In recent years a series of exciting discoveries, largely based on potent global genomic technologies, have profoundly changed our understanding of the mechanisms underlying integrated transcriptional programs. For example, genetic and genomic approaches to study the 3-dimensional organization of the genome at increasing resolution have suggested the impact of locus organization on transcriptional outcome.1 Chromosomes themselves are not randomly distributed in the nucleus, with the suggestion that gene-rich chromosomes (thus most likely transcribed) have a preferred localization in the center of the nucleus. The genome-wide chromosome capture approach (Hi-C) also revealed that interactions between DNA loci are enriched within ∼900kb local domains, and therefore constitute a novel type of architectural entity (topologically associating domains – TADs). Boundaries between these TADs are therefore proposed to correlate with transition between transcriptionally active and inactive genomic regions.1
The nucleus is replete with structures, often referred to as nuclear bodies that have been considered to represent storage granules for various factors involved with gene repression, activation or splicing events, activities now considered to be key functions of gene enhancers. Equally intriguing are the postulated roles of other structures such as nuclear lamina and inner nuclear network in organizing the genome and in transcriptional regulation. While mutations in nuclear lamina proteins have been shown to underlie diseases such as laminopathies,2 which include symptoms of progeria, subtypes of muscular dystrophy, and neuropathy the function of internal structures in development and disease is less well understood with the notable exception of special AT-rich binding protein (SATB1) and its role as a global regulator of gene expression in cancer.3 Matrin-3 protein is a structural component of internal nuclear network also called “nuclear matrix.” The importance of the nuclear matrix in functional organization of chromatin into higher-order loops and “matrix attached” elements has been proposed in 1970s but has been highly debated ever since.4 Matrin-3 has been found in the nuclear matrix/scaffold “central proteome,” regardless of the method of isolation.5
Our investigation6 of a developmentally-required POU-homeodomain transcription factor, Pit1/Pou1f1, has revealed a novel molecular mechanism whereby the binding of Pit1-occupied enhancers to a proteinaceous nuclear structure rich in matrin-3 is a key event in effective activation of the Pit1-regulated enhancers and coding gene transcriptional program (Fig. 1). We have shown that Pit1 association with Satb1 and β-catenin is required to tether Pit 1-occupied enhancers to the matrin-3 network. Two independent mechanisms of target gene activation - recruitment of the transcriptional coactivator p300 and expression of enhancer RNA (eRNA) are both dependent on the interaction of Pit1 bound enhancers with βcatenin/SATB1 and the matrin3-rich nuclear structure. These findings formally establish the critical importance of internal nuclear structures in mediating the formation of gene activation- competent complexes between the matrin-3-network, regulatory DNA sequences and transcriptional machinery, including non-coding RNAs.
Figure 1.
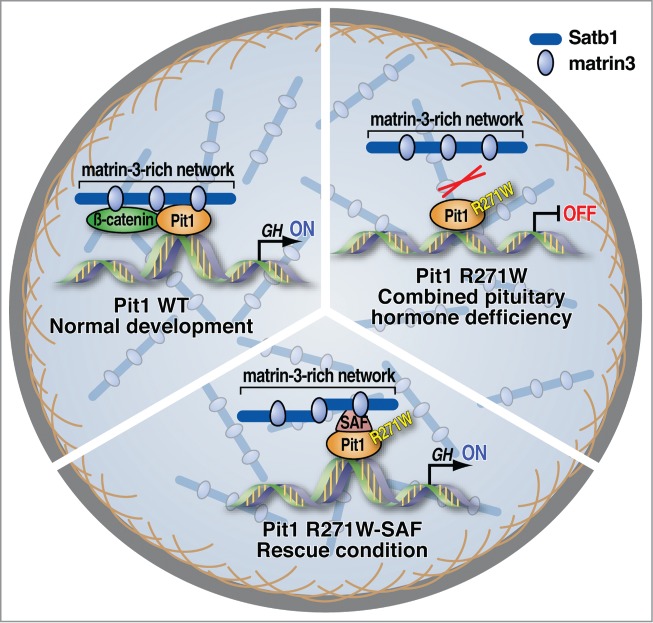
Model. Left: Pit1 association with the matrin-3-rich network is indispensable for its biological activity. Right: A naturally-occurring, dominant negative, point mutation in human Pit1 (R271W), causing combined pituitary hormone deficiency (CPDH), affects Pit1 association with b-catenin and Satb1 and therefore the matrin-3-rich network, blocking Pit1-dependent enhancer/coding target gene activation. Bottom: Artificial tethering mutant Pit1 protein restores its ability to interact with nuclear matrix thus activate target genes.
We have also shown that a naturally-occurring, dominant negative, single point mutation in human Pit1 (R271W) that causes combined pituitary hormone deficiency (CPHD) results in a loss of Pit1 association with β-catenin and Satb1 and, as a consequence, the matrin-3-rich network, resulting in a blockade of Pit1-dependent enhancer/coding target gene activation (Fig. 1). Critically, we demonstrated that this loss of activation can be rescued by artificial tethering of the mutant R271W Pit1 protein to the matrin-3 network, bypassing the otherwise pre-requisite association with β-catenin and Satb1. Indeed, the matrin-3 network-tethered R271W Pit1 mutant, but not the untethered protein, restores Pit1-dependent enhancer activation and recruitment of co-activators such as p300, and induces eRNA transcription and target gene activation. Our studies have thus revealed an unanticipated mechanism whereby the homeodomain factor/β-catenin/Satb1-dependent re-localization of target-gene enhancer regions to the internal nuclear network/nuclear matrix is a key step by which an enhancer-bound homeodomain factor effectively activates developmental gene transcriptional programs. Moreover, our data suggested that the association of subnuclear structures such as the matrin-3-enriched nuclear network with regulatory DNA elements is dynamic and dependent on cell-type specific transcription factors.
What are other components of internal nuclear network? It has been previously noted that nuclear matrix is highly enriched in RNA molecules4 and several proteins identified as a “central proteome” of this substructure are in fact RNA binding proteins, some with suggested role in RNA processing.5 Interestingly, several studies highlight the role of RNA, in particular long non-coding RNAs and repeat-associated RNAs in regulation of chromatin structure and genome organization. For example, spreading of Xist noncoding RNA on X chromosome is imposing repressing chromatin marks. In contrast, C0T-1, repeat rich RNA, is stably associated with euchromatin and its association is suggested to be prerequisite for active chromatin status. Notably, the tight association of C0T-1 RNA with the chromosome is dependent on functionally intact hnRNP-U/SAF-A, another hallmark protein of an internal nuclear network.7
Collectively those studies suggest that interplay between structural proteins in the nucleus and RNA are at the basis of dynamic genome organization alterations that, in turn, have critical impact on gene expression. It will be of great future interest to investigate the precise molecular mechanism of those interactions.
References
- 1. Bickmore WA. Annu Rev Genomics Hum Genet 2013; 14:67-84; PMID:23875797; http://dx.doi.org/ 10.1146/annurev-genom-091212-153515 [DOI] [PubMed] [Google Scholar]
- 2. Cau P, et al. Semin Cell Dev Biol 2014; 29:125-47; PMID:24662892; http://dx.doi.org/ 10.1016/j.semcdb.2014.03.021 [DOI] [PubMed] [Google Scholar]
- 3. Kohwi-Shigematsu T, et al. Semin Canc Biol 2013; 23:72-9; PMID:22771615; http://dx.doi.org/ 10.1016/j.semcancer.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nickerson JJ. Cell Sci 2001; 114:463-74. [DOI] [PubMed] [Google Scholar]
- 5. Engelke R, et al. J Prot Res 2014; 13:3940-56; PMID:25090448; http://dx.doi.org/ 10.1021/pr500218f [DOI] [PubMed] [Google Scholar]
- 6. Skowronska-Krawczyk D, et al. Nature 2014; 514:257-61; PMID:25119036; http://dx.doi.org/ 10.1038/nature13573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hall LL, et al. Cell 2014; 156:907-19; PMID:24581492; http://dx.doi.org/ 10.1016/j.cell.2014.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]


