Abstract
Elongator is a 6 subunit protein complex highly conserved in eukaryotes. The role of this complex has been controversial as the pleiotropic phenotypes of Elongator mutants have implicated the complex in several cellular processes. However, in yeast there is convincing evidence that the primary and probably only role of this complex is in formation of the 5-methoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5) side chains on uridines at wobble position in tRNA. In this review we summarize the cellular processes that have been linked to the Elongator complex and discuss its role in tRNA modification and regulation of translation. We also describe additional gene products essential for formation of ncm5 and mcm5 side chains at U34 and their influence on Elongator activity.
Keywords: elongator complex; KTI genes; SIT4; SAP genes; tRNA wobble uridine modifications,; translation; Kluveromyces lactis γ-toxin
The Yeast Elongator Complex has been Implicated in Many Cellular Processes
The Elongator complex in S. cerevisiae was first described to consist of 3 proteins (Elp1, Elp2, and Elp3), which were found to be associated with the hyperphosphorylated elongating form of RNA polymerase II (Pol II).1 Furthermore, when introducing an elp1Δ strain into new growth conditions, mRNAs encoding gene products required for growth adaptation showed slow induction, supporting a defect in Pol II transcription and a nuclear localization of the complex.1 Additional investigations identified the Elp4, Elp5, and Elp6 as a sub complex of Elongator complex.2-4 In vitro, the Elp3 subunit of the 6-subunit Elongator complex was able to transfer acetyl groups from acetyl-CoA to histones and an Elp3p with amino acid substitutions in the C-terminal acetyl-CoA binding domain (HAT) show reduced histone acetylation.5,6 In vivo, inactivation of the ELP3 gene resulted in decreased H3 and H4 acetylation.7 From these data it was concluded that Elongator complex was important for transcription elongation of Pol II and therefore it was named Elongator complex.1,5-7 However, the involvement of Elongator complex in histone acetylation and transcription elongation was questioned, as chromatin immuno-precipitation experiments failed to detect Elongator on transcribing open reading frames and Elp1 to Elp3 proteins are localized in the cytosol.8
A cytoplasmic role of Elongator complex was suggested from the finding that Elp1p interacted with Sec2p, a protein required for polarized transport of secretory vesicles to the bud tip in S. cerevisiae.9 Transport of secretory vesicles requires the guanine nucleotide exchange factor Sec2p for activation of the vesicle-associated GTPase Sec4p.10,11 Sec2p associates with Elp1p and it was proposed that the Elongator complex is required for regulation of exocytosis by influencing localization of Sec2p.9
A different nuclear function described for the Elongator complex was in telomeric gene silencing and DNA repair as Elongator mutants display partial loss of telomeric gene silencing and increased sensitivity to DNA damage agents.12 In addition to the HAT domain, the Elp3p subunit of the Elongator complex shares sequence homology to proteins from the Radical S-adenosylmethionine (SAM) superfamily, proteins harboring an iron-sulfur cluster that catalyze a variety of radical reactions using SAM.13 Point mutations resulting in amino acid substitutions in the Radical SAM or HAT domains of the ELP3 gene displayed defects in telomeric gene silencing and DNA repair suggesting a role of Elongator complex in these processes.12 A role for the Elongator complex in DNA repair was supported by its interaction with proliferating cell nuclear antigen (PCNA), a protein involved in DNA replication and DNA repair.12
Another function of Elongator complex, linking it to modification of tRNA, was based on the characterization of the Schizosaccharomyces pombe sin3-193 mutant. A sin3-193 mutant shows reduced levels of the modified wobble (U34) nucleoside 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) (Fig. 1)14,15 causing an antisuppressor phenotype, i. e. the ochre serine tRNA suppressor encoded by the sup3-18 gene will no longer suppress the ade7-413 ochre allele.16 A sin3-193 mutant displays slight increase in cell volume, length, and amount of dead cells. This observed increase was independent of the presence or absence of the ochre tRNA suppressor, indicating that the Sin3 protein is required for proper cell cycle regulation.14,15 The sin3+ gene was identified as an uncharacterized open reading frame (ORF).17 A strain with a null allele of the sin3+ gene lacks the wobble uridine nucleosides mcm5s2U in and 5-methoxycarbonylmethyluridine (mcm5U) (Fig. 1) in the sup3-18 encoded ochre suppressor tRNASer.17 The sin3+ gene encodes a conserved protein with 77% identity on the amino acid level to the S. cerevisiae Elp3 protein. The ELP3 gene of Saccharomyces cerevisiae is essential for formation of mcm5 and ncm5 side chains in mcm5s2U, mcm5U, 5-carbamoylmethyluridine (ncm5U) and 5-carbamoylmethyl-2'-O-methyluridine (ncm5Um) at U34 in tRNA (Fig. 1).17 Inactivation of the other S. cerevisiae Elongator genes encoding Elp1-Elp2p and Elp4-Elp6p displayed identical phenotypes as the elp3 null mutant including the tRNA modification defect.17,18 This observation suggested that the Elongator complex is required for synthesis of the first step, an intermediate likely to be 5-carboxymethyluridine (cm5U)19-23, in formation of mcm5U, mcm5s2U, ncm5U and ncm5Um at U34 in tRNA (Fig. 1).17 Additional support for a role in tRNA modification was recently strengthened by the observation that the Elp3p homolog from the archaea Methanocaldococcus infernus in vitro modifies U34 in tRNA to cm5U in the presence of acetyl-CoA.24
Figure 1.
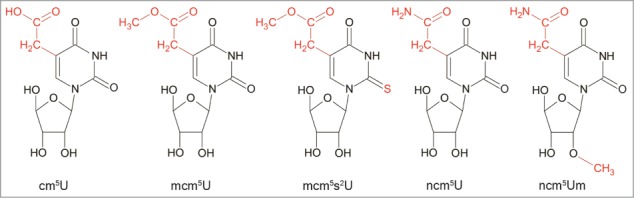
Structure of 5-carboxymethyluridine (cm5U), 5-methoxycarbonylmethyluridine (mcm5U), 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), 5-carbamoylmethyluridine (ncm5U) and 5-carbamoylmethyl-2'-O-methyluridine (ncm5Um). Highlighted in red are uridine side groups cm5, mcm5, ncm5, s2 and the 2'-O-methylation of the ribose.
In Yeast, The Phenotypes of Elongator Deficient Cells are Linked to tRNA Modification
As the Elongator complex in yeast was implicated in 4 different cellular processes, it was important to determine if the complex has 4 distinct functions or if it affects one key process that leads to multiple downstream effects. In elongation of Pol II transcription, Elp3p was suggested to act as a HAT transferring acetyl groups to histones and indeed in an elp3 mutant there is a defect primarily in histone H3 acetylation.5,7 The Elp3 protein was also suggested to be crucial for telomeric gene silencing and DNA repair.12 In exocytosis, an interaction between Elp1p and Sec2p was proposed to be important for correct polarized localization of Sec2p.9 Interestingly, overexpression of various combinations of , and restored acetylation of histone H3, telomeric gene silencing and DNA repair in an elp3 mutant and localization of Sec2p in an elp1 mutant.18,25 In an elp3 mutant, the expression level of the Sir4 protein is decreased at translational level, a phenotype that is corrected if , and are overexpressed.25 The Sir4 protein is involved in assembly of silent chromatin at telomeres and in this gene AAA codons decoded by are overrepresented.25 Based on these observations function of the S. cerevisiae Elongator complex is linked to tRNA modification/ translation and not transcription, exocytosis, telomeric gene silencing and DNA repair. The notion that mcm5s2U is important in , and was further supported by the observation that an ncs2 mutant unable to form the s2 but not the mcm5 group shows identical but weaker phenotypes than Elongator deficient yeast cells. In S.cerevisieae, the Ncs2p in complex with Ncs6p is required for the final step in formation of the 2-thio group at wobble position in tRNAs 26-28. Phenotypes observed in the ncs2 mutant are suppressed by overexpression of various combinations of , and (Table 1).18,25 An explanation for the dependence of the mcm5s2U34 nucleoside is that it promotes a canonical anticodon loop conformation which stabilize codon-anticodon interaction.29,30
Table 1.
Phenotypes of S. cerevisiae and S. pombe mutants lacking wobble uridine modifications.
| A. S. cerevisiae | ||
|---|---|---|
| Growth Defects | Deletion Mutants | Suppressed by tRNA* |
| Slow growth at 30°C | elp3Δ or ncs2Δ | tKUUU, tQUUG18 |
| Ts at 38°C | elp3Δ or ncs2Δ | tKUUU, tQUUG18 |
| Prolonged G1 phase | elp3Δ | tKUUU, tQUUG18 |
| Adaptation to carbon source | elp3Δ or ncs2Δ | tKUUU, tQUUG18 |
| Sodium-chloride sensitivity | elp3Δ or ncs2Δ | tKUUU, tQUUG18 |
| Rapamycin Sensitivity | nsc2Δ, ncs6Δ, urm1Δ or uba4Δ | tKUUU, tQUUG, tEUUC28 |
| Caffeine sensitivity | elp3Δ or ncs2Δ | tKUUU, tQUUG18 |
| nsc2Δ, ncs6Δ, urm1Δ or uba4Δ | tKUUU, tQUUG, tEUUC28 | |
| Diamide Sensitivity | nsc2Δ, ncs6Δ or urm1Δ | tKUUU, tQUUG, tEUUC28 |
| uba4Δ | tKUUU, tQUUG28 | |
| Transcription and Chromatin-Remodelling Defects | Deletion Mutants | Suppressed by tRNA* |
| GAL1 mRNA induction | elp3Δ or ncs2Δ | tKUUU, tQUUG18 |
| ENA1 mRNA induction | elp3Δ | tKUUU, tQUUG18 |
| Lethal in combination with histone H4 (4-19Δ) | elp3Δ or ncs2Δ | tKUUU, tQUUG18 |
| Ts in combination with histone H3 (3-29Δ) | elp3Δ or ncs2Δ | tKUUU, tQUUG18 |
| Ts in combination with histone H3 (K14R) / histone H4 (K8, 16R) | elp3Δ or ncs2Δ | tKUUU, tQUUG18 |
| Synergistic growth defect in combination with gcn5Δ | elp3Δ or ncs2Δ | tKUUU, tQUUG18 |
| Acetylation defect of lys14 in histone H3 | elp3Δ | tKUUU, tQUUG18 |
| Secretion Defects | Deletion Mutants | Suppressed by tRNA* |
| Viable at 34°C in combination with sec2-59 | elp1Δ or ncs2Δ | tKUUU, tQUUG18 |
| Mislocalization of Sec2p | elp1Δ or ncs2Δ | tKUUU, tQUUG18 |
| DNA Repair and Telomere Gene Silencing Defects | Deletion Mutants | Suppressed by tRNA* |
| Hydroxyurea (HU) sensitivity | elp3Δ or tuc2Δ | tKUUU, tQUUG, tEUUC25 |
| Telomere gene silencing | elp3Δ or tuc2Δ | tKUUU, tQUUG, tEUUC25 |
| tRNA Modification Defects | Deletion Mutants | Suppressed by tRNA* |
| mcm5, ncm5 or s2 modification defect | elp3Δ or ncs2Δ | No 18. 25 |
| B. S. pombe | ||
| Growth Defects | Deletion Mutants | Suppressed by tRNA** |
| Ts at 36°C | ctu1Δ | tKUUU, tEUUC31 |
| elp1Δ, elp3/sin3Δ, elp4Δ, elp6Δ or ctu1Δ elp3/sin3Δ | tKUUU32 | |
| Rapamycin Sensitivity | elp1Δ, elp3/sin3Δ, elp4Δ, elp6Δ or ctu1Δ elp3/sin3Δ | tKUUU32 |
| SDS Sensitivity | ctu1Δ elp3/sin3Δ | tKUUU32 |
| H2O2 Sensitivity | elp3/sin3Δ or ctu2Δ | tKUUU33 |
In S. cerevisiae, tRNA tKUUU(Lys), tQUUG(Gln) and tEUUC(Glu) have mcm5s2U at wobble position.
In S. pombe, tEUUC(Glu) has mcm5s2U at wobble position; identity of U34 in tRNA tKUUU(Lys) is unknown.
In S. cerevisiae, null mutations in Elongator genes (ELP1-ELP6) abolish formation of mcm5 side chain; null mutations in the NCS2/TUC2, NCS6, URM1 or UBA4 genes abolish formation of the s2 moiety.
* In S. pombe, null mutations in Elongator genes (elp1+, elp3+, elp4+ or elp6+) abolish formation of mcm5 side chain and the s2 moiety (see text); null mutations in the ctu1+ or ctu2+ genes abolish formation of the s2 moiety.
Additional support that elevated levels of hypomodified tRNA correct phenotypes observed in mutants with defects in wobble uridine modifications came from experiments in Schizosaccharomyces pombe. In S. pombe, elevated levels of hypomodified or or combinations thereof suppress phenotypes observed in the ctu1Δ and ctu2Δ single mutants lacking the s2-group, the elp3/sin3Δ single mutant or the elp3Δ ctu1Δ double mutant lacking both modifications (Table 1).31-33 In S. pombe Ctu1p is the homolog to Ncs6p and Ctu2p is the homolog to Ncs2p 31.
Kluveromyces Lactis γ-toxin a Tool to Identify Genes Required for Wobble Uridine Modifications
Another phenotype of yeast Elongator mutants is resistance to K. lactis killer toxin.34-37 Certain strains of the dairy yeast K. lactis contains “killer DNA," a plasmid pair (k1 and k2) encoding a 3-subunit anti-yeast toxin complex known as zymocin.38-40 Upon secretion, the α- and β-subunits dock the zymocin to the cell wall of susceptible yeasts and facilitate transfer of the cytotoxic γ-subunit, which will arrest the cell before START in the G1 phase of the cell cycle.37,40,41 Two types of S. cerevisiae mutants resistant to zymocin have been described.37 Type I resistant mutants are defective in binding and uptake of zymocin, but sensitive to endogenous expression of the γ-toxin. Type II mutants were believed to be target site mutants as they are resistant to both exogenous zymocin and endogenous expression of γ-toxin.37,40 The cellular target(s) of K. lactis γ-toxin was unsolved for more than 20 y and initially adenylate cyclase was mistakenly identified as the target of γ-toxin.41,42 The γ-toxin turned out to be an endonuclease having , and as substrates.43 These tRNAs have the mcm5s2U modified nucleoside at wobble position and the endonuclease cleaves the tRNAs between U34 and U35.43 Presence of the mcm5 side chain of mcm5s2U is crucial for these tRNAs to be substrates, explaining why Elongator mutants are resistant to zymocin or endogenously expressed γ-toxin.43,44 Thus, the γ-toxin resistance phenotype as well as phenotypes of yeast Elongator mutants suppressed by overexpression of hypomodified tRNAs are explained by an inability to make mcm5 and ncm5 side chains at wobble uridines.18,43
There has been a number of genetic screens to identify mutants resistant to zymocin, generating iki mutants (insensitive to killer toxin), kti mutants (killer toxin insensitive) and tot mutants (toxin target).34-37 Strains with mutations in any of the Elongator subunit genes (ELP1-ELP6), killer toxin insensitive genes (KTI11-KTI14), the TRM9 gene, the SIT4 gene or both the SAP185 and SAP190 genes are type II mutants 34-37,40,45, and these mutants are unable to form mcm5 and ncm5 side chains at wobble position (Table 2).17,46 To identify additional mutants affecting formation of the mcm5 group, a yeast deletion collection containing 4800 strains, with different non-essential gene deletions, was screened for resistance to zymocin.46 In addition to strains deleted for ELP, KTI, SIT4, and TRM9 genes, 5 strains (urm1Δ, uba4Δ, ncs2Δ, ncs6Δ, and tum1Δ) were identified (Table 2). These strains lacked the s2 group in mcm5s2U (Fig. 1), illustrating the importance of both the mcm5 and s2 groups for the action of γ-toxin.
Table 2.
Genes having mutated alleles causing a defect in wobble uridine modifications.
| S. cerevisiae (Gene name/Alias) | S. pombe (Gene name/Alias) | C. elegans (Gene name/Alias) | A. thaliana (Gene name/Alias) | M. musculus (Gene name) | H. sapiens (Gene name) |
|---|---|---|---|---|---|
| ncm5/mcm5 side chains | |||||
| ELP1/IKI3/KTI7/TOT117 | - | elpc-181 | AtELP1/ELO286 | Ikbkap85 | IKBKAP87 |
| ELP2/KTI3/TOT217 | - | . | - | - | - |
| ELP3/KTI8//TOT3/HPA117 | elp3+/sin3+17 | elpc-381 | AtELP3/ELO386 | - | - |
| ELP4/KTI9/TOT7/HAP117 | - | . | - | - | - |
| ELP5/IKI1/TOT5/HAP217 | - | . | - | - | - |
| ELP6/KTI4/ TOT6/HAP317 | - | - | - | - | - |
| KTI11/DPH3/KTI5 * 17, 46 | - | dph-3103 | - | - | - |
| KTI12/TOT417 | - | - | - | - | - |
| KTI13/ATS1/FUN2817 | - | - | - | - | - |
| KTI14/HRR2546 | - | - | - | - | - |
| SIT4/PPH146 | - | - | - | - | - |
| SAP185 ** 46 | - | - | - | - | - |
| SAP190 ** 46 | - | - | - | - | - |
| 5-methoxy residue of mcm5U/mcm5s2U | |||||
| TRM9/KTI120 | - | - | AtTRM9 **** 53 | Alkbh8 *** 22 | - |
| TRM11221, 23 | - | - | - | - | - |
| 2-thio group (s2) of mcm5s2U | |||||
| NFS1/SPL1104 | - | - | - | - | - |
| ISU1/NUA1105 | - | - | - | - | - |
| ISU2/NUA2105 | - | - | - | - | - |
| CFD1/DRE3105 | - | - | - | - | - |
| NBP35 105 | - | - | - | - | - |
| CIA1105 | - | - | - | - | - |
| URM126-28, 46 | - | - | - | - | - |
| UBA4/YHR126-28, 46 | - | moc-3103 | - | - | - |
| NCS6/YGL210W-A/TUC126-28,46, 106 | ctu1+31 | ctu-1/tuc-1/tut-128, 31, 81 | - | - | - |
| NCS2/TUC218, 26-28, 46 | ctu2+31 | - | AtCTU2107 | - | - |
| TUM127, 28, 46 | - | - | - | - | - |
KTI5 is a dominant allele of KTI11.
** A sap185Δ sap190Δ double mutant lacks ncm5/mcm5 side chains.
*** M. musculus Alkbh8 encodes the ALKBH8 protein containing a RNA recognition motif (RRM), AlkB-like dioxygenase domain (AlkB) and methyltransferase (MT) domain. MT domain catalyses methylation of cm5U into mcm5U together with TRM112; RRM/AlkB domains catalyse hydroxylation of mcm5U into (S)-mchm5U.
**** A. thaliana AtTRM9 encodes the AtTRM9 protein catalysing the methylation of cm5U into mcm5U together with AtTRM112a/b; AtALKBH8 catalyses hydroxylation of mcm5U into (S)-mchm5U.
Proteins Required for Formation of Wobble Uridine Modifications
Synthesis of the mcm5 side chain at the wobble position requires 15 gene products and formation of the s2 group in mcm5s2U requires 11 gene products (Fig. 2). Strains with a deletion of any of the ELP1-ELP6, KTI11, KTI12, KTI14, SIT4 or SAP185 and SAP190 genes lack the mcm5U, mcm5s2U and ncm5U nucleosides, whereas a kti13 deletion mutant has severely reduced levels of these nucleosides.17,46 No intermediates of mcm5U, and ncm5U are detected in any of the mutants, whereas s2U is detected in tRNAs normally containing mcm5s2U.17,46 Thus, all these gene products are required for an early step in synthesis of mcm5 and ncm5 groups (Fig. 2).
Figure 2.
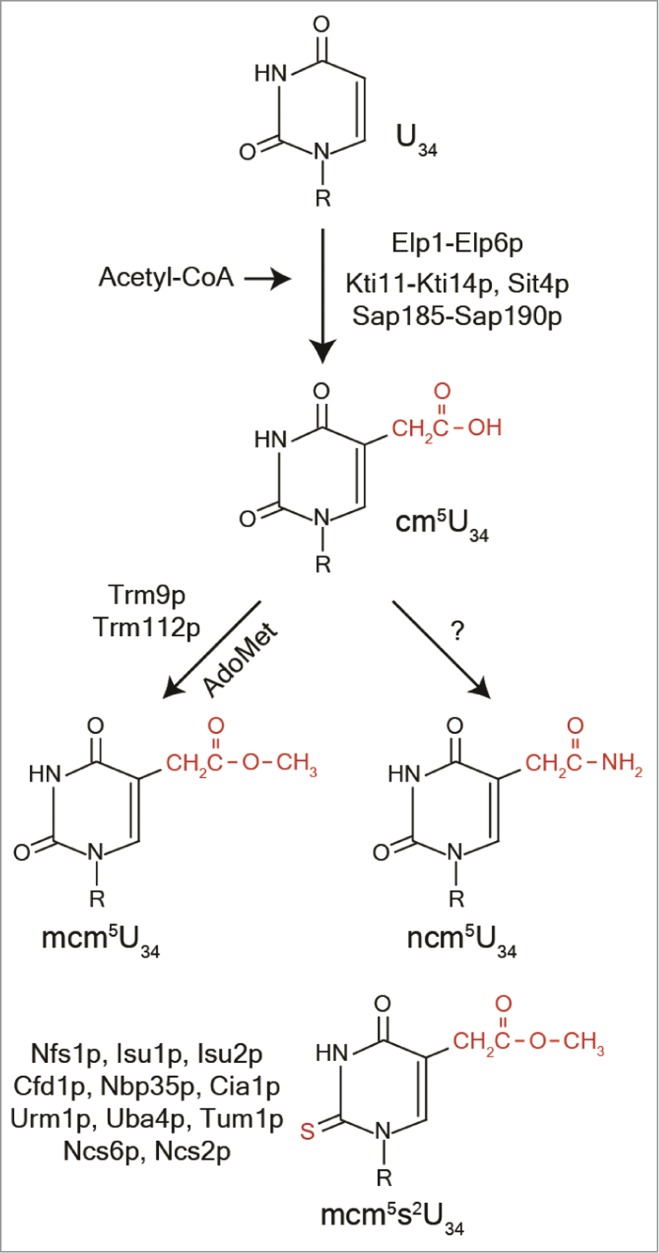
Proteins required for formation of the 5-methoxycarbonylmethyl (mcm5), 5-carbamoylmethyl (ncm5) and the 2-thio (s2) side groups on uridines at wobble position. Acetyl-CoA acts as a donor in formation of the cm5 side group. Trm9p and Trm112p utilize AdoMet (S-Adenosylmethionine) as a methyl donor to form the mcm5 side group. Last step in formation of ncm5 is unknown (?). R represents ribose and highlighted in red are uridine side groups cm5, mcm5, ncm5 and s2. For details, see text.
The Elongator complex consists of 2 sub complexes, Elp1-Elp3p and Elp4-Elp6p 1–4, and recently the crystal structure of the Elp4-Elp6p sub complex was solved at 2.1Å resolution.47 The crystal structure revealed that all 3 subunits share a RecA-like fold and 2 heterotrimers form a hexameric ring like structure. A functional Elongator complex was proposed to contain 2 copies of the Elp1-Elp3p core complex on each hexameric Elp4-Elp6p ring.47 Hexameric RecA-like ATPases of ring-translocases have the ability to bind specific DNA or RNA substrates.48 Experiments supporting a direct role of Elongator complex in tRNA binding are; (i) a tRNA that should obtain a mcm5 group at wobble position co-precipitated with Elp1p or Elp3p17, (ii) in electrophoretic mobility shift assay (EMSA) experiments, the Elp4-Elp6p sub complex binds tRNA47, (iii) the C-terminal domain (CTD) of Elp1 binds tRNA in an EMSA experiment.49 The catalytic activity of Elongator complex is believed to reside in the Elp3 protein due to the presence of 2 domains. One sharing homology to acetyl-CoA binding (HAT) domains and the other to Radical S-adenosylmethionine (SAM) domains harboring an iron-sulfur (Fe-S) cluster that catalyze a variety of radical reactions by using SAM.5,13,50 The presence of a Fe-S cluster in the Radical SAM domain and ability to bind SAM has been verified for the archaeal M. jannaschii Elp3p homologue.51 Homologues to the Elp3 protein are found in essentially all archaea but not the other subunits of the Elongator complex. 24, 51 This suggests that archaea require only Elp3p for formation of cm5U. In vitro, recombinant Elp3p from Methanocaldococcus infernus catalyze the transfer of an acetyl radical forming cm5U in the presence of acetyl-CoA, the reducing agent sodium dithionite and S-adenosylmethionine.24 Thus, this unique enzymatic reaction mechanism explains the requirement of the SAM and HAT domains in Elp3p.
The last step in formation of mcm5U requires the tRNA methyltransferase Trm9 and the Trm112 protein, whereas no gene product responsible for the last step in formation of ncm5U is known (Fig. 2).20-23,52,53 Other than being a subunit required for Trm9p activity, Trm112p is also a subunit required for the activity of 3 other methyltransferases, the tRNA methyltransferase Trm11p forming m2G10 in tRNA, the ribosomal methyl transferase Bud23p methylating G1575 in 18S rRNA54 and Mtq2p methylating the release factor eRF1.52,55
A set of proteins associated with Elongator complex are suggested to regulate its activity. These are the Kti11-Kti14, Sit4, Sap185 and Sap190 proteins (Fig. 3). The Sit4p is a type 2A protein phosphatase that associates with members of a protein family termed SAPs.56,57 Deletion of all 4 SAP genes (SAP4, SAP155, SAP185 and SAP190) confers the same phenotypes as loss of SIT4.56 SAPs fall into 2 groups based on their sequence similarity: the SAP4/ SAP155 group and the SAP185/ SAP190 group.56 The 2 groups of SAPs are believed to be either effectors or activators of Sit4p.56 Sit4p, Sap185p, and Sap190p have been shown to physically interact with Kti14p.58 KTI14 (HRR25) encodes a homologue to the mammalian casein kinase 1δ (CK1δ).59 Kti14p interacts with Elongator and the interaction is dependent on Kti12p.60 The kinase Kti14p and the phosphatase Sit4p seem to antagonistically regulate activity of Elongator complex by phosphorylation/ de- phosphorylation of the largest Elongator subunit Elp1p.60,61 In addition to phosphorylation/ de- phosphorylation, the activity of Elongator complex also seems to be regulated by proteolysis of Elp1p.62 The Kti11p interacts with Elongator complex through its C-termini and loss of Kti11p enhances the proteolysis of Elp1p.62,63 Except for being crucial for ncm5 and mcm5 side chain formation, Kti11p/Dph3p is also required for biosynthesis of the posttranslational modification diphtamide, a unique target on translation elongation factor 2 (eEF2) for bacterial ADP-ribosylating toxins.64 In diphthamide biosynthesis, Kti11p is an electron donor for the Fe-S clusters in the Dph1-Dph2p65, and it is conceivable that in the tRNA modification reaction Kti11p also acts as an electron donor for the Fe-S cluster in Elp3p. The Kti11p also interacts with Kti13p a protein with structural features similar to the guanine exchange factors.66 However, the role of Kti13p in yeast is not yet elucidated.
Figure 3.
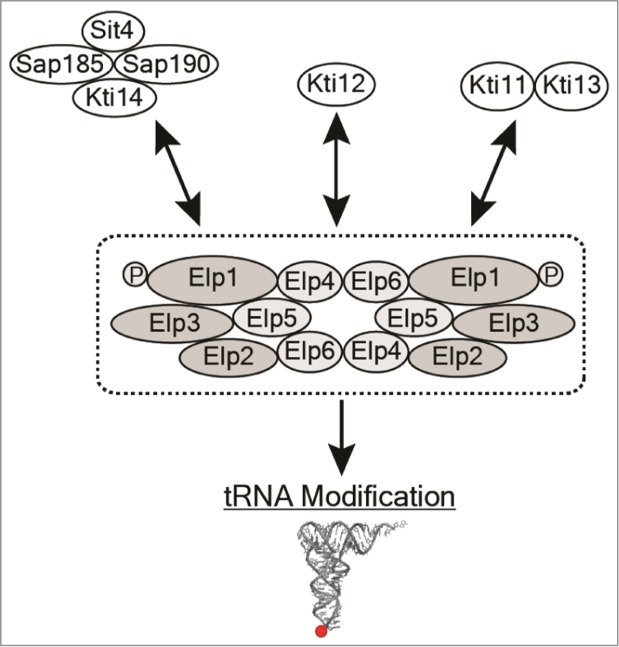
Elongator and Elongator associated proteins. Elongator complex is suggested to contain 2 copies of the Elp1-Elp3p core complex (dark gray) on each hexameric Elp4-Elp6p ring.47 Elongator associated proteins are clustered according to interaction studies. P indicates phosphorylation of Elp1p. For details, see text.
Formation of the 2-thio group present in , and , requires 11 gene products (Fig. 2) and the thiolation reaction has been successfully reconstituted in vitro.26-28 In S. cerevisiae, presence of a mcm5 side chain at U34 is a prerequisite for efficient 2-thio group formation and in an S. pombe elp3/sin3 mutant the 2-thio group formation is abolished.26-28,32
Recently it was suggested that levels of certain modified nucleosides in tRNA, among them ncm5U, mcm5U and mcm5s2U could be altered in response to various stress conditions.67 However, the levels of the ncm5U, mcm5U and mcm5s2U nucleosides were not quantified in individual tRNA isoacceptors rather they were quantified in bulk tRNA. Therefore, it is not possible to distinguish between regulation of modification on individual tRNA isoacceptors or if the levels of the tRNA isoacceptors are altered.
Role of Wobble Uridine Modifications in Translation
The wobble uridine nucleosides mcm5U and ncm5U were believed to either restrict pairing to A 68 or to allow efficient interaction with both A and G.68 Presence of the s2 group in the wobble nucleoside mcm5s2U has been suggested to restrict reading to A-ending codons.68,69 In S. cerevisiae, there are 42 different cytoplasmic tRNA species and 11 of these contain ncm5U, ncm5Um, mcm5U, or mcm5s2U at wobble position (Fig. 4).70-73 Thus in yeast mutants affecting formation of ncm5 and mcm5 side chains, like Elongator mutants, these side chains are abolished in about 25% of the tRNA population which is a likely explanation for the pleiotropic phenotypes. As mutants defective in formation of mcm5/ncm5 or s2 groups at wobble uridines are available, the in vivo roles of these modifications have been investigated using a set of different strategies.
Figure 4.
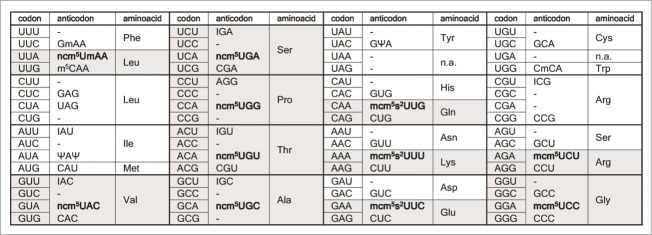
The genetic code and distribution of cytoplasmic S. cerevisiae tRNAs. The anticodon sequences of the 42 different tRNA species (1 initiator and 41 elongator tRNAs) are indicated.43,69-72 For anticodons with an uncharacterized RNA sequence, the primary sequence is shown. The initiator and elongator tRNAMet species have identical anticodon sequences. The wobble rules suggest that an inosine (I34) residue allows paring with U, C, and sometimes A. A tRNA with a G or its 2′-O-methyl derivative (Gm) at the wobble position should read U- and C-ending codons. Presence of a C34 residue or its 5-methyl (m5C) or 2′-O-methyl (Cm) variant should only allow pairing with G. The pseudouridine (Ψ)-containing tRNAIle is presumably unable to pair with the methionine AUG codon. The anticodons containing mcm5U, mcm5s2U, ncm5U and ncm5Um derivatives are shown in bold. Copyright © American Society for Microbiology, [Molecular and Cellular Biology, 28, 2008, 3301–3312 and doi:10.1128/MCB.01542-07].72
In one strategy, strains with specific tRNA gene deletions in combination with deletion of genes responsible for formation of mcm5/ ncm5 or s2 groups were constructed and the growth properties of the strains were studied.73 These analyses revealed that mcm5 and ncm5 side chains promote pairing with G-ending codons in most codon boxes and that concurrent mcm5 and s2 groups improve reading of both A- and G- ending codons.73
A second in vivo strategy is to quantify the role of wobble uridine modifications in a tRNA isoacceptor decoding the cognate A or the near cognate G ending codon using reporter systems. The SUP4-encoded ochre suppressor tRNA, where the primary anticodon sequence is UUA, has a mcm5 side chain at U34.17 In an elp3 mutant lacking the entire mcm5 side chain resulting in an unmodified U34, efficiency of decoding the cognate ochre UAA and the near cognate amber UAG codons by the SUP4 suppressor tRNA is reduced.73 Also having the anticodon sequence UCU has a mcm5 side chain at U34. In a trm9 mutant lacking the methyl esterification of cm5U34, efficiency of decoding of multiple cognate AGA and multiple near cognate AGG codons by is decreased.74
The proteome expression in S. cerevisiae urm1 and uba4 mutants lacking the s2 group of mcm5s2U or an elp3 mutant lacking ncm5 and mcm5 side chains has been investigated using stable isotope-labeling of amino acids in cell culture (SILAC) technology.75 In urm1 and uba4 mutants about 270 proteins were either up- or down-regulated. The analysis revealed that 85% of up regulated proteins and 75% of down regulated proteins from the elp3 mutant overlap with the results from the urm1 and uba4 mutants. Codons AAA, CAA and GAA read by tRNAs having the mcm5s2U34 modified nucleoside were enriched in the downregulated genes. Reporter constructs having any of these codons in multiple copies show reduced expression in urm1Δ, uba4Δ and elp3Δ backgrounds.75 However, no correlation to mRNA levels of down regulated genes was reported.
Ribosome profiling (Ribo-seq) is a method to determine genome wide distribution of ribosomes at the codon level on mRNA.76 Ribo-seq and RNA-seq (RNA Sequencing) analysis were done in S. cerevisiae strains lacking the mcm5 and ncm5 side chains (elp3 mutant), the s2 group (ncs2, ncs6 and uba4 mutants), and a wild type strain.77 Mutants with a defect in s2 group formation show an accumulation of ribosomes when AAA and CAA codons were in the A-site, whereas the elp3 mutant lacking the mcm5 and ncm5 side chain, show ribosome accumulation at CAA and GAA codons. Although accumulation of ribosomes at these codons suggests a slowdown in translation, a correlation with a reduction in protein output could not be established.
Another tool for genome wide analysis in Schizosaccharomyces pombe is the fission yeast integrated ORFeome library, which is a library of 4910 open reading frames having a Flag2-His6 tag, where expression of constructs can be determined using anti-His-antibody.78 A null allele of elp3+/ sin3+ gene was introduced into the ORFeome library and expression levels of the tagged ORF's were analyzed in elp3/ sin3Δ and wild type background.32 About 500 genes enriched in AAA and GAA codons showed reduced expression in elp3/ sin3Δ compared to wild type background. To investigate the relevance of AAA codons for expression, a candidate gene cdr2+ was chosen, where the AAA codons were mutagenized to AAG codons read by having no Elongator dependent tRNA modification. In elp3/ sin3Δ background, expression of the protein encoded by the modified cdr2 gene improved significantly, suggesting that the expression was no longer Elongator dependent.32 With a similar strategy, in the S. pombe atf1+ gene AAA codons were changed to AAG codons and a similar result was obtained.33
Elongator Complex in Multicellular Eukaryotes
Orthologues of the Elongator complex proteins can be found in multicellular eukaryotes and 6 subunit protein complexes have been purified from humans and plants.3,79-85 Consistent with yeast, mutations in Elongator genes in the mouse M. musculus, the worm C. elegans, the plant A. thaliana and human cause defects in formation of wobble uridine modifications (Table 2).82,86-88 In vitro, Elongator complex purified from HeLa cells have histone H3 and H4 acetyltransferase activity and depletion of the human homolog hELP1, IKAP (I kappa B kinase complex associated protein) in fibroblasts, leads to hypoacetylation of histone H3, transcription impairment and cell migration defects.79,89,90 Mutations in genes encoding Elongator subunit proteins are associated to human diseases.91 Point mutations in the IKBKAP gene encoding IKAP cause familial dysautonomia (FD), a severe human hereditary neurodegenerative disorder.92,93 Levels of the mcm5s2U nucleoside in tRNA isolated from brain tissue and fibroblast cell lines were 64–71% in FD patients compared to non-FD individuals.88 As mice homozygous ikbkap-/- knockouts are embryonic lethal, a complete loss of modified wobble uridine nucleosides in FD patients was not expected. 94 Furthermore, a conditional inactivation of Ikbkap in mice leads to meiotic defects during spermatogenesis.86 Also observed in mice is that knockdown of the Elp1p, the Elp3p or the Elp4p homologues in oocytes impairs zygotic paternal genome demethylation.81 In the fruit fly D. melanogaster, the Elongator complex has been implicated in several processes such as, larval- and neuro-development.95,96 An explanation for the neurodevelopmental defects in D. melanogaster could be that Elongator complex acetylate Bruchpilot, a protein important for neuronal differentiation.97 Both in mice and C. elegans, the Elp3p homolog has been implicated in acetylating lysine 40 (K40) in α-tubulin.98,99 However, the C. elegans elpc-3 mutant did not have a defect in α-tubulin K40 acetylation.82 Later studies revealed MEC-17 and its paralogue ATAT-2 as the sole α-tubulin K40 acetyltransferases in C. elegans as single mec-17 and atat-2 mutants show reduced levels of α-tubulin K40 acetylation, whereas the double mec-17 and atat-2 mutants lack the K40 acetylation of α-tubulin.100,101 The elpc-1, elpc-3 and tuc-1 mutants cause defects in translation in C. elegans.82 ELPC-1::GFP and ELPC-3::GFP reporters are strongly expressed in a subset of chemosensory neurons required for salt chemotaxis learning.82 The elpc-1 or elpc-3 gene inactivation causes a defect in this process, associated with a posttranscriptional reduction of neuropeptide and a decreased accumulation of acetylcholine in the synaptic cleft. The elpc-1 and elpc-3 mutations are synthetic lethal with the tuc1 mutation at 25°C and double mutants display developmental defects.82 In the plant Arabidopsis thaliana, mutations in the ELP1, ELP3, ELP4, or KTI12 gene homologues cause cell proliferation defects.83,102
Conclusions and Perspectives
Independent of organism, Elongator mutants show very pleiotropic phenotypes. This has led to a debate whether Elongator complex has multiple functions or if its participation in one cellular process results in multiple downstream phenotypes. As Elp3 protein contains Radical SAM and HAT domains binding S-adenosylmethionine and acetyl-CoA, one theme is that Elongator complex is important for acetylation. Five substrates targeted for acetylation have been described, histones H3 and H4, α-tubulin, the neural differentiation protein Bruchpilot and lately a unique transfer of an acetyl radical to form cm5U34 in tRNA. In yeast there is convincing evidence that the only function of the Elongator complex is in formation of the intermediate cm5U in biosynthesis of ncm5U and mcm5U side chains present at wobble position in 11 out of 42 tRNA isoacceptors.73 This means that a substantial amount of the tRNA pool is missing wobble uridine modifications, implicating a defect in decoding during translation. All investigated phenotypes except the wobble uridine modification defect of Elongator mutants are efficiently suppressed by overexpressing tRNAs that in the wild type strain have the modified nucleoside mcm5s2U. This is an example of high copy suppression, i. e. when these tRNAs are overexpressed they compensate for the defect of the initial mutation. In this case higher levels of hypomodified tRNAs most likely compensate for the reduced codon/ anticodon interaction during translation due to lack of modifications at wobble position. Consistent with a Elongator tRNA dependent defect in translation, changing codons in a gene that are read by tRNAs containing modified wobble uridines to synonymous codons read by tRNAs not containing modified wobble uridines, restore protein expression, i.e. protein expression becomes Elongator independent.
In multicellular organisms, there has been no experiment where phenotypes observed in Elongator mutants have been suppressed by tRNA high copy suppression. However, as the role of Elongator in tRNA modification is conserved in eukaryotes it is likely that observed phenotypes are secondary to its primary function in tRNA modification.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Drs Marcus Johansson and Jan Larsson for comments on the manuscript.
Funding
A.S.B. is supported by grants from the Swedish Cancer Foundation (13 0301), Swedish Research Council (621-2012-3576) and Karin and Harald Silvanders Foundation (223-2808-12).
References
- 1. Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell 1999; 3:109-18; PMID:10024884; http://dx.doi.org/ 10.1016/S1097-2765(00)80179-3 [DOI] [PubMed] [Google Scholar]
- 2. Krogan NJ, Greenblatt JF. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol Cell Biol 2001; 21:8203-12; PMID:11689709; http://dx.doi.org/ 10.1128/MCB.21.23.8203-8212.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Winkler GS, Petrakis TG, Ethelberg S, Tokunaga M, Erdjument-Bromage H, Tempst P, Svejstrup JQ. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J Biol Chem 2001; 276:32743-9; PMID:11435442; http://dx.doi.org/ 10.1074/jbc.M105303200 [DOI] [PubMed] [Google Scholar]
- 4. Li Y, Takagi Y, Jiang Y, Tokunaga M, Erdjument-Bromage H, Tempst P, et al. A multiprotein complex that interacts with RNA polymerase II elongator. J Biol Chem 2001; 276:29628-31; PMID:11390369; http://dx.doi.org/ 10.1074/jbc.C100274200 [DOI] [PubMed] [Google Scholar]
- 5. Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell 1999; 4:123-8; PMID:10445034; http://dx.doi.org/ 10.1016/S1097-2765(00)80194-X [DOI] [PubMed] [Google Scholar]
- 6. Wittschieben BO, Fellows J, Du W, Stillman DJ, Svejstrup JQ. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J 2000; 19:3060-8; PMID:10856249; http://dx.doi.org/ 10.1093/emboj/19.12.3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci U S A 2002; 99:3517-22; PMID:11904415; http://dx.doi.org/ 10.1073/pnas.022042899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell 2002; 9:799-809; PMID:11983171; http://dx.doi.org/ 10.1016/S1097-2765(02)00502-6 [DOI] [PubMed] [Google Scholar]
- 9. Rahl PB, Chen CZ, Collins RN. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell 2005; 17:841-53; PMID:15780940; http://dx.doi.org/ 10.1016/j.molcel.2005.02.018 [DOI] [PubMed] [Google Scholar]
- 10. Salminen A, Novick PJ. A ras-like protein is required for a post-Golgi event in yeast secretion. Cell 1987; 49:527-38; PMID:3552249; http://dx.doi.org/ 10.1016/0092-8674(87)90455-7 [DOI] [PubMed] [Google Scholar]
- 11. Walch-Solimena C, Collins RN, Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J cell biol 1997; 137:1495-509; PMID:9199166; http://dx.doi.org/ 10.1083/jcb.137.7.1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Q, Fazly AM, Zhou H, Huang S, Zhang Z, Stillman B. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet 2009; 5:e1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res 2001; 29:1097-106; PMID:11222759; http://dx.doi.org/ 10.1093/nar/29.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Heyer WD, Thuriaux P, Kohli J, Ebert P, Kersten H, Gehrke C, Kuo KC, Agris PF. An antisuppressor mutation of Schizosaccharomyces pombe affects the post-transcriptional modification of the "wobble" base in the anticodon of tRNAs. J Biol Chem 1984; 259:2856-62; PMID:6559822 [PubMed] [Google Scholar]
- 15. Grossenbacher AM, Stadelmann B, Heyer WD, Thuriaux P, Kohli J, Smith C, et al. Antisuppressor mutations and sulfur-carrying nucleosides in transfer RNAs of Schizosaccharomyces pombe. J Biol Chem 1986; 261:16351-5; PMID:3782124 [PubMed] [Google Scholar]
- 16. Thuriaux P, Minet M, Hofer F, Leupold U. Genetic analysis of antisuppressor mutants in the fission yeast Schizosaccharomyces pombe. Mol G Genetics 1976; 142:251-61. [DOI] [PubMed] [Google Scholar]
- 17. Huang B, Johansson MJO, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005; 11:424-36; PMID:15769872; http://dx.doi.org/ 10.1261/rna.7247705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Esberg A, Huang B, Johansson MJ, Byström AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell 2006; 24:139-48; PMID:17018299; http://dx.doi.org/ 10.1016/j.molcel.2006.07.031 [DOI] [PubMed] [Google Scholar]
- 19. Tumaitis TD, Lane BG. Differential labelling of the carboxymethyl and methyl substituents of 5-carboxymethyluridine methyl ester, a trace nucleoside constituent of yeast transfer RNA. Biochim Biophys Acta 1970; 224:391-403; PMID:5498072; http://dx.doi.org/ 10.1016/0005-2787(70)90572-1 [DOI] [PubMed] [Google Scholar]
- 20. Kalhor HR, Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol 2003; 23:9283-92; PMID:14645538; http://dx.doi.org/ 10.1128/MCB.23.24.9283-9292.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mazauric MH, Dirick L, Purushothaman SK, Björk GR, Lapeyre B. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J Biol Chem 2010; 285:18505-15; PMID:20400505; http://dx.doi.org/ 10.1074/jbc.M110.113100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PØ, Klungland A. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol Cell Biol 2010; 30:1814-27; PMID:20123966; http://dx.doi.org/ 10.1128/MCB.01602-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen C, Huang B, Anderson JT, Byström AS. Unexpected accumulation of ncm(5)U and ncm(5)S(2) (U) in a trm9 mutant suggests an additional step in the synthesis of mcm(5)U and mcm(5)S(2)U. PloS one 2011; 6:e20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Selvadurai K, Wang P, Seimetz J, Huang RH. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat Chem Biol 2014; 10:810-2; In Press.; PMID:25151136; http://dx.doi.org/ 10.1038/nchembio.1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen C, Huang B, Eliasson M, Ryden P, Byström AS. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet 2011; 7:e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakai Y, Nakai M, Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem 2008; 283:27469-76; PMID:18664566; http://dx.doi.org/ 10.1074/jbc.M804043200 [DOI] [PubMed] [Google Scholar]
- 27. Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res 2009; 37:1335-52; PMID:19151091; http://dx.doi.org/ 10.1093/nar/gkn1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, et al. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 2009; 458:228-32; PMID:19145231; http://dx.doi.org/ 10.1038/nature07643 [DOI] [PubMed] [Google Scholar]
- 29. Durant PC, Bajji AC, Sundaram M, Kumar RK, Davis DR. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 2005; 44:8078-89; PMID:15924427; http://dx.doi.org/ 10.1021/bi050343f [DOI] [PubMed] [Google Scholar]
- 30. Vendeix FA, Murphy FVT, Cantara WA, Leszczynska G, Gustilo EM, Sproat B, Malkiewicz A, Agris PF. Human tRNA(Lys3)(UUU) is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J Mol Biol 2012; 416:467-85; PMID:22227389; http://dx.doi.org/ 10.1016/j.jmb.2011.12.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dewez M, Bauer F, Dieu M, Raes M, Vandenhaute J, Hermand D. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc Natl Acad Sci U S A 2008; 105:5459-64; PMID:18391219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bauer F, Matsuyama A, Candiracci J, Dieu M, Scheliga J, Wolf DA, Yoshida M, Hermand D. Translational control of cell division by Elongator. Cell reports 2012; 1:424-33; PMID:22768388; http://dx.doi.org/ 10.1016/j.celrep.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fernandez-Vazquez J, Vargas-Perez I, Sanso M, Buhne K, Carmona M, Paulo E, Hermand D, Rodríguez-Gabriel M, Ayté J, Leidel S, et al. Modification of tRNA(Lys) UUU by Elongator Is Essential for Efficient Translation of Stress mRNAs. PLoS Genet 2013; 9:e1003647; PMID:23874237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kishida M, Tokunaga M, Katayose Y, Yajima H, Kawamura-Watabe A, Hishinuma F. Isolation and genetic characterization of pGKL killer-insensitive mutants (iki) from Saccharomyces cerevisiae. Biosci Biotechnol Biochem 1996; 60:798-801; PMID:8704309; http://dx.doi.org/ 10.1271/bbb.60.798 [DOI] [PubMed] [Google Scholar]
- 35. Butler AR, White JH, Folawiyo Y, Edlin A, Gardiner D, Stark MJ. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol Cell Biol 1994; 14:6306-16; PMID:8065362; http://dx.doi.org/ 10.1128/MCB.14.9.6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J 2001; 20:1993-2003; PMID:11296232; http://dx.doi.org/ 10.1093/emboj/20.8.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Butler AR, Porter M, Stark MJ. Intracellular expression of Kluyveromyces lactis toxin gamma subunit mimics treatment with exogenous toxin and distinguishes two classes of toxin-resistant mutant. Yeast 1991; 7:617-25; PMID:1767590; http://dx.doi.org/ 10.1002/yea.320070610 [DOI] [PubMed] [Google Scholar]
- 38. Gunge N, Tamaru A, Ozawa F, Sakaguchi K. Isolation and characterization of linear deoxyribonucleic acid plasmids from Kluyveromyces lactis and the plasmid-associated killer character. J Bacteriol 1981; 145:382-90; PMID:6257636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Stark MJ, Boyd A. The killer toxin of Kluyveromyces lactis: characterization of the toxin subunits and identification of the genes which encode them. EMBO J 1986; 5:1995-2002; PMID:3758030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schaffrath R, Meinhardt F. Kluveromyces lactis zymocin and other plasmid-encoded yeast killer toxins. Topics Curr Genetics 2005; 11:133-55. [Google Scholar]
- 41. Sugisaki Y, Gunge N, Sakaguchi K, Yamasaki M, Tamura G. Kluyveromyces lactis killer toxin inhibits adenylate cyclase of sensitive yeast cells. Nature 1983; 304:464-6; PMID:6192345; http://dx.doi.org/ 10.1038/304464a0 [DOI] [PubMed] [Google Scholar]
- 42. White JH, Butler AR, Stark MJR. Kluyveromyces lactis toxin does not inhibit yeast adenylyl cyclase. Nature 1989; 341:666-8. [Google Scholar]
- 43. Lu J, Huang B, Esberg A, Johansson MJO, Byström AS. The Kluyveromyces lactis g-toxin targets tRNA anticodons. RNA 2005; 11:1648-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lu J, Esberg A, Huang B, Byström AS. Kluyveromyces lactis gamma-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res 2008; 36:1072-80; PMID:18096622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jablonowski D, Butler AR, Fichtner L, Gardiner D, Schaffrath R, Stark MJ. Sit4p protein phosphatase is required for sensitivity of Saccharomyces cerevisiae to Kluyveromyces lactis zymocin. Genetics 2001; 159:1479-89; PMID:11779790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang B, Lu J, Byström AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 2008; 14:2183-94; PMID:18755837; http://dx.doi.org/ 10.1261/rna.1184108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Glatt S, Létoquart J, Faux C, Taylor NM, Séraphin B, Müller CW. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat Struct Mol Biol 2012; 19:314-20; PMID:22343726; http://dx.doi.org/ 10.1038/nsmb.2234 [DOI] [PubMed] [Google Scholar]
- 48. Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Structu Biol 2011; 21:240-8; PMID:21282052; http://dx.doi.org/ 10.1016/j.sbi.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Di Santo R, Bandau S, Stark MJ. A conserved and essential basic region mediates tRNA binding to the Elp1 subunit of the Saccharomyces cerevisiae Elongator complex. Mol Microbiol 2014; 92:1227-42; PMID:24750273; http://dx.doi.org/ 10.1111/mmi.12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chinenov Y. A second catalytic domain in the Elp3 histone acetyltransferases: a candidate for histone demethylase activity? Trends Biochem Sci 2002; 27:115-7; PMID:11893502; http://dx.doi.org/ 10.1016/S0968-0004(02)02058-3 [DOI] [PubMed] [Google Scholar]
- 51. Paraskevopoulou C, Fairhurst SA, Lowe DJ, Brick P, Onesti S. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol 2006; 59:795-806; PMID:16420352; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04989.x [DOI] [PubMed] [Google Scholar]
- 52. Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol Cell Biol 2005; 25:4359-70; PMID:15899842; http://dx.doi.org/ 10.1128/MCB.25.11.4359-4370.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Leihne V, Kirpekar F, Vagbo CB, van den Born E, Krokan HE, Grini PE, Meza TJ, Falnes PØ. Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in Arabidopsis tRNA. Nucleic Acids Res 2011; 39:7688-701; PMID:21653555; http://dx.doi.org/ 10.1093/nar/gkr406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. White J, et al. , Mol. Cell. Biol. 2008; 28:10: 3151-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heurgue-Hamard V, Graille M, Scrima N, Ulryck N, Champ S, van Tilbeurgh H, Buckingham RH. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J Biol Chem 2006; 281:36140-8; PMID:17008308; http://dx.doi.org/ 10.1074/jbc.M608571200 [DOI] [PubMed] [Google Scholar]
- 56. Luke MM, Della Seta F, Di Como CJ, Sugimoto H, Kobayashi R, Arndt KT. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol Cell Biol 1996; 16:2744-55; PMID:8649382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sutton A, Immanuel D, Arndt KT. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol 1991; 11:2133-48; PMID:1848673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 2002; 415:180-3; PMID:11805837; http://dx.doi.org/ 10.1038/415180a [DOI] [PubMed] [Google Scholar]
- 59. DeMaggio AJ, Lindberg RA, Hunter T, Hoekstra MF. The budding yeast HRR25 gene product is a casein kinase I isoform. Proc Natl Acad Sci U S A 1992; 89:7008-12; PMID:1495994; http://dx.doi.org/ 10.1073/pnas.89.15.7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mehlgarten C, Jablonowski D, Breunig KD, Stark MJ, Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol Microbio 2009; 73:869-81; PMID:19656297; http://dx.doi.org/ 10.1111/j.1365-2958.2009.06811.x [DOI] [PubMed] [Google Scholar]
- 61. Jablonowski D, Fichtner L, Stark MJ, Schaffrath R. The yeast elongator histone acetylase requires sit4dependent dephosphorylation for toxin-target capacity. Mol Biol Cell 2004; 15:1459-69; PMID:14718557; http://dx.doi.org/ 10.1091/mbc.E03-10-0750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fichtner L, Jablonowski D, Schierhorn A, Kitamoto HK, Stark MJ, Schaffrath R. Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol 2003; 49:1297-307; PMID:12940988; http://dx.doi.org/ 10.1046/j.1365-2958.2003.03632.x [DOI] [PubMed] [Google Scholar]
- 63. Bar C, Zabel R, Liu S, Stark MJ, Schaffrath R. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. Mol Microbiol 2008; 69:1221-33; PMID:18627462 [DOI] [PubMed] [Google Scholar]
- 64. Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol 2004; 24:9487-97; PMID:15485916; http://dx.doi.org/ 10.1128/MCB.24.21.9487-9497.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dong M, Su X, Dzikovski B, Dando EE, Zhu X, Du J, Freed JH, Lin H. Dph3 is an electron donor for Dph1-Dph2 in the first step of eukaryotic diphthamide biosynthesis. J Am Chem Soc 2014; 136:1754-7; PMID:24422557; http://dx.doi.org/ 10.1021/ja4118957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zabel R, Bar C, Mehlgarten C, Schaffrath R. Yeast alpha-tubulin suppressor Ats1/Kti13 relates to the Elongator complex and interacts with Elongator partner protein Kti11. Mol Microbiol 2008; 69:175-87; PMID:18466297; http://dx.doi.org/ 10.1111/j.1365-2958.2008.06273.x [DOI] [PubMed] [Google Scholar]
- 67. Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 2010; 6:e1001247; PMID:21187895; http://dx.doi.org/ 10.1371/journal.pgen.1001247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yokoyama S, Nishimura S. Modified nucleosides and codon recognition. tRNA: Structure, Biosynthesis, and Function. Washington, DC: ASM Press, 1995:207-23. [Google Scholar]
- 69. Lim VI. Analysis of action of wobble nucleoside modifications on codon-anticodon pairing within the ribosome. J Molecular Biol 1994; 240:8-19; PMID:8021943 [DOI] [PubMed] [Google Scholar]
- 70. Hani J, Feldmann H. tRNA genes and retroelements in the yeast genome. Nucleic Acids Res 1998; 26:689-96; PMID:9443958; http://dx.doi.org/ 10.1093/nar/26.3.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Percudani R, Pavesi A, Ottonello S. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J Mol Biol 1997; 268:322-30; PMID:9159473; http://dx.doi.org/ 10.1006/jmbi.1997.0942 [DOI] [PubMed] [Google Scholar]
- 72. Johansson MJO, Byström AS. Transfer RNA modifications and modifying enzymes in Saccharomyces cerevisiae. In: Grosjean H, ed. Fine-tuning of RNA functions by modification and editing. New York: Springer-Verlag, 2005. [Google Scholar]
- 73. Johansson MJO, Esberg A, Huang B, Björk GR, Byström AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 2008; 28:3301-12; PMID:18332122; http://dx.doi.org/ 10.1128/MCB.01542-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell 2007; 28:860-70; PMID:18082610; http://dx.doi.org/ 10.1016/j.molcel.2007.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rezgui VA, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PG. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci U S A 2013; 110:12289-94; PMID:23836657; http://dx.doi.org/ 10.1073/pnas.1300781110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet 2014; 15:205-13; PMID:24468696 [DOI] [PubMed] [Google Scholar]
- 77. Zinshteyn B, Gilbert WV. Loss of a Conserved tRNA Anticodon Modification Perturbs Cellular Signaling. PLoS Genet 2013; 9:e1003675; PMID:23935536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotec 2006; 24:841-7; PMID:16823372; http://dx.doi.org/ 10.1038/nbt1222 [DOI] [PubMed] [Google Scholar]
- 79. Hawkes NA, Otero G, Winkler GS, Marshall N, Dahmus ME, Krappmann D, Scheidereit C, Thomas CL, Schiavo G, Erdjument-Bromage H, et al. Purification and characterization of the human elongator complex. J Biol Chem 2002; 277:3047-52; PMID:11714725; http://dx.doi.org/ 10.1074/jbc.M110445200 [DOI] [PubMed] [Google Scholar]
- 80. Close P, Gillard M, Ladang A, Jiang Z, Papuga J, Hawkes N, Nguyen L, Chapelle JP, Bouillenne F, Svejstrup J, et al. DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator. J Biol Chem 2012; 287:32535-45; PMID:22854966; http://dx.doi.org/ 10.1074/jbc.M112.402727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Okada Y, Yamagata K, Hong K, Wakayama T, Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature 2010; 463:554-8; PMID:20054296; http://dx.doi.org/ 10.1038/nature08732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chen C, Tuck S, Byström AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet 2009; 5:e1000561; PMID:19593383; http://dx.doi.org/ 10.1371/journal.pgen.1000561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nelissen H, Fleury D, Bruno L, Robles P, De Veylder L, Traas J, Micol JL, Van Montagu M, Inzé D, Van Lijsebettens M. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci U S A 2005; 102:7754-9; PMID:15894610; http://dx.doi.org/ 10.1073/pnas.0502600102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nelissen H, De Groeve S, Fleury D, Neyt P, Bruno L, Bitonti MB, Vandenbussche F, Van der Straeten D, Yamaguchi T, Tsukaya H, et al. Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc Natl Acad Sci U S A 2010; 107:1678-83; PMID:20080602; http://dx.doi.org/ 10.1073/pnas.0913559107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Simpson CL, Lemmens R, Miskiewicz K, Broom WJ, Hansen VK, van Vught PW, Landers JE, Sapp P, Van Den Bosch L, Knight J, et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet 2009; 18:472-81; PMID:18996918; http://dx.doi.org/ 10.1093/hmg/ddn375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lin FJ, Shen L, Jang CW, Falnes PO, Zhang Y. Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet 2013; 9:e1003516; PMID:23717213; http://dx.doi.org/ 10.1371/journal.pgen.1003516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mehlgarten C, Jablonowski D, Wrackmeyer U, Tschitschmann S, Sondermann D, Jager G, Gong Z, Byström AS, Schaffrath R, Breunig KD. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol 2010; 76:1082-94; PMID:20398216; http://dx.doi.org/ 10.1111/j.1365-2958.2010.07163.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Karlsborn T, Tükenmez H, Chen C, Byström AS. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm5s2U in tRNA. Biochem Biophys Res Commun 2014. 454:441-5; in Press; PMID:25450681 [DOI] [PubMed] [Google Scholar]
- 89. Kim JH, Lane WS, Reinberg D. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc Natl Acad Sci U S A 2002; 99:1241-6; PMID:11818576; http://dx.doi.org/ 10.1073/pnas.251672198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Close P, Hawkes N, Cornez I, Creppe C, Lambert CA, Rogister B, Siebenlist U, Merville MP, Slaugenhaupt SA, Bours V, et al. Transcription impairment and cell migration defects in elongator-depleted cells: implication for familial dysautonomia. Mol Cell 2006; 22:521-31; PMID:16713582; http://dx.doi.org/ 10.1016/j.molcel.2006.04.017 [DOI] [PubMed] [Google Scholar]
- 91. Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trend Mol Med 2014; PMID:24581449 [DOI] [PubMed] [Google Scholar]
- 92. Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, Ekstein J, Rubin BY. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet 2001; 68:753-8; PMID:11179021; http://dx.doi.org/ 10.1086/318808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, Liebert CB, Chadwick B, Idelson M, Reznik L, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet 2001; 68:598-605; PMID:11179008; http://dx.doi.org/ 10.1086/318810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chen YT, Hims MM, Shetty RS, Mull J, Liu L, Leyne M, Slaugenhaupt SA. Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol Cell Biol 2009; 29:736-44; PMID:19015235; http://dx.doi.org/ 10.1128/MCB.01313-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Walker J, Kwon SY, Badenhorst P, East P, McNeill H, Svejstrup JQ. Role of elongator subunit Elp3 in Drosophila melanogaster larval development and immunity. Genetics 2011; 187:1067-75; PMID:21288872; http://dx.doi.org/ 10.1534/genetics.110.123893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Singh N, Lorbeck MT, Zervos A, Zimmerman J, Elefant F. The histone acetyltransferase Elp3 plays in active role in the control of synaptic bouton expansion and sleep in Drosophila. J Neurochem 2010; 115:493-504; PMID:20626565; http://dx.doi.org/ 10.1111/j.1471-4159.2010.06892.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Miskiewicz K, Jose LE, Bento-Abreu A, Fislage M, Taes I, Kasprowicz J, Swerts J, Sigrist S, Versées W, Robberecht W, et al. ELP3 controls active zone morphology by acetylating the ELKS family member Bruchpilot. Neuron 2011; 72:776-88; PMID:22153374; http://dx.doi.org/ 10.1016/j.neuron.2011.10.010 [DOI] [PubMed] [Google Scholar]
- 98. Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 2009; 136:551-64; PMID:19185337; http://dx.doi.org/ 10.1016/j.cell.2008.11.043 [DOI] [PubMed] [Google Scholar]
- 99. Solinger JA, Paolinelli R, Kloss H, Scorza FB, Marchesi S, Sauder U, Mitsushima D, Capuani F, Stürzenbaum SR, Cassata G. The Caenorhabditis elegans Elongator complex regulates neuronal alpha-tubulin acetylation. PLoS Genet; 6:e1000820; PMID:20107598; http://dx.doi.org/ 10.1371/journal.pgen.1000820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci U S A 2010; 107:21517-22; PMID:21068373; http://dx.doi.org/ 10.1073/pnas.1013728107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. MEC-17 is an alpha-tubulin acetyltransferase. Nature 2010; 467:218-22; PMID:20829795; http://dx.doi.org/ 10.1038/nature09324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Nelissen H, Clarke JH, De Block M, De Block S, Vanderhaeghen R, Zielinski RE, Dyer T, Lust S, Inzé D, Van Lijsebettens M. DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 2003; 15:639-54; PMID:12615938; http://dx.doi.org/ 10.1105/tpc.007062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 2009; 37:D159-62; PMID:18957446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kim S, Johnson W, Chen C, Sewell AK, Byström AS, Han M. Allele specific suppressors of lin-1(R175Opal) identify functions of MOC-3 and DPH-3 in tRNA modification complexes in caenorhabditis elegans. Genetics 2010; 185:1235-47; PMID:20479142; http://dx.doi.org/ 10.1534/genetics.110.118406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nakai Y, Umeda N, Suzuki T, Nakai M, Hayashi H, Watanabe K, Kagamiyama H. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J Biol Chem 2004; 279:12363-8; PMID:14722066; http://dx.doi.org/ 10.1074/jbc.M312448200 [DOI] [PubMed] [Google Scholar]
- 106. Nakai Y, Nakai M, Lill R, Suzuki T, Hayashi H. Thio modification of yeast cytosolic tRNA is an iron-sulfur protein-dependent pathway. Mol Cell Biol 2007; 27:2841-7; PMID:17283054; http://dx.doi.org/ 10.1128/MCB.01321-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Björk GR, Huang B, Persson OP, Byström AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 2007; 13:1245-55; PMID:17592039; http://dx.doi.org/ 10.1261/rna.558707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Philipp M, John F, Ringli C. The cytosolic thiouridylase CTU2 of Arabidopsis thaliana is essential for posttranscriptional thiolation of tRNAs and influences root development. BMC Plant Biol 2014; 14:109; PMID:24774365; http://dx.doi.org/ 10.1186/1471-2229-14-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. van den Born E, Vagbo CB, Songe-Moller L, Leihne V, Lien GF, Leszczynska G, Malkiewicz A, Krokan HE, Kirpekar F, Klungland A, et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun 2011; 2:172; PMID:21285950; http://dx.doi.org/ 10.1038/ncomms1173 [DOI] [PubMed] [Google Scholar]


