Abstract
MicroRNAs (miRNAs) are evolutionarily conserved small noncoding RNAs found in most plants and animals. The miRNA pathway regulates posttranscriptional gene expression through the deadenylation and translation repression of target mRNAs. Recent studies revealed that the early step of translation initiation is the target of “pure” translation repression by the miRNA pathway. Moreover, particularly in animals, the miRNA pathway is required for neuronal development, differentiation, and plasticity. In addition, some functions of miRNAs are regulated by RNA-binding proteins (RBPs) in neuronal cells. This review summarizes new insights about the molecular mechanisms of pure translation repression by miRNA pathway and the communication between the miRNA pathway and RBPs in neuronal local translation.
Keywords: deadenylation, local translation, microRNA, pure translation, RNA-binding protein
Introduction
In mammalian cells, translation of most cellular mRNAs occurs in a 5′ cap-dependent manner. The initiation step of mRNA translation is orchestrated by a set of eukaryotic translation initiation factors (eIFs): (1) assembly of eIF4F complex (consists of the cap-binding protein eIF4E, the RNA helicase eIF4A, the large scaffolding protein eIF4G) on the mRNA, (2) recruitment of the 43S preinitiation complex (PIC) to the mRNA, (3) scanning along 5′ untranslated region (UTR) to the start codon, (4) joining of the 60S ribosomal subunit, and forming 80S ribosome.1 In general, cellular mRNA translation is facilitated to form a closed-loop structure through the interaction between eIF4G at the 5′ end and poly(A)-binding protein (PABP) at the 3′ poly(A) tail.2 Translation initiation is the rate-limiting step of mRNA translation and is the functional target of translational control.3,4
MicroRNAs (miRNAs) have emerged as a key mediator for the regulation of posttranscriptional gene expression in mammals. Most studies of miRNAs indicate that they repress translation and cause deadenylation and destabilization of target mRNAs.5,6 miRNAs form miRNA-induced silencing complex (miRISC) with Argonaute (Ago) proteins for the regulation of target genes.7 In animals, Ago proteins bind to TNRC6/GW182, which is a scaffolding protein for recruiting the CCR4/NOT deadenylation complex to the target mRNA.5,6 In addition, TNRC6/GW182 can bind to PABP directly, and this binding facilitates miRISC loading to the target mRNA.8-11 In addition, the CCR4/NOT complex recruits DDX6, an RNA helicase that represses translation and enhances decapping, to the target mRNA.12,13 Although the destabilization of target mRNAs through deadenylation and decapping causes translation repression, recent biochemical studies indicate that “pure” translation repression by miRISC occurs even in the absence of deadenylation. These results have been reported in flies,14 zebrafish,15 and humans.16 Fukaya and Tomari demonstrated that miRISC hampers the recruitment of ribosomal 43S PIC to the target mRNA.17 Their work suggests that the main target of pure translation repression by miRISC is the early step of translation initiation.
Recently, Meijer and colleagues have reported that eIF4AII is a key factor for miRNA-mediated translation repression in mammal. They also showed that CAF1/CNOT7, a component of CCR4/NOT deadenylation complex, can interact with eIF4AII but not eIF4AI, and suggested a model of translation repression by miRISCs in which eIF4AII is recruited by miRISC to the target mRNA through the interaction with CCR4/NOT complex.18 This model implied that eIF4AII is the negative regulator in translation, unlike the well-characterized function of eIF4AI in stimulating translation. However, the difference of functions between eIF4AI and eIF4AII is still uncharacterized. In addition, it is difficult to adapt this model of miRNA-mediated translation repression for flies because they have only one eIF4A.19 Therefore, the detailed molecular mechanism of translation repression by miRISC via eIF4A should be elucidated.
Here we focus on the novel mechanism of “pure” translation repression via miRISC. In addition, we will discuss new insights into posttranscriptional gene regulation through the communication between the miRNA pathway and translational control via RNA-binding proteins (RBPs).
The Molecular Mechanism of Translation Repression by miRISC
RNA-binding proteins (RBPs) are essential for accurate spatial and temporal gene expression during cell proliferation, development, and differentiation.20,21 Several RBPs, including neuronal Hu proteins, are specifically expressed in neurons and essential for neuron development and plasticity.22 HuD is a member of neuronal Hu proteins and functions via binding to adenine/uridine-rich elements (AREs) on target mRNAs.23 Our previous work revealed that the function of HuD is not only target mRNA stabilization but also the translation stimulation of cap-poly(A) mRNA.24 We also showed that HuD can associate with cap-binding eIF4F complex through the direct interaction with eIF4A and the stimulatory effect of HuD on translation depends on the binding of HuD to eIF4A and poly(A) tail.24 Thus, the target of translational control by HuD is the early step of translation initiation, similarly to miRISC. These facts prompted us to hypothesize that HuD can potentially counteract translation repression by miRISC. Therefore, to dissect the molecular mechanism of translation repression by miRISC, we utilized HuD as the best “tool” of explorer.
Our latest study revealed that the pure translation repression by miRISC occurs in a deadenylation-independent manner in humans.16 Moreover, we proved that miRISC repressed the target mRNA translation by the dissociation of eIF4AI and eIF4AII from translation initiation complex, using the method of mRNP including miRISC purification from in vitro translation system arranged with GRNA affinity chromatography.16,25 We also showed that miRISC releases eIF4As from translation initiation complex on the target mRNA at the early time point of translation reaction, before the displacement of other eIF4F components and PABP (Fig. 1A). In addition, at the middle time point, the displacement of PABP occurs in deadenylation-independent manner, consistent with 2 recent studies.10,11 At the late time point, PABP and also other eIF4F components are released from the target mRNA, following mRNA destabilization by miRISC. To confirm that the release of eIF4As from the target mRNA is critical for pure translation repression by miRISC, we utilized the neuronal RNA binding protein HuD which stimulates cap-dependent translation in a eIF4A- and poly(A)-dependent manner. HuD expectedly inhibited translation repression by miRISC via protecting the interaction of eIF4AI and eIF4AII with translation initiation complex on mRNAs (Fig. 1B). Moreover, we also confirmed that molecular mechanism of miRISC utilizing pharmacological inhibitor of eIF4A, “silvestrol,” which locks eIF4A on the mRNA.26-28 Silvestrol inhibited translation repression and the release of eIF4As from the target mRNA by miRISC same as HuD. Therefore, our study revealed that the dissociation of eIF4As from translation initiation complex is a key for pure translation repression by miRISC. Supporting this idea, Fukaya et al. also demonstrated that fly Ago1-RISC induces the dissociation of eIF4A from translation initiation complex in Drosophila, using site-specific UV crosslinking analysis, which is completely different from our approach.29 These reports suggest that eIF4A is a target of miRNA-mediated pure translation repression and this appears to be conserved among species.
Figure 1.
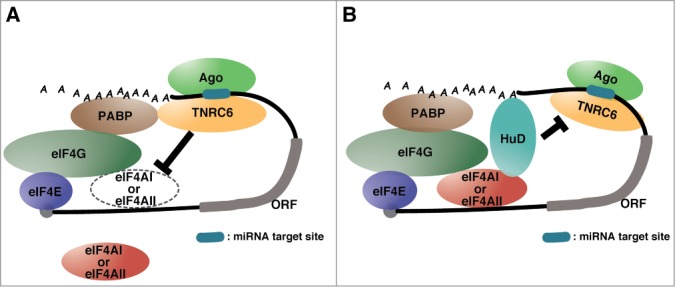
(A) The mechanism of pure translation repression by miRISC in mammal miRISC loading to the target mRNA releases both eIF4AI and eIF4AII from translation initiation complex without deadenylation. Supporting this model, Fukaya et al. also showed that fly Ago1-RISC induces the dissociation of eIF4A from translation initiation complex. (B) The function of HuD for miRNA pathway. The neuronal RNA-binding protein HuD impedes pure translation repression by miRISC via protecting the interaction of eIF4AI and eIF4AII with translation initiation complex on mRNAs.
The Deadenylation and Translation Repression by ARE-BPs
In miRNA pathway, the target gene silencing is regulated through the interplay of both deadenylation and “pure” translation repression. However, the regulation of gene expression via deadenylation is not only miRNA pathway. Tristetraprolin (TTP) is one of the ARE-binding proteins (ARE-BPs), and induces deadenylation and destabilization of the target mRNA.30 Recently, Fabian et al. revealed that TTP recruited CCR4/NOT complex to the target mRNA via direct interaction between TTP and CNOT1, which is a scaffolding subunit of the deadenylation complex.31 In contrast, other ARE-BPs, such as KSRP and AUF1, recruit PARN to regulate the deadenylation of target mRNAs.32 Thus, ARE-BPs are well understood as the regulators of deadenylation and mRNA decay. However, it is still unknown about how these ARE-BPs repress translation during mRNA destabilization. Several studies have revealed that ARE-BPs can regulate translation of target mRNAs via direct interaction with translational machinery. Musashi1 (Msi1) is one ARE-BP that is highly expressed in neuronal stem cells. Kawahara et al. revealed that Msi1 represses translation of m-Numb mRNA through the interaction with PABP, competing with eIF4G at translation initiation complex.33 In addition, HuD stimulates translation through the binding to eIF4A.24 Therefore, we considered the possibility that ARE-BPs such as TTP and AUF1 are also regulators involved in the translational machinery. In fact, CAF1/CNOT7, the main enzyme of deadenylase activity, can repress translation in Xenopus laevis oocytes. In addition, it also represses the cap dependent translation in a deadenylation independent manner.34 This finding suggests that gene silencing accompanying deadenylation such as ARE-BPs and miRNA pathway is regulated by “pure” translation repression using the function of a deadenylase complex.
Local Translation in Neuron Through the Communication between RBPs and miRNA Pathway
Many studies of miRNA indicate that miRNA pathway is required for neuronal development and differentiation. For example, miR-124 is a neuron-specific miRNA, and its expression is stimulated during neuronal differentiation.35 In addition, miR-124 regulates neuronal differentiation through the suppression of Sox9 expression in neural stem cells.36 miR-137 is also a well-characterized miRNA that regulates an early step in neuronal differentiation37 and a late step in developmental plasticity,38 neuronal maturation.39 These miRNA functions imply that the local translational control through the miRNA pathway is necessary for spatial and temporal gene regulation in neurons. Moreover, recent studies indicated that neuron-specific RBPs enable more ingenious gene expression in neurons. Fragile-X mental retardation protein (FMRP), one of the neuron-specific RBPs, associates with core components of the miRNA pathway, such as Dicer and Ago2, and with specific miRNAs. In addition, miR-125b and miR-132 are associated with FMRP and have opposite effects on dendritic spine morphology in hippocampal neurons. Furthermore, the different effect of these miRNAs is regulated by the function of FMRP.40 Muddashetty et al. revealed that FMRP promotes the formation of an Ago2-miR-125a complex on postsynaptic density protein 95 (PSD-95) mRNA and this translation inhibitory complex is regulated by the phosphorylation status of FMRP.41 Thus, the interplay among the miRNA pathway, RBPs and posttranslational modification appears to be important for the local translation regulation in neurons. As mentioned above, HuD is essential for neuronal differentiation and has an inhibitory effect of miRISC in an eIF4A-dependent manner. In addition, the function of HuD is regulated by phosphorylation and methylation.42,43 Therefore, we hypothesize that HuD is involved in local translation through miRNA pathway like FMRP. Indeed, It was proposed that HuD promotes the translation of voltage-gated potassium channel Kv1.1 mRNA at neuron, and this translational control competes with miR-129 through mTORC1 activity.44 Interestingly, HuD directly binds to the active/phosphorylated Akt1, one of the mTOR activators, although HuD is not a substrate of active Akt1. We also have shown that the interaction between HuD and active Akt1 is essential for neuronal differentiation.45 Therefore, we considered the local translation system that active Akt1 on target mRNAs through HuD-binding regulates mTORC1 activity and following miR-129 function at neuron.
Future Investigation
Our most recent study showed that HuD impedes pure translation repression by miRISC. However, we cannot determine the effect of HuD for the deadenylation through the CCR4/NOT complex recruited to the target mRNA by miRISC. Although pure translation repression by miRISC occurs in a deadenylation-independent manner in vitro, in fact, the translation repression co-works with the deadenylation in vivo. Therefore, future studies are needed to investigate whether HuD also inhibits the deadenylation of target mRNA by miRISC. We also revealed that the release of eIF4A from translation initiation complex on mRNAs is essential for the translation repression via miRNA pathway in humans. What is the factor responsible for eIF4A dissociation? CAF1/CNOT7 represses the cap-dependent translation in a deadenylation-independent manner.34 This suggests that CCR4/NOT complex might control the pure translation repression by miRNA pathway.
An interesting report showed that Akt1 stabilizes mRNAs containing ARE by phosphorylating butyrate response factor (BRF1), which binds to ARE-containing mRNAs and promotes their deadenylation, followed by rapid degradation of the mRNAs.46 In Akt1-HuD-mRNP complexes, such inhibition of mRNA-destabilizing factors may occur by means of the associated Akt1 activity, leading to synergistic and localized translational upregulation of mRNAs which are bound by HuD. This notion is supported by the fact that HuD associates with polysome engaged in translation,24 which depends on its RNA-binding activity. Identification of mRNA and protein components of the Akt1-Hu-mRNP complex should provide important clues to elucidate the coupling mechanism between translation and mRNA degradation mediated by RNA-binding protein HuD that regulates neuronal differentiation in vertebrates.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Yukihide Tomari for comments on the manuscript.
Funding
This work was supported in part by a Grant-in-Aid for Scientific Research on Innovative Areas (“Functional machinery for noncoding RNAs”), a Grant-in-Aid for Scientific Research (B) from Japan Ministry of Education, Culture, Sports, Science and Technology. Part of this study was supported by the Uehara memorial foundation and the Naito Foundation (to TF).
References
- 1.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb Perspect Biol 2012; 4:29-53; PMID:22815232; http://dx.doi.org/ 10.1101/cshperspect.a011544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 1998; 2:135-40; PMID:9702200; http://dx.doi.org/ 10.1016/S1097-2765(00)80122-7 [DOI] [PubMed] [Google Scholar]
- 3.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 1999; 68:913-63; PMID:10872469; http://dx.doi.org/ 10.1146/annurev.biochem.68.1.913 [DOI] [PubMed] [Google Scholar]
- 4.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 2004; 5:827-35; PMID:15459663; http://dx.doi.org/ 10.1038/nrm1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell 2008; 132:9-14; PMID:18191211; http://dx.doi.org/ 10.1016/j.cell.2007.12.024 [DOI] [PubMed] [Google Scholar]
- 6.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol 2012; 19:586-93; PMID:22664986; http://dx.doi.org/ 10.1038/nsmb.2296 [DOI] [PubMed] [Google Scholar]
- 7.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 2004; 15:185-97; PMID:15260970; http://dx.doi.org/ 10.1016/j.molcel.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 8.Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J 2010; 29:4146-60; PMID:21063388; http://dx.doi.org/ 10.1038/emboj.2010.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huntzinger E, Kuzuoglu-Ozturk D, Braun JE, Eulalio A, Wohlbold L, Izaurralde E. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res 2013; 41:978-94; PMID:23172285; http://dx.doi.org/ 10.1093/nar/gks1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol 2012; 19:603-8; PMID:22635249; http://dx.doi.org/ 10.1038/nsmb.2309 [DOI] [PubMed] [Google Scholar]
- 11.Zekri L, Kuzuoglu-Ozturk D, Izaurralde E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J 2013; 32:1052-65; PMID:23463101; http://dx.doi.org/ 10.1038/emboj.2013.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, Dziembowski A, Nowotny M, Conti E, Filipowicz W. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell 2014; 54:751-65; PMID:24768538; http://dx.doi.org/ 10.1016/j.molcel.2014.03.036 [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Boland A, Kuzuoglu-Ozturk D, Bawankar P, Loh B, Chang CT, Weichenrieder O, Izaurralde E. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell 2014; 54:737-50; PMID:24768540; http://dx.doi.org/ 10.1016/j.molcel.2014.03.034 [DOI] [PubMed] [Google Scholar]
- 14.Fukaya T, Tomari Y. PABP is not essential for microRNA-mediated translational repression and deadenylation in vitro. EMBO J 2011; 30:4998-5009; PMID:22117217; http://dx.doi.org/ 10.1038/emboj.2011.426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishima Y, Fukao A, Kishimoto T, Sakamoto H, Fujiwara T, Inoue K. Translational inhibition by deadenylation-independent mechanisms is central to microRNA-mediated silencing in zebrafish. Proc Natl Acad Sci U S A 2012; 109:1104-9; PMID:22232654; http://dx.doi.org/ 10.1073/pnas.1113350109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukao A, Mishima Y, Takizawa N, Oka S, Imataka H, Pelletier J, Sonenberg N, Thoma C, Fujiwara T. MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol Cell 2014; 56:79-89; PMID:25280105; http://dx.doi.org/ 10.1016/j.molcel.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 17.Fukaya T, Tomari Y. MicroRNAs mediate gene silencing via multiple different pathways in drosophila. Mol Cell 2012; 48:825-36; PMID:23123195; http://dx.doi.org/ 10.1016/j.molcel.2012.09.024 [DOI] [PubMed] [Google Scholar]
- 18.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 2013; 340:82-5; PMID:23559250; http://dx.doi.org/ 10.1126/science.1231197 [DOI] [PubMed] [Google Scholar]
- 19.Dorn R, Morawietz H, Reuter G, Saumweber H. Identification of an essential Drosophila gene that is homologous to the translation initiation factor eIF-4A of yeast and mouse. Mol Gen Genet 1993; 237:233-40 [DOI] [PubMed] [Google Scholar]
- 20.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; http://dx.doi.org/ 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113-27; PMID:20094052; http://dx.doi.org/ 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci 2008; 65:3168-81; PMID:18581050; http://dx.doi.org/ 10.1007/s00018-008-8252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bronicki LM, Jasmin BJ. Emerging complexity of the HuD/ELAVl4 gene; implications for neuronal development, function, and dysfunction. Rna 2013; 19:1019-37; PMID:23861535; http://dx.doi.org/ 10.1261/rna.039164.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, Thoma C, Fujiwara T. The ELAV protein HuD stimulates cap-dependent translation in a Poly(A)- and eIF4A-dependent manner. Mol Cell 2009; 36:1007-17; PMID:20064466; http://dx.doi.org/ 10.1016/j.molcel.2009.11.013 [DOI] [PubMed] [Google Scholar]
- 25.Duncan KE, Strein C, Hentze MW. The SXL-UNR corepressor complex uses a PABP-mediated mechanism to inhibit ribosome recruitment to msl-2 mRNA. Mol Cell 2009; 36:571-82; PMID:19941818; http://dx.doi.org/ 10.1016/j.molcel.2009.09.042 [DOI] [PubMed] [Google Scholar]
- 26.Bordeleau ME, Robert F, Gerard B, Lindqvist L, Chen SM, Wendel HG, Brem B, Greger H, Lowe SW, Porco JA Jr., et al.. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J Clin Invest 2008; 118:2651-60; PMID:18551192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, Brem B, Teodoro JG, Greger H, Tremblay ML, et al.. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PloS one 2009; 4:e5223; PMID:19401772; http://dx.doi.org/ 10.1371/journal.pone.0005223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malina A, Cencic R, Pelletier J. Targeting translation dependence in cancer. Oncotarget 2011; 2:76-88; PMID:21378410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukaya T, Iwakawa HO, Tomari Y. MicroRNAs block assembly of eIF4F translation initiation complex in Drosophila. Mol Cell 2014; 56:67-78; PMID:25280104; http://dx.doi.org/ 10.1016/j.molcel.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 30.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 2005; 19:351-61; PMID:15687258; http://dx.doi.org/ 10.1101/gad.1282305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabian MR, Frank F, Rouya C, Siddiqui N, Lai WS, Karetnikov A, Blackshear PJ, Nagar B, Sonenberg N. Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat Struct Mol Biol 2013; 20:735-9; PMID:23644599; http://dx.doi.org/ 10.1038/nsmb.2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell 2004; 14:571-83; PMID:15175153; http://dx.doi.org/ 10.1016/j.molcel.2004.05.002 [DOI] [PubMed] [Google Scholar]
- 33.Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol 2008; 181:639-53; PMID:18490513; http://dx.doi.org/ 10.1083/jcb.200708004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem 2010; 285:28506-13; PMID:20634287; http://dx.doi.org/ 10.1074/jbc.M110.150763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol 2002; 12:735-9; PMID:12007417; http://dx.doi.org/ 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- 36.Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 2009; 12:399-408; PMID:19287386; http://dx.doi.org/ 10.1038/nn.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, et al.. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 2008; 6:14; PMID:18577219; http://dx.doi.org/ 10.1186/1741-7015-6-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, Santistevan NJ, Li W, Zhao X, Jin P. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol 2010; 189:127-41; PMID:20368621; http://dx.doi.org/ 10.1083/jcb.200908151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, Teng ZQ, Luo Y, Peng J, Bordey A, et al.. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 2010; 28:1060-70; PMID:20506192; http://dx.doi.org/ 10.1002/stem.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 2010; 65:373-84; PMID:20159450; http://dx.doi.org/ 10.1016/j.neuron.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell 2011; 42:673-88; PMID:21658607; http://dx.doi.org/ 10.1016/j.molcel.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pascale A, Amadio M, Scapagnini G, Lanni C, Racchi M, Provenzani A, Govoni S, Alkon DL, Quattrone A. Neuronal ELAV proteins enhance mRNA stability by a PKCalpha-dependent pathway. Proc Natl Acad Sci U S A 2005; 102:12065-70; PMID:16099831; http://dx.doi.org/ 10.1073/pnas.0504702102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujiwara T, Mori Y, Chu DL, Koyama Y, Miyata S, Tanaka H, Yachi K, Kubo T, Yoshikawa H, Tohyama M. CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol Cell Biol 2006; 26:2273-85; PMID:16508003; http://dx.doi.org/ 10.1128/MCB.26.6.2273-2285.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sosanya NM, Huang PP, Cacheaux LP, Chen CJ, Nguyen K, Perrone-Bizzozero NI, Raab-Graham KF. Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J Cell Biol 2013; 202:53-69; PMID:23836929; http://dx.doi.org/ 10.1083/jcb.201212089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujiwara T, Fukao A, Sasano Y, Matsuzaki H, Kikkawa U, Imataka H, Inoue K, Endo S, Sonenberg N, Thoma C, et al.. Functional and direct interaction between the RNA binding protein HuD and active Akt1. Nucleic Acids Res 2012; 40:1944-53; PMID:22075994; http://dx.doi.org/ 10.1093/nar/gkr979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidlin M, Lu M, Leuenberger SA, Stoecklin G, Mallaun M, Gross B, Gherzi R, Hess D, Hemmings BA, Moroni C. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J 2004; 23:4760-9; PMID:15538381; http://dx.doi.org/ 10.1038/sj.emboj.7600477 [DOI] [PMC free article] [PubMed] [Google Scholar]


