx.doi.org/ 10.1038/sj.onc.1207545 [DOI] [PubMed] [Google Scholar]
- 61. Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 1990; 345:544-7; PMID:2348862; http://dx.doi.org/ 10.1038/345544a0 [DOI] [PubMed] [Google Scholar]
- 62. Rinker-Schaeffer CW, Graff JR, De Benedetti A, Zimmer SG, Rhoads RE. Decreasing the level of translation initiation factor 4E with antisense RNA causes reversal of ras-mediated transformation and tumorigenesis of cloned rat embryo fibroblasts. Int J Cancer 1993; 55:841-7; PMID:8244582; http://dx.doi.org/ 10.1002/ijc.2910550525 [DOI] [PubMed] [Google Scholar]
- 63. De Benedetti A, Joshi-Barve S, Rinker-Schaeffer C, Rhoads RE. Expression of antisense RNA against initiation factor eIF-4E mRNA in HeLa cells results in lengthened cell division times, diminished translation rates, and reduced levels of both eIF-4E and the p220 component of eIF-4F. Mol Cell Biol 1991; 11:5435-45; PMID:1922056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 2004; 428:332-7; PMID:15029198; http://dx.doi.org/ 10.1038/nature02369 [DOI] [PubMed] [Google Scholar]
- 65. Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med 2004; 10:484-6; PMID:15098029; http://dx.doi.org/ 10.1038/nm1042 [DOI] [PubMed] [Google Scholar]
- 66. Avdulov S, Li S, Michalek V, Burrichter D, Peterson M, Perlman DM, Manivel JC, Sonenberg N, Yee D, Bitterman PB, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell 2004; 5:553-63; PMID:15193258; http://dx.doi.org/ 10.1016/j.ccr.2004.05.024 [DOI] [PubMed] [Google Scholar]
- 67. Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E–from translation to transformation. Oncogene 2004; 23:3172-9; PMID:15094766; http://dx.doi.org/ 10.1038/sj.onc.1207549 [DOI] [PubMed] [Google Scholar]
- 68. Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J 1992; 11:4153-8; PMID:1396596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 2001; 7:382-94; PMID:11333019; http://dx.doi.org/ 10.1017/S135583820100108X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J 1994; 13:1205-15; PMID:8131750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hiremath LS, Webb NR, Rhoads RE. Immunological detection of the messenger RNA cap-binding protein. J Biol Chem 1985; 260:7843-9; PMID:3891747 [PubMed] [Google Scholar]
- 72. Duncan R, Milburn SC, Hershey JW. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J Biol Chem 1987; 262:380-8; PMID:3793730 [PubMed] [Google Scholar]
- 73. Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci U S A 2013; 110:13339-44; PMID:23901100; http://dx.doi.org/ 10.1073/pnas.1303781110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Larsson O, Morita M, Topisirovic I, Alain T, Blouin MJ, Pollak M, Sonenberg N. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc Natl Acad Sci U S A 2012; 109:8977-82; PMID:22611195; http://dx.doi.org/ 10.1073/pnas.1201689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Blackinton JG, Keene JD. Post-transcriptional RNA regulons affecting cell cycle and proliferation. Semin Cell Dev Biol 2014; 34:44-54; PMID:24882724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dahan O, Gingold H, Pilpel Y. Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet 2011; 27:316-22; PMID:21763027; http://dx.doi.org/ 10.1016/j.tig.2011.05.008 [DOI] [PubMed] [Google Scholar]
- 77. Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 2007; 8:533-43; PMID:17572691; http://dx.doi.org/ 10.1038/nrg2111 [DOI] [PubMed] [Google Scholar]
- 78. Cunningham JT, Rodgers JT, Arlow DH, Vazquez F, Mootha VK, Puigserver P. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature 2007; 450:736-40; PMID:18046414; http://dx.doi.org/ 10.1038/nature06322 [DOI] [PubMed] [Google Scholar]
- 79. Chaveroux C, Eichner LJ, Dufour CR, Shatnawi A, Khoutorsky A, Bourque G, Sonenberg N, Giguere V. Molecular and genetic crosstalks between mTOR and ERRalpha are key determinants of rapamycin-induced nonalcoholic fatty liver. Cell Metab 2013; 17:586-98; PMID:23562079; http://dx.doi.org/ 10.1016/j.cmet.2013.03.003 [DOI] [PubMed] [Google Scholar]
- 80. Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 2009; 139:149-60; PMID:19804760; http://dx.doi.org/ 10.1016/j.cell.2009.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Goo CK, Lim HY, Ho QS, Too HP, Clement MV, Wong KP. PTEN/Akt signaling controls mitochondrial respiratory capacity through 4E-BP1. PLoS One 2012; 7:e45806; PMID:23049865; http://dx.doi.org/ 10.1371/journal.pone.0045806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yamamoto K, Gandin V, Sasaki M, McCracken S, Li W, Silvester JL, Elia AJ, Wang F, Wakutani Y, Alexandrova R, et al. Largen: a molecular regulator of mammalian cell size control. Mol Cell 2014; 53:904-15; PMID:24656129; http://dx.doi.org/ 10.1016/j.molcel.2014.02.028 [DOI] [PubMed] [Google Scholar]
- 83. Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 2010; 328:1172-6; PMID:20508131; http://dx.doi.org/ 10.1126/science.1187532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Duvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 2010; 39:171-83; PMID:20670887; http://dx.doi.org/ 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhu Y, Soto J, Anderson B, Riehle C, Zhang YC, Wende AR, Jones D, McClain DA, Abel ED. Regulation of fatty acid metabolism by mTOR in adult murine hearts occurs independently of changes in PGC-1alpha. Am J Physiol Heart Circ Physiol 2013; 305:H41-51; PMID:23624629; http://dx.doi.org/ 10.1152/ajpheart.00877.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Polak P, Cybulski N, Feige JN, Auwerx J, Ruegg MA, Hall MN. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell Metab 2008; 8:399-410; PMID:19046571; http://dx.doi.org/ 10.1016/j.cmet.2008.09.003 [DOI] [PubMed] [Google Scholar]
- 87. Robitaille AM, Christen S, Shimobayashi M, Cornu M, Fava LL, Moes S, Prescianotto-Baschong C, Sauer U, Jenoe P, Hall MN. Quantitative phosphoproteomics reveal mTORC1 activates de novo pyrimidine synthesis. Science 2013; 339:1320-3; PMID:23429704; http://dx.doi.org/ 10.1126/science.1228771 [DOI] [PubMed] [Google Scholar]
- 88. Ben-Sahra I, Howell JJ, Asara JM, Manning BD. Stimulation of de novo pyrimidine synthesis by growth signaling through mTOR and S6K1. Science 2013; 339:1323-8; PMID:23429703; http://dx.doi.org/ 10.1126/science.1228792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell 2014; 157:1088-103; PMID:24855946; http://dx.doi.org/ 10.1016/j.cell.2014.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bhaskar PT, Nogueira V, Patra KC, Jeon SM, Park Y, Robey RB, Hay N. mTORC1 hyperactivity inhibits serum deprivation-induced apoptosis via increased hexokinase II and GLUT1 expression, sustained Mcl-1 expression, and glycogen synthase kinase 3beta inhibition. Mol Cell Biol 2009; 29:5136-47; PMID:19620286; http://dx.doi.org/ 10.1128/MCB.01946-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Dodd KM, Yang J, Shen MH, Sampson JR, Tee AR. mTORC1 drives HIF-1alpha and VEGF-A signalling via multiple mechanisms involving 4E-BP1, S6K1 and STAT3. Oncogene 2014; PMID:24931163; http://dx.doi.org/10.1038/onc.2014.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 2013; 153:840-54; PMID:23663782; http://dx.doi.org/ 10.1016/j.cell.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hagiwara A, Cornu M, Cybulski N, Polak P, Betz C, Trapani F, Terracciano L, Heim MH, Ruegg MA, Hall MN. Hepatic mTORC2 activates glycolysis and lipogenesis through Akt, glucokinase, and SREBP1c. Cell Metab 2012; 15:725-38; PMID:22521878; http://dx.doi.org/ 10.1016/j.cmet.2012.03.015 [DOI] [PubMed] [Google Scholar]
- 94. Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 2008; 283:10892-903; PMID:18281291; http://dx.doi.org/ 10.1074/jbc.M800102200 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95. Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012; 31:1095-108; PMID:22343943; http://dx.doi.org/ 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012; 5:ra42; PMID:22692423; http://dx.doi.org/ 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012; 8:903-14; PMID:22576015; http://dx.doi.org/ 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Blagosklonny MV, Hall MN. Growth and aging: a common molecular mechanism. Aging 2009; 1:357-62; PMID:20157523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hands SL, Proud CG, Wyttenbach A. mTOR's role in ageing: protein synthesis or autophagy? Aging 2009; 1:586-97; PMID:20157541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wang X, Chrysovergis K, Kosak J, Kissling G, Streicker M, Moser G, Li R, Eling TE. hNAG-1 increases lifespan by regulating energy metabolism and insulin/IGF-1/mTOR signaling. Aging 2014; 6:690-704; PMID:25239873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Miwa S, Jow H, Baty K, Johnson A, Czapiewski R, Saretzki G, Treumann A, von Zglinicki T. Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice. Nat Commun 2014; 5:3837; PMID:24815183; http://dx.doi.org/ 10.1038/ncomms4837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Martineau Y, Azar R, Bousquet C, Pyronnet S. Anti-oncogenic potential of the eIF4E-binding proteins. Oncogene 2013; 32:671-7; PMID:22508483; http://dx.doi.org/ 10.1038/onc.2012.116 [DOI] [PubMed] [Google Scholar]
- 103. Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, Zammit D, Marcus V, Metrakos P, Voyer LA, et al. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res 2012; 72:6468-76; PMID:23100465; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-2395 [DOI] [PubMed] [Google Scholar]
- 104. Martineau Y, Azar R, Muller D, Lasfargues C, El Khawand S, Anesia R, Pelletier J, Bousquet C, Pyronnet S. Pancreatic tumours escape from translational control through 4E-BP1 loss. Oncogene 2014; 33:1367-74; PMID:23563181; http://dx.doi.org/ 10.1038/onc.2013.100 [DOI] [PubMed] [Google Scholar]
- 105. Ilic N, Utermark T, Widlund HR, Roberts TM. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc Natl Acad Sci U S A 2011; 108:E699-708; PMID:21876152; http://dx.doi.org/ 10.1073/pnas.1108237108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Cope CL, Gilley R, Balmanno K, Sale MJ, Howarth KD, Hampson M, Smith PD, Guichard SM, Cook SJ. Adaptation to mTOR kinase inhibitors by amplification of eIF4E to maintain cap-dependent translation. J Cell Sci 2014; 127:788-800; PMID:24363449; http://dx.doi.org/ 10.1242/jcs.137588 [DOI] [PubMed] [Google Scholar]
- 107. Mallya S, Fitch BA, Lee JS, So L, Janes MR, Fruman DA. Resistance to mTOR kinase inhibitors in lymphoma cells lacking 4EBP1. PLoS One 2014; 9:e88865; PMID:24586420; http://dx.doi.org/ 10.1371/journal.pone.0088865 [DOI] [PMC free article] [PubMed] [Google Scholar]