Abstract
The first step in executing the genetic program of a cell is production of mRNA. In yeast, almost every gene is transcribed as multiple distinct isoforms, differing at their 5′ and/or 3′ termini. However, the implications and functional significance of the transcriptome-wide diversity of mRNA termini remains largely unexplored. In this paper, we show that the GAT1 gene, encoding a transcriptional activator of nitrogen-responsive catabolic genes, produces a variety of mRNAs differing in their 5′ and 3′ termini. Alternative transcription initiation leads to the constitutive, low level production of 2 full length proteins differing in their N-termini, whereas premature transcriptional termination generates a short, highly nitrogen catabolite repression- (NCR-) sensitive transcript that, as far as we can determine, is not translated under the growth conditions we used, but rather likely protects the cell from excess Gat1.
Keywords: yeast, transcription, initiation, termination, GAT1
Introduction
Metabolic adaptation to environmental cues occurs in a variety of ways to enable the most appropriate responses: fast or delayed, short-or long-lasting, moderate or strong. To achieve this wide range of responses, metabolic reprogramming can occur at all steps from the information contained in the genomic DNA to the production of a mature, active, protein: chromatin remodelling, transcription initiation and elongation, mRNA stability and translation, protein cleavage, modification, localization and/or degradation. In many instances, the production of different mRNA isoforms from a single genetic locus has been shown to generate mRNA species characterized by different stabilities, localizations or translation efficiencies1, also possibly resulting in the production of protein isoforms with distinct localizations and/or functions.2,3 More recently, whole-genome mRNA isoform profiling has determined that variations in the 5′ and 3′ termini of mRNAs is the rule rather than the exception.4-6 Widespread phenotypic consequences are to be expected due to changes in the content of RNA-binding protein sites,7,8 uORFs,9,10 as well as N- and C-terminal truncations.2,3,11,12
Saccharomyces cerevisiae cells face extremely changing nutritional environments and have evolved very diverse and efficient mechanisms to cope with them. Fine tuning of nitrogen metabolism, allowing yeast cells to make the most of a plentiful nitrogen supply or cope with a very poor one, is achieved at the transcriptional and posttranslational levels. Posttranslational control targets the activity of amino acid permeases by controlling their modification, internalization and vacuolar degradation (13,14; reviewed in15,16), whereas transcriptional control restrains the production of enzymes and permeases needed to utilize non-preferred, poor nitrogen sources when readily usable, good nitrogen sources are available (for recent reviews, see17-19). Four GATA-family transcription factors are central to this latter control: 2 activators, Gln3 and Gat1/Nil1 and 2 repressors, Dal80/Uga43 and Gzf3/Deh1/Nil2.20-38 When no preferred nitrogen source is available, Gln3 and Gat1 activate the expression of a range of nitrogen catabolite repression (NCR)-sensitive genes, enabling the yeast to use alternative nitrogen sources in its environment.39 Interestingly, recent data strongly suggest that Gat1 and Gln3 are not regulated similarly: the Ure2 negative regulator and TORC1-regulated phosphatases impinge differently on Gln3 and Gat1 activities.40-42 Their respective sensitivities to the TORC1 inhibitor rapamycin, nitrogen catabolite repression, nitrogen starvation and the glutamine synthetase inhibitor methionine sulfoximine also differ markedly.41,42 Although the Gln3 activator was identified first and was long considered as the primary effector of NCR, in part due to its activation of GAT1 expression, additional work in several laboratories has positioned Gat1 as another key factor for the integrated control of NCR-sensitive gene expression in yeast.31-38,43 Indeed, Gat1 appears to be a limiting factor for the expression of some NCR-sensitive genes, with examples of Gat1-dependent Gln3 binding to DNA.40,43 Further, GAT1 expression is regulated by the 4 GATA factors in response to nitrogen availability and, finally, the negative GATA factors hamper Gat1 and Gln3 binding to DNA.31-38,43 Consequently, the levels of Gat1 in yeast cells, when controlled artificially through an inducible promoter, are known to impact on the strength of the nitrogen derepressive response.38,43
In light of this background, our objective in the present work was to investigate 2 paradoxical observations: (i) full length Gat1 protein levels are unaffected by the cell's environmental nitrogen status, i.e., glutamine vs. proline, rather than being produced in the NCR-sensitive manner observed for steady state GAT1 mRNA levels,33 and (ii) NCR-sensitive Gat1 protein production is observed when translation is artificially prematurely terminated, about midway through the protein.44 To this end, we have characterized the GAT1 mRNA levels across the locus and identified an unexpected decrease in those levels using 3′ vs. 5′ GAT1 probes, suggesting the existence of a premature transcription termination that could account for the lack of correlation between GAT1 mRNA and Gat1 protein levels. Remarkably, synthesis of all GAT1 mRNA species, both constitutive and NCR-sensitive, was Gln3-dependent. RACE PCR analyses identified different termini for the GAT1 transcripts: (i) 3 major 5′ GAT1 mRNA termini, correlating with the detection of 2 full-length, constitutively produced protein species beginning at 2 different translation start sites,44 and (ii) 2 major 3′ GAT1 mRNA termini, correlating with one small, NCR-sensitive GAT1 and one full length, constitutive GAT1 mRNA species. The site for premature transcription termination at the GAT1 locus has been defined, and the possible physiological significance investigated. Given the elevated toxicity of a high copy number of GAT145 and the impact of GAT1 over-expression on cell growth, we suggest that premature termination at the GAT1 locus may exist to prevent the over-production of Gat1, from its Gln3-dependent, NCR-sensitive promoter, in conditions of nitrogen limitation.
Material and Methods
Yeast strains and culture conditions
The Saccharomyces cerevisiae strains used are listed in Supplemental Table 1 and the structures of their GAT1 loci are depicted in Supplemental Figure 1. The Saccharomyces paradoxus, bayanus and mikatae strains are Q32.3, CLIB283 and CLIB1352, respectively. Deletion of SKI7 (FV739, Table S1) has been performed according to Wach et al.46 using primers listed in Supplemental Table 2. All allele modifications of GAT1 have been carried out at the chromosomal locus, under the native GAT1 promoter and terminator sequences, unless indicated otherwise. Chromosomal GAT1 was truncated by the addition of 13 copies of the c-myc epitope (Myc13) at positions indicated in Figure 1 (IS1-5, FV743-7; MS, FV655 and VS, FV654) or tagged at its C-terminus by the addition of Myc13 (FV034, FV063 and FV291) or with 3 copies of the HA epitope (HA3; FV446) as described by Longtine et al.,47 using primers listed in Supplemental Tables 1 and 2. The PGAL1-HA-GAT1 and PGAL1-GAT1 alleles in strains FV685 and FV666 were created as described by Longtine et al.47, using primers listed in Supplemental Tables 1 and 2. The PGAT1-HA-GAT1 alleles in strains FV723 and FV726 and the deletion allele in strain FV797 were created by pop in – pop out of the following plasmids (strategy depicted in Fig. S2). For strain FV723 (Fig. S2A), an EcoRI-PstI PCR insert obtained by PCR with OIG0475 and OIG0478 on plasmid pFA6a-3HA-KanMX647 and 2 PCR fragments generated on TB50 genomic DNA (OIG0475 and OIG0476; OIG0477 and OIG0478) were cloned into the corresponding restriction sites of pFL34.48 For strain FV726, the strategy was the same, but using different primers (Tables S1 and S2). To create the GAT1Δ60 deletion allele (FV797; Fig. S1), an EcoRI-BamHI insert obtained by PCR with OIG0374 and OIG0442 on 2 PCR fragments generated on TB50 genomic DNA using primers OIG0374, OIG0442, OIG0487 and OIG0488 (Fig. S2B and Tables S1 and S2) were cloned into the corresponding restriction sites of pFL3448. After sequence confirmation of the absence of mutations in the inserts, the resulting plasmids were linearized within the GAT1 coding sequence (XhoI for FV723 and FV726, BtrI for FV797) and transformed into yeast. Integration events resulting from homologous recombination were determined by PCR of URA3 clones and excision was subsequently carried out by 5-FOAR selection. The excision events that generated the deletion or the HA-insertion in translational fusion to the GAT1 coding sequence were assessed by PCR.
Figure 1.
For figure legend, see page 827. Figure 1 (See previous page). Nucleotide sequence of the GAT1 locus. The GATWAG consensus sequences for GATA factor binding are written in green. The sites for translation initiation are written in orange. Transcription initiation and termination sites detected using RACE-PCR are highlighted in green and red, respectively. The sites where the c-myc tag was inserted for premature transcription termination are indicated by a dot and highlighted in black. Forward qPCR primers are highlighted in gray and reverse primers are in bold and underlined. The nucleotides deleted in strain FV797 are underlined. The sites of insertion of the 117nt encoding the HA tag in fusion with GAT1 ATGM40 and ATGM95 in strains FV723 and FV726 are located by a purple crossed square.
Cultures were grown to mid-log phase (A660nm = 0.5) in YNB (without amino acids or ammonia) minimal medium containing the indicated nitrogen source at a final concentration of 0.1%. As carbon source, glucose 3% or galactose 1% was added. Appropriate supplements (120 µg/ml leucine, 20 µg/ml uracil, 20 µg/ml histidine and 20 µg/ml tryptophan) were added to the medium as necessary to cover auxotrophic requirements. Where indicated, cells were treated with 200 ng/ml rapamycin for 20 mins. or 2 mM Msx for 30 mins. as described earlier.49
5′ RACE PCR
The strategy followed for 5′-RACE PCR is depicted in Supplemental Figure 3. RNA extracts prepared as decribed in43 from Msx-treated ammonia-grown TB50 cells were subjected to reverse transcription with the RevertAid H Minus first-strand cDNA synthesis kit with Fermentas using phosphorylated GAT1O2, GAT1O10 and GAT1O18 primers (Table S2) following the manufacturer's recommended protocol. RNase H and RNA ligase treatments were applied (Fermentas) to the samples and 2 successive PCR amplifications were carried out on the single-stranded cDNA, the first with Gat1O8 and OA429, and the second with OIG0376 and OIG0374. The resulting PCR fragments were cloned into the corresponding restriction sites of pFL3448 and the inserts sequenced.
3′ RACE PCR
The strategy followed for 3′-RACE PCR is depicted in Supplemental Figure 4. RNA extracts prepared as described in43 from Msx-treated ammonia-grown TB50 cells were subjected to reverse transcription with the RevertAid H Minus first-strand cDNA synthesis kit (Fermentas) using primer OIG0372 (Table S2) following the manufacturer's recommended protocol. Subsequent PCR amplification was performed using primer pairs OIG0375 or OIG0377 and OIG0373. The generated fragments were cloned into the EcoRI and BamHI sites of pFL34 and the inserts sequenced.
Quantitative RT-PCR
qRTPCR was performed as described previously43 using primers described in Supplemental Table 2.
Western Blots. Western blotting was carried out as described previously43,44 using anti-myc (Santa Cruz Biotechnology, 9E10), anti-ha (Santa Cruz Biotechnology, F-7) and anti-pgk1 (Invitrogen) antibodies.
Northern Blots
Northern blot experiments were performed as described previously40 using Digoxigenine-labeled probes (Roche) that were synthesized by PCR or in vitro transcription using T7-DNA polymerase and primers described in Supplemental Table 2 following the manufacturer's recommendations.
Results
Two different mRNAs are produced from the GAT1 locus
Our earlier data demonstrated unexpected complexity in the production of the Gat1 protein, with 2 isoforms being produced. Interestingly, neither of these isoforms required the first in-frame methionine for their production.44 Indeed, using amino acid substitutions, we showed that translation of Gat1 is initiated at M40 and M95 relative to the beginning of the open reading frame.44 Phylogenetic analysis was carried out by comparing the amino acid sequences deduced from the open reading frames of Gat1 orthologs from 25 S. cerevisiae strains available on Saccharomyces Genome Database (Fig. 2; http://www.yeastgenome.org/cgi-bin/FUNGI/alignment.pl?locus=YFL021W). ClustalW multiple alignment analysis showed that only 14 of the 25 sequences had the GAT1 ORF starting at ATGM1. ATGM40, ATGM95 and ATGM102 were conserved in all but one sequence, that of strain Vin13, which also differed from the 24 others by an N-terminal 133 amino acid truncation (Fig. S5). Together with the observation that altering ATGM1 did not lead to a detectable phenotype,44 the lack of overall conservation of the full-length open reading frame among S. cerevisiae S288C's closest relatives suggested that the 1st in-phase methionine in the S288C sequence of Gat1 did not function in vivo under the conditions we used in our experiments.
Figure 2.
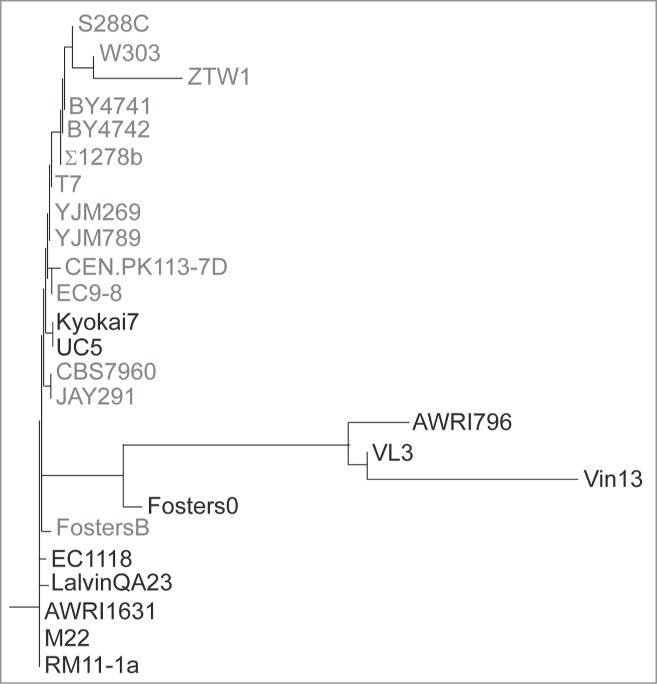
Phylogenetic analysis of Gat1 sequences of 25 S. cerevisiae strains. A multiple alignment was carried out using ClustalW by comparing the amino acid sequences deduced from the open reading frames of the Gat1 orthologs from the 25 sequenced S. cerevisiae strains available on Saccharomyces Genome Database. The sequences highlighted in gray are the ones with an ORF starting at ATGM1.
These observations prompted us to determine the sites for GAT1 transcription initiation using 5′ RACE PCR analysis (Fig. S3). Our results identified GAT1 transcripts with 3 major 5′ termini: +74 to +82, +120 to +126 and +170 to +176 relative to the 1st in phase ATG (highlighted in green in Fig. 1). The observed transcription initiation sites, one downstream of ATGM1 and 2 downstream of ATGM40, correlated with and explained the observation that full length Gat1-Myc13 protein starting at M1 could not be detected. Rather two isoforms, one starting at M40, and the other largely at M95 were produced. The remarkable correlation between mRNA and protein isoforms produced from the GAT1 locus suggested that different mRNA isoforms were responsible for the observed patterns of Gat1 protein production.
Short, inducible and long, constitutive GAT1 transcripts are produced from the GAT1 locus
In addition to 2 full length, constitutively produced Gat1 isoforms being detected in our earlier study of this protein, we also observed production of additional, C-terminal truncated Gat1-Myc13 isoforms that was highly dependent on the growth conditions employed.44 The existence of truncated Gat1-Myc13 isoforms was expected because the Myc13 tag was introduced within the GAT1 coding sequence, creating an artificially shortened open reading frame, thereby leading to the production of an equivalently artificially shortened protein.44 In nitrogen-rich glutamine medium, these truncated isoforms were produced at low levels as was wild type full length Gat1-Myc13. In contrast, in nitrogen-poor proline medium, production of the truncated isoforms was highly derepressed.44 Yet Gat1 production under all circumstances was Gln3-dependent and earlier studies from multiple laboratories had demonstrated GAT1 gene expression to be NCR-sensitive.31-38,44
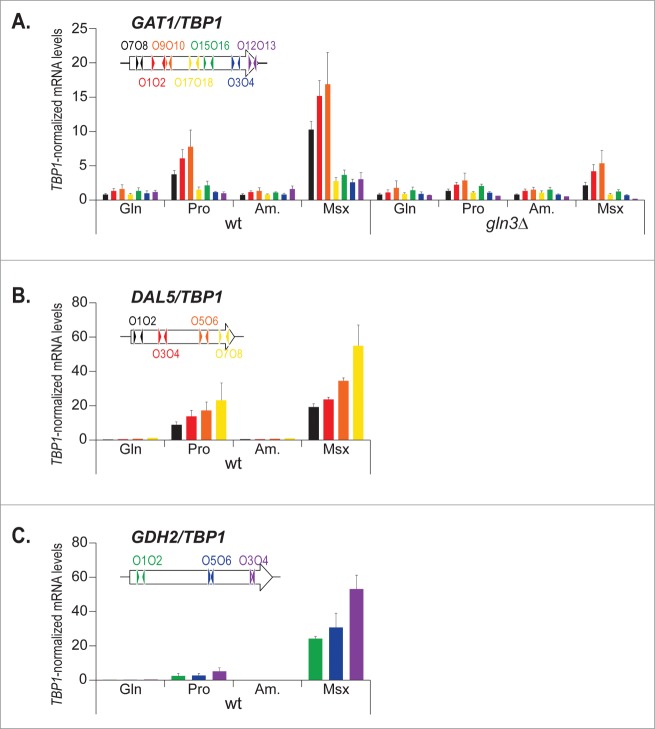
To better understand these observations, and assuming the possibility that the different Gat1 protein levels observed here might be due to RNA polII processivity defects, we investigated GAT1 mRNA levels derived from different sections of the gene, from 5′ to 3′, in wild type and gln3Δ cells grown under repressing (glutamine [Gln], ammonium [Am.]) and derepressing (proline [Pro], conditions as well as in ammonia-grown cells treated with methionine sulfoximine [Msx]) (Fig. 3A). When GAT1 expression was measured using qRTPCR with primers probing the 5′ half of the gene (GAT1O7-O8, GAT1O1-O2, GAT1O9-O10), high NCR sensitivity and Gln3 dependence were observed in keeping with earlier reports. However, when GAT1 levels were assayed with primers located in the 3′ half of GAT1 (GAT1O17-O18, GAT1O15-O16, GAT1O3-O4, GAT1O12-O13), very little derepressed (Pro) and weak Msx-elicited (Msx), Gln3-dependent mRNA production was observed. To ensure that this observation was not caused by experimental bias, we performed similar analyses with 2 well-characterized NCR-sensitive genes, DAL5 and GDH2 (Fig. 3B, C). In the case of DAL5 and GDH2 expression, the profile was clearly different from that of GAT1. The mRNA levels did not decrease from the 5′ to the 3′ regions of these genes. They even tended to progressively rise from the 5′ to the 3′ regions, probably due to decreased efficiency of the reverse transcriptase during the PolyT-driven RT step of qRTPCR.
Figure 3.
The decrease in GAT1 mRNA levels from the 5′ to the 3′ region is gene-specific. Total RNA was isolated from wild type (TB50) and gln3Δ (FV005) mutant cells grown in YNB medium with glutamine (Gln), proline (Pro) or ammonium (Am.) as the nitrogen source and treated with methionine sulfoximine (Am. + Msx). mRNA levels were quantified by quantitative RT-PCR as described in “Materials and Methods.” The values reported represent the averages of at least 2 experiments from independent cultures; error bars indicate standard errors. (A). GAT1 expression was assayed using 7 pairs of primers along the open reading frame as shown on the insert. (B). DAL5 expression was assayed using 4 pairs of primers along the open reading frame as shown on the insert. (C). GDH2 expression was assayed using 3 pairs of primers along the open reading frame as shown on the insert.
Northern Blot analyses reveal the presence of 2 transcripts, the smallest one being NCR-sensitive
The strong decrease in transcription between the 5′ and 3′ regions of GAT1 in derepressive conditions suggested that, in some cases, transcription stopped in the middle of the gene. To evaluate this possibility, we performed Northern blotting experiments. Using a probe covering the 5′ half of the gene (GAT1O7-O10), we detected 2 GAT1 mRNA species (Fig. 4A). The largest one (Full Length, FL GAT1-MYC13) was constitutively produced at low levels in both repressive and derepressive conditions and correlated with the size expected for a full length GAT1-MYC13 transcript (∼2200 nt). In contrast, the smallest species (Short, SH GAT1) was produced in a highly NCR-sensitive manner, occurring in greatest amounts in derepressive conditions, i.e., in Pro-grown cells or when cells were treated with Msx. Its size (∼600 nt) corresponded to what could be expected if transcription prematurely arrested in a region between primers GAT1O10 and GAT1O17 (Positions 587 and 775 relative to ATGM1). Confirming this expectation, when a probe covering the 3′ half of the gene (GAT1O15-O4) was used in a similar Northern blotting experiment, the full length (FL GAT1-MYC13) transcript was detected but the short one (SH GAT1) was not (Fig. 4B).
Figure 4.
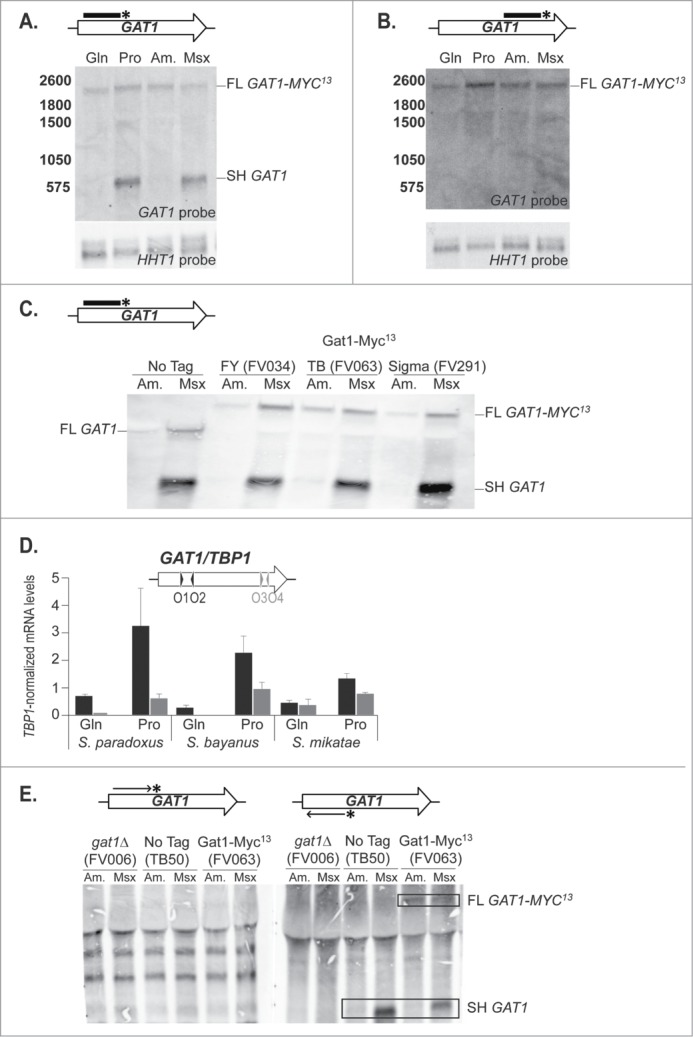
Two mRNAs are produced from the GAT1 locus. Total RNA was isolated from wild type (TB50), gat1Δ (FV006) and GAT1-MYC13 cells of the TB (FV063), FY (FV034) and Sigma (FV291) genetic backgrounds that were grown in YNB medium with glutamine (Gln), proline (Pro) or ammonium (Am.) as the nitrogen source and treated with methionine sulfoximine (Am. + Msx). 30 μg of total RNA from each sample were subjected to Northern blot analysis. HHT1 was used as the loading and transfer efficiency control. (A). GAT1 mRNA analysis in wild type GAT1-MYC13 cells using a double stranded probe covering the 5′ region of the gene (GAT1O7-GAT1O10). (B). GAT1 mRNA analysis in wild type GAT1-MYC13 cells using a double stranded probe covering the 3′ region of the gene (GAT1O15-GAT1O4). (C). GAT1 mRNA analysis in wild type untagged and GAT1-MYC13 cells from 3 different genetic backgrounds using a double stranded probe covering the 5′ region of the gene (GAT1O7-GAT1O10). (D). Wild type cells of Saccharomyces paradoxus, bayanus and mikatae were grown in YNB medium with glutamine (Gln) or proline (Pro) as the nitrogen source. Total RNA was isolated and GAT1 mRNA levels were assessed using qRTPCR as described in Figure 3 using GAT1O1-O2 and GAT1O3-O4 primer pairs specific for each species. (E). GAT1 mRNA analysis in wild type untagged (TB50), gat1Δ (FV006) and GAT1-MYC13 (FV063) cells using strand-specific probes covering the 5′ region of the gene (GAT1O7-GAT1O10).
Northern analysis was also carried out in several different strain backgrounds using a 5′ GAT1 probe (GAT1O7-O10) (Fig. 4C). The short, derepressible, Msx-elicited, mRNA species (SH GAT1) was produced in all strain backgrounds assayed. Moreover, qRTPCR performed on the ortholougous GAT1 loci from 3 Saccharomyces species (Fig. 4D) suggested that production of the the smallest, inducible species was evolutionary conserved. Additionally, using strand-specific probes, we demonstrated the short RNA (SH GAT1) was produced from the same template strand as the full length (FL GAT1-MYC13) transcript, the one encoding the full length Gat1 protein (Fig. 4E), ruling out the possibility of production of a regulatory antisense RNA. The multiple bands appearing in all lanes of Figure 4E are probably due to unspecific hybridization, since they are common to all lanes, including those with cells lacking GAT1.
Altogether, these results strongly suggested that 2 major transcripts were produced from the GAT1 locus: a long, constitutive and weakly-produced one, covering the full length gene and a short one produced in a highly NCR-sensitive manner, which potentially corresponded to a transcript generated by premature transcription termination.
Identification of the sequences responsible for the premature transcription termination
In order to identify the sequences responsible for production of the short GAT1 mRNA, we analyzed the GAT1 sequence (from ATGM1 to 1000 bp after the stop codon) with a 3′-processing site prediction program50 (Fig. S6). In addition to detecting a termination region at the probable 3′ terminus of the full-length transcript (∼1650bp after ATGM1), the software revealed 2 other zones of potential termination, one was in the location expected (∼670bp after ATGM1) to produce the short, derepressible transcript and the other at ∼150 bp after ATGM1. 3′ RACE PCR identified 2 major 3′ termini (+701 to +708 and +1541 after ATGM1; indicated in red in Fig. 1), indicating the locations of transcription termination. The fact that 3′ RACE PCR was performed using polyT primers suggested the short, prematurely terminated transcript was, like the full length transcript, polyadenylated.
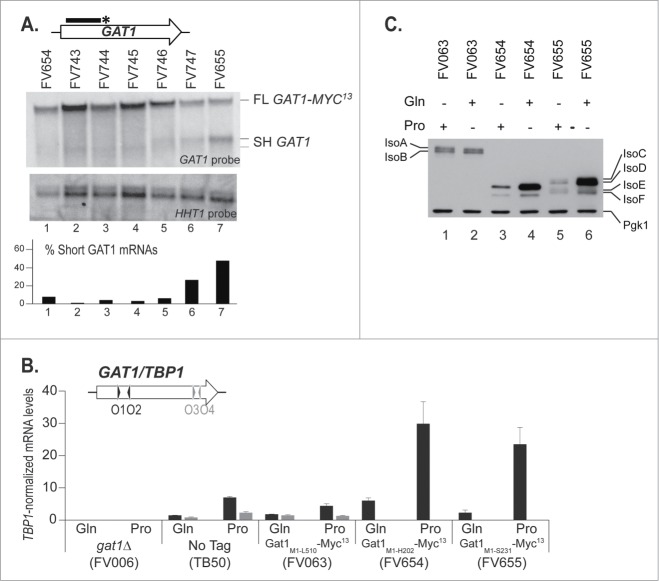
With the objective of locating the elements controlling premature transcription termination at the GAT1 locus, we created 7 strains in which the MYC13 tag was inserted in 7 different positions between nucleotides 606 and 693 in the GAT1 sequence (Fig. 1; VS, IS1–5, MS). The 7 strains were cultured in proline medium and subjected to Northern Blot analysis using the GAT1 5′ probe, GAT1O7-O10 (Fig. 5A). The upper band (Fig. 5A, FL GAT1-MYC13), corresponding to truncated GAT1-MYC13 mRNA, was detected in all strains although in lesser amounts in strains FV747 and FV655. The lower band, indicative of premature termination (SH GAT1), did not disappear sharply from one strain to another, but rather progressively fainted from FV655 to FV746, and was hardly detectable afterwards (Fig. 5A, SH GAT1).
Figure 5.
C-terminal deletions to identify the site for premature transcription termination at the GAT1 locus. gat1Δ (FV023), wild type untagged (TB50), GAT1-MYC13 (FV063), GAT11–606-MYC13 (FV654), GAT11–621-MYC13 (FV743), GAT11–636-MYC13 (FV744), GAT11–651-MYC13 (FV745), GAT11–666-MYC13 (FV746), GAT11–681-MYC13 (FV747) and GAT11–693-MYC13 (FV655) cells were grown in YNB medium with glutamine (Gln) or proline (Pro) as the nitrogen source. (A). Total RNA was isolated and 30 μg from each sample were subjected to Northern blot analysis using a double stranded GAT1-specific probe covering the 5′ region of the gene. HHT1 was used as the loading and transfer efficiency control. Myc13-tagged mRNAs and prematurely terminated transcripts were quantified. Histograms in the lower part of the panel indicate the proportion of prematurely terminated transcript versus total GAT1 mRNAs within each experiment. (B). Total RNA was isolated and GAT1 mRNA levels were assessed using qRTPCR as described in Figure 3 using GAT1O1-O2 and GAT1O3-O4 primer pairs. (C). Gat1 protein species were analyzed with anti-myc western blotting as described in material and methods. Loading uniformity was assessed using anti-pgk1 antibodies.
GAT1 mRNA production from the truncated GAT1-MYC13 loci (FV654 and FV655) was compared to wild type using northern blotting (data not shown) and qRTPCR (Fig. 5B). The expression of GAT11–693-MYC13 and GAT11–606-MYC13 mRNAs was NCR-sensitive and strikingly higher than the levels observed in the wild type strains carrying either the wild type GAT1 gene (TB50), or its MYC13-tagged version (FV063) (Fig. 5B).
Gat1 protein production from the truncated GAT1-MYC13 loci was characterized in strains FV654 and FV655 and compared to wild type. In strain FV654, 2 truncated, NCR-sensitive, Gat1-Myc13 protein isoforms were produced (Fig. 5C, lanes 3–4), whereas in strain FV655, 4 truncated, NCR-sensitive, Gat1-Myc13 isoforms (IsoC-F) were produced from M40 and M95/M102 (Fig. 5C, lanes 5–6 and44). Thus, NCR-sensitive production of the truncated, tagged proteins correlated with the observed NCR sensitivity of the short untagged GAT1 transcript (Fig. 4A) and of the truncated GAT1-MYC13 transcripts (Fig. 5B, FV654 and FV655). Moreover, the absence of 2 minor isoforms in strain FV654 compared to FV655 (compare lanes 3–4 with lanes 5–6 in Fig. 5C) suggests that the region from aa 202 to 231 could be important for posttranslational modification of Gat144 or, alternatively, the deleted 29 amino acids could be required for the proper Gat1 conformation. As expected given the absence of their zinc finger DNA binding domain, the truncated Gat1 proteins produced in strains FV654 and FV655 were unable to contribute to NCR-sensitive gene activation (data not shown).
We further generated a deletion strain lacking 60bp upstream and covering the presumed premature termination sites (the region is underlined in Fig. 1). The functionality of the resulting mutant protein, lacking 20 amino acids (Gat1 aa218 to 237, relative to ATGM1), expressed from GAT1′s native promoter at its native locus, was tested for its capacity to support derepressed DAL5 expression in proline-grown or rapamycin-treated cultures (Fig. 6A). The DAL5 transcription levels supported by wild type (FV063) and deletion mutant proteins (FV797) were similar in glutamine-grown and rapamycin-treated cells; the level of derepressed DAL5 expression in proline medium was modestly lower in mutant cells, although much higher than when GAT1 was fully deleted (FV006; Fig. 6A), showing that the 20 deleted amino acids are not required for the regulated transcriptional activation capability of Gat1.
Figure 6.
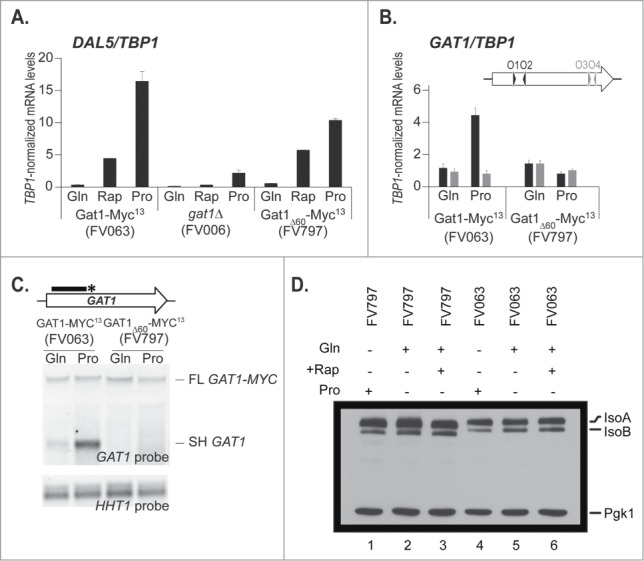
Internal deletion of a 60nt region responsible for premature transcription termination. Panel A. Total RNA was isolated from gat1Δ (FV006), GAT1-MYC13 (FV063) and GAT1Δ60-MYC13 (FV797) cells grown in YNB medium with proline (Pro) or glutamine (Gln) as the nitrogen source, with or without rapamycin treatment. DAL5 mRNA levels were quantified by quantitative RT-PCR with primer pair DAL5O1-DAL5O2. Panel B. Total RNA was isolated from GAT1-MYC13 (FV063) and GAT1Δ60-MYC13 (FV797) mutant cells grown in YNB medium with glutamine (Gln) or proline (Pro). GAT1 mRNA levels were quantified by quantitative RT-PCR with primer pairs GAT1O1-O2 and GAT1O3-O4. Panel C. Total RNA was isolated from GAT1-MYC13 (FV063) and GAT1Δ60-MYC13 (FV797) mutant cells grown in YNB medium with glutamine (Gln) or proline (Pro). 30 μg of total RNA from each sample were subjected to Northern blot analysis using a double stranded GAT1-specific probe covering the 5′ region of the gene. HHT1 was used as the loading and transfer efficiency control. Panel D. Proteins were isolated from GAT1-MYC13 (FV063) and GAT1Δ60-MYC13 (FV797) cells grown in YNB medium with proline (Pro) or glutamine (Gln) as the nitrogen source, with or without rapamycin treatment. Gat1 protein species were analyzed with anti-myc western blotting as described in material and methods. Loading uniformity was assessed using anti-pgk1 antibodies.
We expected that deleting all of the sequences required for premature transcription termination would lead to increased production of the full length GAT1 mRNA in derepressed conditions and hence, increased Gat1 protein. However, when GAT1 expression was analyzed, the GAT1 mRNA levels in the 5′ region of the gene (GAT1O1-O2) reached on proline were near basal levels in the mutants relative to wild type (Fig. 6B). This suggested that instead of increasing the amounts of full length mRNA due to elimination of impaired premature transcription termination, it strikingly reduced the GAT1 levels in derepressive proline medium, as if abolishing premature transcription termination resulted in the simple loss of the short transcript, with no concomitant increase in the production of the full length mRNA. Northern blot analysis confirmed the impairment of premature transcription termination, as demonstrated by the absence of the short, proline-derepressed, untagged fragment (SH GAT1) in FV797 cells compared to wild type (Fig. 6C). Consistently, the protein levels were all at the level observed in the glutamine-grown cells regardless of the conditions assayed, though there appeared to be somewhat greater amounts of the isoforms in FV797 than in FV063 (Fig. 6D).
Functional significance for premature transcription termination at the GAT1 locus
No detectable protein is produced from the short transcript
To investigate the potential functional significance of premature transcription termination at the GAT1 locus, we first addressed the question of whether or not a short protein, equivalent to that observed when the Myc13 tag was fused after Gat1S233 (Fig. 5C, Gat1IsoC-F),44 was produced from the short GAT1 mRNA. To this end, we constructed strains with N-terminal HA tags inserted at ATGM40 of the GAT1 locus: one with its expression driven by the GAL1 promoter (strain FV685), and the other by the native GAT1 promoter, at ATGM40 (FV723; Fig. 7A, Fig. 1, purple crossed squares and Fig. S1). As a control, a third strain was also constructed in which the HA tag was fused to the C-terminus of the GAT1 ORF (strain FV446). Although the expression levels in the HA-tagged strains are much lower than in wild type or GAT1-MYC13 strains (compare Fig. 7B with Fig. 1 of ref. 40), N-terminal HA-tagging did not impact on Gat1′s ability to participate in rapamycin-induced DAL5 expression compared to C-ter HA tagging (Fig. 7B).
Figure 7.
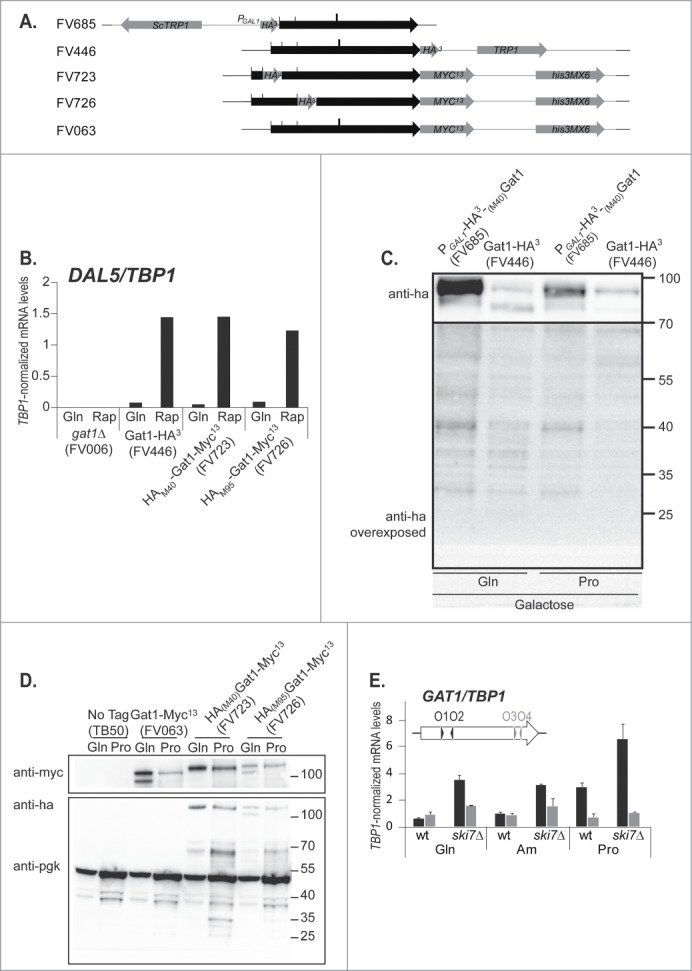
N-terminal tagging of GAT1 does not allow the detection of a truncated protein produced from the prematurely terminated transcript. Panel A. Scale representation of the GAT1 locus in the strains used in this figure. The GAT1 ORF is symbolized by a black arrow. C-terminal tagging cassettes are in orange and N-terminal tagging cassettes are in blue. The sites for transcription termination are indicated by a bold line. The sites for transcription initiation are indicated by a thin line. Panel B. Total RNA was isolated from gat1Δ (FV006), GAT1-HA3 (FV446), HA3-(M40)GAT1-MYC13 (FV723) and HA3-(M95)GAT1-MYC13 (FV726) cells grown in YNB medium with glutamine with (Rap) or without (Gln) rapamycin treatment. DAL5 mRNA levels were quantified by quantitative RT-PCR with primer pair DAL5O1-O2. Panel C. Total protein extracts were prepared from PGAL1-HA3-(M40)GAT1 (FV685) and GAT1-HA3 (FV446) cells grown in YNB galactose medium with glutamine (Gln) or proline (Pro) as the nitrogen source, and subjected to Western blot analysis using anti-ha antibodies. Ponceau staining ensured proper loading and transfer efficiency (not shown). At regular exposure (like the upper panel, from 100kDa to 70kDa), no band was detected in the mass ranges under 70Kda. The lower panel (from 70kDa to 25kDa) is a higher exposure in the lower mass ranges. Panel D. Total protein extracts were prepared from wild type untagged (TB50), GAT1-MYC13 (FV063), GAT1-HA3 (FV446), HA3-(M40)GAT1-MYC13 (FV723) and HA3-(M95)GAT1-MYC13 (FV726) cells grown in YNB medium with glutamine (Gln) or proline (Pro) as the nitrogen source, and subjected to Western blot analysis using anti-ha or anti-myc antibodies. Pgk1 was used as the loading standard. Panel E. Total RNA was isolated from GAT1-MYC13 (FV063) and ski7Δ GAT1-MYC13 (FV739) cells grown in YNB medium with glutamine, ammonium or proline. GAT1 mRNA levels were quantified by quantitative RT-PCR with primer pairs GAT1O1-O2.and GAT1O3-O4.
In these strains expressing N-terminal HA tagged Gat1 (FV685 in Fig. 7C and FV723 in Fig. 7D), a single protein isoform was detected, indicating that HA fusion to M40 prevents isoform production from M95, probably by preventing transcription initiation downstream of ATGM40 (Fig. 1). Another shorter protein band could be detected in FV685 cells (Fig. 7C), but it does not fit the expected size for the (M95)HA3-Gat1 protein isoforms (Fig. 7C, glutamine-grown FV446), suggesting that these probably correspond to degradation products, possibly due to excessive amounts of the proteins in galactose-grown cells. No protein sized around 25kDa was detected in galactose-induced PGAL1-HA3-(M40)GAT1 cells even after overexposure (FV685; Fig. 7C). There was no detectable 25kDa protein in PGAT1-HA3-(M40)GAT1-MYC13 cells either (FV723; Fig. 7D). However, there were several proline-elicited 35kDa HA-reactive protein species (Fig. 7D). These protein species probably correspond to cross reactivity, since they were also detected in the untagged (TB50) and GAT1-MYC13 (FV063) strains. Internal HA tagging at ATGM95 (strain FV726) allowed the detection of the 2 characteristic isoforms of Gat1, expressed from the native GAT1 promoter. Reactive species were also detected between 55 and 70kDa in FV723 and FV726 cells, but they also probably result from aspecific degradation. Altogether, these results suggest that the short native GAT1 transcript does not lead to detectable protein production.
A possible explanation for the absence of protein production from the short transcript could be that the prematurely terminated transcript does not contain a stop codon, and would therefore be targeted by nonstop mRNA decay mechanisms.51 In order to test this possibility, we deleted the SKI7 gene, which codes for an adaptor recruiting the exosome to nonstop mRNAs.51-54 Deleting SKI7 led to increased short GAT1 transcript mRNA levels (Fig. 7E, GAT1O1-O2). This effect was not observed using primers covering the 3′ region of GAT1 (Fig. 7E, GAT1O3-O4), indicating that long GAT1 transcripts were unaffected. Deletion of SKI7 did not affect DAL5 expression either (data not shown), suggesting that the stop codon-containing transcripts are unaffected by the SKI7 deletion.
Excess GAT1 is toxic for the cell
Aware that GAT1 transcription is subjected to tight control by the other GATA factors, we hypothesized that its overexpression might have deleterious effects on the cell. Therefore, the growth rate was analyzed in cells expressing GAT1-MYC13 from the GAL1 promoter (FV666) and compared to cells expressing GAT1-MYC13 from its native promoter (FV063). As expected, growth rates in proline-grown cells were lower than in glutamine-grown cells when glucose was the carbon source, irrespective of the promoter governing GAT1 expression (Fig. 8A, doubling times, g, appear below the abscissa). In these conditions, GAT1 expression coincided with our expectations: it was NCR-sensitive when the promoter was PGAT1 and, repressed by glucose when the promoter was PGAL1(Fig. 8A). When galactose was the carbon source, the growth rates were as expected slightly lower for the wild type FV063 cells, but the growth rate of PGAL1-GAT1-MYC13 cells was much more severely impacted, correlating with the elevated full length GAT1 mRNA levels (Fig. 8A, GAT1O3-O4) and protein levels as well (Fig. 8B). These observations are consistent with the suggestion that high Gat1 levels in the cell are harmful.
Figure 8.
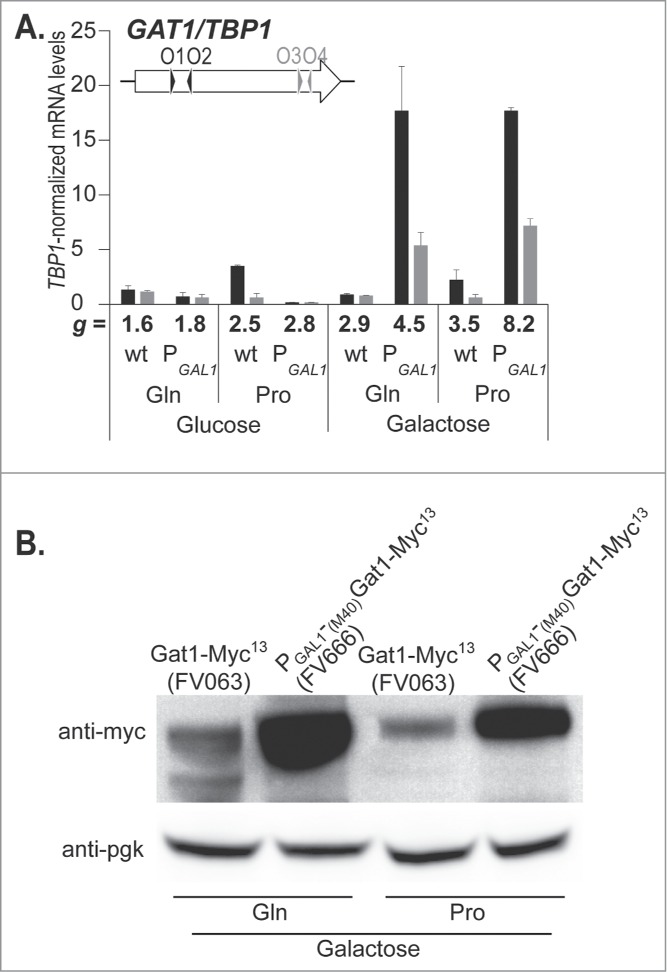
Gat1 overproduction impairs cell growth. GAT1-MYC13 (FV063, wt) and PGAL1-(M40)GAT1-MYC13 (FV666, PGAL1) cells were grown in YNB with glucose (Glu) or galactose (Gal) as the carbon source, and glutamine (Gln) or proline (Pro) as the nitrogen source. The doubling time (g) was measured and is indicated for each strain and culture condition. (A). Total RNA was extracted and GAT1 mRNA levels were quantified by quantitative RT-PCR with primer pairs GAT1O1-O2 and GAT1O3-O4. (B). Total protein extracts were prepared and subjected to Western blot analysis using anti-myc antibodies. Loading uniformity was assessed using anti-pgk1 antibodies.
Discussion
The data presented in this paper demonstrate the complexity in the mRNAs produced from the GAT1 locus. A large variety of transcripts have already been described in previous large scale studies, and the GAT1 3′ ends that we have identified using RACE-PCR are consistent with the high-throughput data provided in reference5. However, due to sensitivity limitations inherent to the techniques used in this study, the high complexity revealed using RNA-seq was not reached here. Nonetheless, due to the conditions used in this study, we could demonstrate that the production of 3′ ends was subjected to nutritional control (NCR-sensitive) whereas 5′ were not, that could be indicative of a possible functional role.
Further, we have explained the striking paradox that overall GAT1 gene expression and Gat1 protein production are not coordinately regulated. SH GAT1 mRNA is NCR-sensitive and, when carrying a stop codon (in strain FV655), is translated into a short Gat1 protein that is NCR-sensitive as well. FL GAT1 mRNA is weakly (if at all) NCR-sensitive and full sized Gat1 protein production is regulated similarly.
Variations in transcription termination at the GAT1 locus
The mRNA expression analyses described in this paper demonstrate the presence of 2 GAT1-specific sense transcripts, one large and constitutive, carrying the complete GAT1 ORF and the other, small and NCR-sensitive, ending within the GAT1 coding sequence. Several observations suggest that the short transcript results from premature termination and not degradation or cleavage: 1) in the 3′ region, GAT1 RNA levels were low Fig. 3A; 2) no other band was detected in the Gat1 3′ region Fig. 4B; 3) the short transcript was polyadenylated; 4) a consensus sequence for transcription termination was detected within the corresponding region where the short GAT1 transcript was terminated.
In order to understand the possible functions for the premature termination of GAT1 transcription, we tried to detect a protein product for the short GAT1 transcript. All of our attempts failed (Fig. 7C, D). A possible explanation for that observation is that the prematurely terminated transcript does not contain a stop codon, leading to stalling of ribosomes when they translate into the 3′ poly-A tail region. Hence, short GAT1 transcripts are therefore likely targeted by nonstop mRNA decay mechanisms, that free the stalled ribosomes and mark the nonstop mRNA for nuclease degradation51. Interestingly, deleting SKI7, coding for an adaptor recruiting the exosome to nonstop mRNAs, led to increased short GAT1 transcript mRNA levels (Fig. 7E). This effect was not observed using primers covering the 3′ region of GAT1 (long GAT1 transcripts, containing a stop codon, were unaffected). An alternative possibility, which was not investigated in this study, is a hypothetical function that would be played by the short mRNA itself. We discarded this possibility because the short RNA detected is in the sense orientation relative to GAT1, and no antisense was detected in our experiments (Fig. 4D). However, the role played by a sense ncRNA cannot be formally ruled out.
Another tenable hypothesis that we did test was the possibility that premature termination of GAT1 transcription during derepressing conditions would protect the cells against harmful amounts of the Gat1 transcriptional activator. Indeed, a certain level of expression increase of the Gat1 protein is sometimes detected in proline- vs. glutamine-grown cells, and Gat1 production requires both the UASGATA sites in PGAT1 and the Gln3 transcriptional activator44. A very small, potentially undetectable, increase in Gat1 amounts could be sufficient for NCR-sensitive induction, the latter being considered a potential bistable (ON/OFF) switch, characteristic of many regulatory cascades. The presence of GATA sites in PGAT1 could enable this NCR-sensitive induction, and premature transcription termination would help reduce the strength of induction, limiting the amounts of GAT1 within the cell. In line with this speculation, Gat1 function in the cell is tightly controlled, by multiple mechanisms: cytoplasmic retention by Ure240, nuclear sequestration by Gzf3 and competition with Dal80 for its binding to target promoters43. Moreover, Gat1 and Gln3 cannot be over-expressed without adverse consequences45. This is the case for a vast majority of transcriptional activators45, 55. In line with these observations, we could also show that GAT1 overproduction from the GAL1 promoter severely impaired the growth rate (Fig. 8A, B).
Aiming at unravelling the functional consequences of impaired premature transcription termination, we have identified the zone where premature termination occurs, using polyadenylation prediction tools as well as progressive deletions of the 3′ side of GAT1. A 60nt sequence was removed, eliminating premature termination: no more short transcript was detected in the GAT1Δ60 cells (Fig. 6C). Very surprisingly, however, eradicating premature termination did not lead to increased production of full length GAT1 transcripts under derepressed conditions. Rather, the GAT1 expression levels in the 5′ half of the gene did not exceed those in the 3′ region in the GAT1Δ60 mutant; no more NCR-sensitive derepression of GAT1 was observed in this mutant (Fig. 6B, C). This striking observation indicates that altering the nucleotide sequence of the GAT1 ORF, located in a region involved in premature transcription termination, affected NCR-sensitive transcription from the GAT1 promoter, without altering FL Gat1 production (Fig. 6D) or impairing Gat1 function (Fig. 6A). Conversely, strengthening of premature termination, by insertion of a bona fide termination site (TADH1 downstream of the MYC13 tag in strains FV654 and FV655), markedly increased the production of the truncated GAT1-MYC13 transcript (Fig. 5B) and the corresponding proteins (Fig. 5B and ref. 44), compared to the wild type. An attractive explanation for this phenomenon derives from the “gene looping” model56, with recent data showing that a point mutation in a polyadenylation site decreased transcription initiation, suggesting that 3′ end processing likely facilitates the recycling of transcription factors from 3′ ends to promoter regions57. A tentative model depicts this hypothesis in Figure 9.
Figure 9.
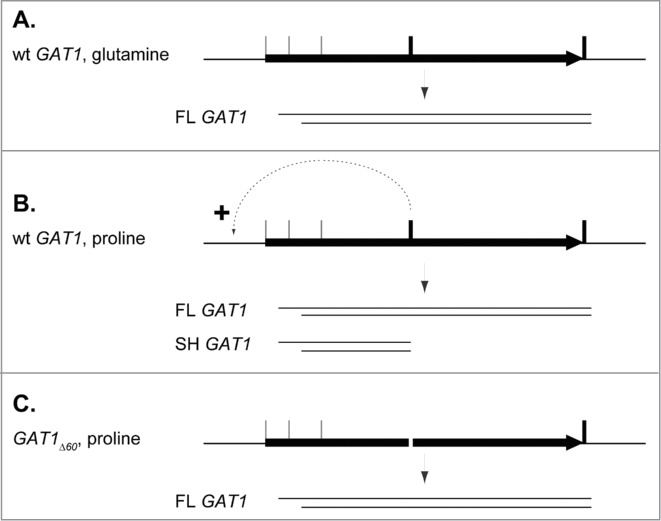
Tentative model depicting the possible mRNA production regulation by gene looping at the GAT1 locus. (A). In rich nitrogen conditions, wild type cells produce a wide variety of mRNAs differing in their 5′ and 3′ ends, but the major species is a full length RNA (FL GAT1) spanning from the 2nd in phase ATGMet codon (ATGM40) to the 1st Stop codon. (B). In nitrogen derepressing conditions, the production of the full length GAT1 mRNA is unchanged, although DNA-bound GATA factors activate the expression from the GAT1 promoter. Premature transcription termination occurs to generate the most abundant mRNA species, SH GAT1, and the transcription machinery is recycled at the GAT1 promoter, as indicated by the dotted arrow. (C). In proline-grown GAT1Δ60 cells, no termination and no recycling occurs, resulting in the absence of SH GAT1. FL GAT1 production remains unchanged.
Variations in transcription initiation at the GAT1 locus
GAT1 transcripts have been identified with a large number of different 5′ termini5 and only some of which were detected in our 5′-RACE PCR experiment. This experiment was not carried out at saturation, and hence we speculate that the 5′ ends that were identified in our work are likely the most abundant ones. These 5′ termini can be grouped into 2 zones, one located between the 1st and 2nd in phase ATGMet (ATGM1 and ATGM40), and the other between the 2nd and the 3rd ones (ATGM40 and ATGM95). This observation nicely correlates with the detection of the 2 Gat1-Myc13 protein species that result from distinct translation initiation sites44. Although we cannot formally rule out leaky ribosome scanning58,59 or shunting60,61 as a possible cause for the production of the 2 Gat1 protein isoforms, alternative transcription initiation combined with translation from the 1st ATGMet encountered on both GAT1 mRNA populations appears to be the simplest explanation. Our failure to detect GAT1 mRNAs starting upstream of ATGM1 suggests that our inability to demonstrate its function (ATGM1 mutation had no consequences on Gat1 protein production;44) is likely due to insufficient production of an mRNA that would encode it. Consistently, large scale analyses could demonstrate the production of such a transcript, but it was largely underrepresented5. Phylogenetic analyses indicate that the 1st ATGMet is being progressively lost through evolution, suggesting that it may have had a functional role early on, but that this is no longer the case.
The functional significance of producing 2 different transcripts, leading to 2 different protein isoforms has been raised. The detection of the Gat1-Myc13 species starting at Met95 was reproducibly altered in proline-grown cells (Figs. 7A, B and 8B). This behavior was not, however, observed in earlier western blots44 or in Figure 5C and 6D. This observation may result from selective derepression of the promoter governing the production of the Gat1-Myc13 species starting at Met40. However, the resolution of our Northern experiments was not sufficient to distinguish between the 2 mRNA species. Alternatively, differential stability of the mRNAs, their ability to be translated, or the extraction properties for the protein isoforms are other possible explanations that we cannot rule out. Localization of the 2 Gat1 isoforms, in the different methionine mutants, showed an altered cytoplasmic retention44, probably due to alteration of a predicted nuclear export signal, N-terminal of Gat1 (NetNES 1.1). However, all isoforms supported normally regulated DAL5 transcription44. In sum, the different isoforms could well carry out distinct functions in the yeast cells, but we were unable to pinpoint these in our working conditions.
In sum, alternative transcription initiation, premature transcription termination and potentially gene looping are responsible of the unanticipated complexity in mRNA production across the GAT1 gene locus. As previously shown, diversity of mRNAs produced from a single locus is the rule for most genes, leading to various protein isoforms and unexpected as well as unrecognized regulatory properties. However, the functional consequences of such a complexity are far from being fully understood and still require further investigation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Lars Steinmetz for sharing data prior to publication. We thank Claudine Bleystaken and Jean-Luc Souciet for providing the S. paradoxus strains.
Funding
This work was supported by NIH grant GM-35642 (to J.J.T. and T.G.C.), by the Commission Communautaire Française (COCOF, to I.G. and E.D.) and the Fonds de la Recherche Fondamentale Collective (FRFC 2.4547.11) (to I.G. and E.D.).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Molecular Cell 2011; 43:853-66; PMID:21925375; http://dx.doi.org/ 10.1016/j.molcel.2011.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 1982; 28:145-54; PMID:7039847; http://dx.doi.org/ 10.1016/0092-8674(82)90384-1 [DOI] [PubMed] [Google Scholar]
- 3.Chatton B, Walter P, Ebel JP, Lacroute F, Fasiolo F. The yeast VAS1 gene encodes both mitochondrial and cytoplasmic valyl-tRNA synthetases. J Biol Chem 1988; 263:52-7; PMID:3275649 [PubMed] [Google Scholar]
- 4.Moqtaderi Z, Geisberg JV, Jin Y, Fan X, Struhl K. Species-specific factors mediate extensive heterogeneity of mRNA 3′ ends in yeasts. Proc Natl Acad Sci U S A 2013; 110:11073-8; PMID:23776204; http://dx.doi.org/ 10.1073/pnas.1309384110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 2013; 497:127-31; PMID:23615609; http://dx.doi.org/ 10.1038/nature12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 2013; 14:496-506; PMID:23774734; http://dx.doi.org/ 10.1038/nrg3482 [DOI] [PubMed] [Google Scholar]
- 7.Riordan DP, Herschlag D, Brown PO. Identification of RNA recognition elements in the Saccharomyces cerevisiae transcriptome Nucleic Acids Res 2011; 39:1501-9; PMID:20959291; http://dx.doi.org/ 10.1093/nar/gkq920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freeberg MA, Han T, Moresco JJ, Kong A, Yang YC, Lu ZJ, Yates JR, Kim JK. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae Genome Biol 2013; 14:R13; PMID:23409723; http://dx.doi.org/ 10.1186/gb-2013-14-2-r13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hood HM, Neafsey DE, Galagan J, Sachs MS. Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Microbiol 2009; 63:385-409; PMID:19514854; http://dx.doi.org/ 10.1146/annurev.micro.62.081307.162835 [DOI] [PubMed] [Google Scholar]
- 10.Waern K, Snyder M. Extensive transcript diversity and novel upstream open reading frame regulation in yeast. G3 2013; 3:343-52; PMID:23390610; http://dx.doi.org/full_text [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu Y, Yan J, Saidel GM, Fox PL. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell 2012; 149: 88-100; PMID:22386318; http://dx.doi.org/ 10.1016/j.cell.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majumdar S, Ghatak J, Mukherji S, Bhattacharjee H, Bhaduri A. UDPgalactose 4-epimerase from Saccharomyces cerevisiae. A bifunctional enzyme with aldose 1-epimerase activity. Eur J Biochem/FEBS 2004; 271:753-9; PMID:14764091; http://dx.doi.org/ 10.1111/j.1432-1033.2003.03974.x [DOI] [PubMed] [Google Scholar]
- 13.Helliwell SB, Losko S, Kaiser CA. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J Cell Biol 2001; 153:649-62; PMID:11352928; http://dx.doi.org/ 10.1083/jcb.153.4.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Springael JY, Andre B. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol Biol Cell 1998; 9:1253-63; PMID:9614172; http://dx.doi.org/ 10.1091/mbc.9.6.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haguenauer-Tsapis R, Andre B. Membrane trafficking of yeast transporters: mechanisms and physiological control of downregulation. Topics Curr Genet 2004; 9:273-323. [Google Scholar]
- 16.Schothorst J, Kankipati HN, Conrad M, Samyn DR, Van Zeebroeck G, Popova Y, Rubio-Texeira M, Persson BL, Thevelein JM. Yeast nutrient transceptors provide novel insight in the functionality of membrane transporters. Curr Genet 2013; 59:197-206; PMID:24114446; http://dx.doi.org/ 10.1007/s00294-013-0413-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broach JR. Nutritional control of growth and development in yeast. Genetics 2012; 192:73-105; PMID:22964838; http://dx.doi.org/ 10.1534/genetics.111.135731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 2014; 38:254-299; PMID:24483210; http://dx.doi.org/ 10.1111/1574-6976.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ljungdahl PO, Daignan-Fornier B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012; 190:885-929; PMID:22419079; http://dx.doi.org/ 10.1534/genetics.111.133306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chisholm G, Cooper TG, Isolation and characterization of mutants that produce the allantoin-degrading enzymes constitutively in Saccharomyces cerevisiae. Mol Cell Biol 1982; 2:1088-95; PMID:6757722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell AP, Magasanik B. Regulation of glutamine-repressible gene products by the GLN3 function in Saccharomyces cerevisiae. Mol Cell Biol 1984; 4:2758-66; PMID:6152012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell AP, Magasanik B. Three regulatory systems control production of glutamine synthetase in Saccharomyces cerevisiae. Mol Cell Biol 1984; 4:2767-73; PMID:6152013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Courchesne WE, Magasanik B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J Bacteriol 1988, 170:708-13; PMID:2892826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper TG, Ferguson D, Rai R, Bysani N. The GLN3 gene product is required for transcriptional activation of allantoin system gene expression in Saccharomyces cerevisiae. J Bacteriol 1990; 172:1014-8; PMID:2153652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cunningham TS, Cooper TG. Expression of the DAL80 gene, whose product is homologous to the GATA factors and is a negative regulator of multiple nitrogen catabolic genes in Saccharomyces cerevisiae, is sensitive to nitrogen catabolite repression. Mol Cellular Biol 1991; 11:6205-15; PMID:1944286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minehart PL, Magasanik B. Sequence and expression of GLN3, a positive nitrogen regulatory gene of Saccharomyces cerevisiae encoding a protein with a putative zinc finger DNA-binding domain. Mol Cell Biol 1991; 11, 6216-28; PMID:1682800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coornaert D, Vissers S, Andre B, Grenson M. The UGA43 negative regulatory gene of Saccharomyces cerevisiae contains both a GATA-1 type zinc finger and a putative leucine zipper. Curr Genet 1992; 21, 301-7; PMID:1525858; http://dx.doi.org/ 10.1007/BF00351687 [DOI] [PubMed] [Google Scholar]
- 28.Cunningham TS, Cooper TG. The Saccharomyces cerevisiae DAL80 repressor protein binds to multiple copies of GATAA-containing sequences (URSGATA). J Bacteriol 1993; 175:5851-5861; PMID:8376332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daugherty JR, Rai R, el Berry HM, Cooper TG. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J Bacteriol 1993; 175:64-73; PMID:8416910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andre B, Talibi D, Soussi Boudekou S, Hein C, Vissers S, Coornaert D. Two mutually exclusive regulatory systems inhibit UASGATA, a cluster of 5′-GAT(A/T)A-3′ upstream from the UGA4 gene of Saccharomyces cerevisiae. Nucl Acids Res 1995; 23:558-64; PMID:7899075; http://dx.doi.org/ 10.1093/nar/23.4.558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coffman JA, Rai R, Cooper TG. Genetic evidence for Gln3p-independent, nitrogen catabolite repression-sensitive gene expression in Saccharomyces cerevisiae. J Bacteriol 1995; 177:6910-8; PMID:7592485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanbrough M, Rowen DW, Magasanik B. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc Natl Acad Sci U S A 1995; 92:9450-4; PMID:7568152; http://dx.doi.org/ 10.1073/pnas.92.21.9450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coffman JA, Rai R, Cunningham T, Svetlov V, Cooper TG. Gat1p, a GATA family protein whose production is sensitive to nitrogen catabolite repression, participates in transcriptional activation of nitrogen-catabolic genes in Saccharomyces cerevisiae. Mol Cell Biol 1996; 16, 847-858; PMID:8622686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coffman JA, Rai R, Loprete DM, Cunningham T, Svetlov V, Cooper TG. Cross regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J Bacteriol 1997; 179:3416-29; PMID:9171383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowen DW, Esiobu N, Magasanik B. Role of GATA factor Nil2p in nitrogen regulation of gene expression in Saccharomyces cerevisiae. J Bacteriol 1997, 179:3761-6; PMID:9171427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soussi-Boudekou S, Vissers S, Urrestarazu A, Jauniaux JC, Andre B. Gzf3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol Microbiol 1997, 23:1157-68; PMID:9106207; http://dx.doi.org/ 10.1046/j.1365-2958.1997.3021665.x [DOI] [PubMed] [Google Scholar]
- 37.Cunningham TS, Rai R, Cooper TG. The level of DAL80 expression down-regulates GATA factor-mediated transcription in Saccharomyces cerevisiae. J Bacteriol 2000; 182, 6584-91; PMID:11073899; http://dx.doi.org/ 10.1128/JB.182.23.6584-6591.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cunningham TS, Andhare R, Cooper TG. Nitrogen catabolite repression of DAL80 expression depends on the relative levels of Gat1p and Ure2p production in Saccharomyces cerevisiae. J Biol Chem 2000; 275:14408-14; PMID:10799523; http://dx.doi.org/ 10.1074/jbc.275.19.14408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooper TG. (1982) In Strathern JN, JEW, Broach JR (ed.), Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, pp. 39-99. [Google Scholar]
- 40.Georis I, Tate JJ, Cooper TG, Dubois E. Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae. J Biol Chem 2008; 283, 8919-29; PMID:18245087; http://dx.doi.org/ 10.1074/jbc.M708811200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tate JJ, Georis I, Dubois E, Cooper TG. Distinct phosphatase requirements and GATA Factor responses to nitrogen catabolite repression and rapamycin treatment in saccharomyces cerevisiae. J Biol Chem 2010; 285:17880-95; PMID:20378536; http://dx.doi.org/ 10.1074/jbc.M109.085712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Georis I, Tate JJ, Cooper TG, Dubois E. Nitrogen-responsive regulation of GATA protein family activators Gln3 and Gat1 occurs by two distinct pathways, one inhibited by rapamycin and the other by methionine sulfoximine. J Biol Chem, 286, 44897-44912; PMID:22039046; http://dx.doi.org/ 10.1074/jbc.M111.290577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Georis I, Feller A, Vierendeels F, Dubois E. The yeast GATA factor Gat1 occupies a central position in nitrogen catabolite repression-sensitive gene activation. Mol Cell Biol 2009; 29, 3803-15; PMID:19380492; http://dx.doi.org/ 10.1128/MCB.00399-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rai R, Tate JJ, Georis I, Dubois E, Cooper TG. Constitutive and nitrogen catabolite repression-sensitive production of Gat1 isoforms. The J Biol Chem 2014; 289:2918-33; PMID:24324255; http://dx.doi.org/ 10.1074/jbc.M113.516740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Makanae K, Kintaka R, Makino T, Kitano H, Moriya H. Identification of dosage-sensitive genes in Saccharomyces cerevisiae using the genetic tug-of-war method. Gen Res 2013; 23:300-11; PMID:23275495; http://dx.doi.org/ 10.1101/gr.146662.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 1996; 12:259-65; PMID:8904338; http://dx.doi.org/ 10.1002/(SICI)1097-0061(19960315)12:3%3c259::AID-YEA901%3e3.0.CO;2-C [DOI] [PubMed] [Google Scholar]
- 47.Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14:953-61; PMID:9717241; http://dx.doi.org/ 10.1002/(SICI)1097-0061(199807)14:10%3c953::AID-YEA293%3e3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- 48.Bonneaud N, Ozier-Kalogeropoulos O, Li GY, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast 1991, 7:609-15; PMID:1767589; http://dx.doi.org/ 10.1002/yea.320070609 [DOI] [PubMed] [Google Scholar]
- 49.Tate JJ, Georis I, Feller A, Dubois E, Cooper TG. Rapamycin-induced Gln3 dephosphorylation is insufficient for nuclear localization: Sit4 and PP2A phosphatases are regulated and function differently. J Biol Chem 2009; 284, 2522-34; PMID:19015262; http://dx.doi.org/ 10.1074/jbc.M806162200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graber JH, McAllister GD, Smith TF. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites Nucl Acids Res 2002; 30:1851-8; PMID:11937640; http://dx.doi.org/ 10.1093/nar/30.8.1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 2002; 295:2262-4; PMID:11910110; http://dx.doi.org/ 10.1126/science.1067272 [DOI] [PubMed] [Google Scholar]
- 52.Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J 2001; 20:4684-93; PMID:11532933; http://dx.doi.org/ 10.1093/emboj/20.17.4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi S, Araki Y, Sakuno T, Katada T. Interaction between Ski7p and Upf1p is required for nonsense-mediated 3′-to-5′ mRNA decay in yeast. EMBO J 2003; 22, 3951-9; PMID:12881429; http://dx.doi.org/ 10.1093/emboj/cdg374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Hoof A, Staples RR, Baker RE, Parker R. Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol Cell Biol 2000; 20:8230-43; PMID:11027292; http://dx.doi.org/ 10.1128/MCB.20.21.8230-8243.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR et al.. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 2006; 21:319-30; PMID:16455487; http://dx.doi.org/ 10.1016/j.molcel.2005.12.011 [DOI] [PubMed] [Google Scholar]
- 56.O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 2004; 36, 1014-8; PMID:15314641; http://dx.doi.org/ 10.1038/ng1411 [DOI] [PubMed] [Google Scholar]
- 57.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′ end processing and transcription initiation. Mol Cell 2010; 40:410-422; PMID:21070967; http://dx.doi.org/ 10.1016/j.molcel.2010.10.012 [DOI] [PubMed] [Google Scholar]
- 58.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA 2010; 12:1755-85; PMID:16957278; http://dx.doi.org/ 10.1261/rna.157806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene 2010. 299:1-34; PMID:12459250; http://dx.doi.org/ 10.1016/S0378-1119(02)01056-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mauro VP, Chappell SA, Dresios J. Analysis of ribosomal shunting during translation initiation in eukaryotic mRNAs. Methods Enzymol 2010; 429:323-54; PMID:17913630 [DOI] [PubMed] [Google Scholar]
- 61.Fournier CT, Cherny JJ, Truncali K, Robbins-Pianka A, Lin MS, Krizanc D, Weir MP. Amino termini of many yeast proteins map to downstream start codons. J Proteome Res 2012; 11:5712-9; PMID:23140384 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.