Abstract
Probiotics are live microorganisms that, when administered in sufficient doses, provide health benefits on the host. The United States Food and Drug Administration (FDA) requires phase I safety studies for probiotics when the intended use of the product is as a drug. The purpose of the study was to determine the safety of Bifidobacterium animalis subsp lactis (B. lactis) strain BB-12 (BB-12)-supplemented yogurt when consumed by a generally healthy group of adults who were prescribed a 10-day course of antibiotics for a respiratory infection. Secondary aims were to assess the ability of BB-12 to affect the expression of whole blood immune markers associated with cell activation and inflammatory response. A phase I, double-blinded, randomized controlled study was conducted in compliance with FDA guidelines for an Investigational New Drug (IND). Forty participants were randomly assigned to consume 4 ounces of either BB-12 -supplemented yogurt or non-supplemented control yogurt daily for 10 d. The primary outcome was to assess safety and tolerability, assessed by the number of reported adverse events. A total of 165 non-serious adverse events were reported, with no differences between the control and BB-12 groups. When compared to the control group, B lactis fecal levels were modestly higher in the BB-12-supplemented group. In a small subset of patients, changes in whole blood expression of genes associated with regulation and activation of immune cells were detected in the BB-12-supplemented group. BB-12-supplemented yogurt is safe and well tolerated when consumed by healthy adults concurrently taking antibiotics. This study will form the basis for future randomized clinical trials investigating the potential immunomodulatory effects of BB-12-supplemented yogurt in a variety of disease states.
Keywords: antibiotics, clinical trial, gut microbiota, probiotics, safety
Abbreviations
- BB-12
Bifidobacterium animalis subsp lactis strain BB-12
- IND
investigational new drug
- FDA
Food and Drug Administration
- CBER
Center for Biologics Evaluation and Research
- NIH
National Institutes of Health
- NCCAM
National Center for Complementary and Alternative Medicine
Introduction
Probiotics are live microorganisms which, when administered in sufficient amounts, may improve health.1 Probiotics have shown potential benefits in the treatment and prevention of varied diseases, including diarrhea, asthma, necrotizing enterocolitis, respiratory infections and allergies.2-5
The gastrointestinal tract contains a complex commensal microbiota that contributes to homeostasis of the gut. Probiotics may help regulate the microbiota of a disrupted gastrointestinal tract.6 The balance of the resident microbiota can be disturbed by medical interventions such as antibiotics, resulting in, among other effects, decreased short chain fatty acid metabolism with accumulation of luminal carbohydrate, subsequent pH changes, and water absorption.7 While many studies have examined the role of probiotics for the prevention of antibiotic-associated diarrhea, the majority of these studies were conducted outside the United States and none were conducted under Investigational New Drug (IND) regulatory policies of the Food and Drug Administration, Center for Biologics Evaluation and Research (FDA/CBER).8
Probiotics marketed as nutritional supplements or found in functional foods are principally members of the genera Bifidobacterium and Lactobacillus. Bifidobacterium species, particularly Bifidobacterium animalis subsp lactis (B. lactis), strain BB-12 (BB-12), the principal focus of this study, can be found in the gastrointestinal tract as both autochthonous (indigenous) and allochthonous (derived from outside a system) residents.9 Newborns, especially those that are breast-fed, are colonized with bifidobacteria within days after birth. Once the infant is weaned, the population of this genus in the colon appears to be relatively stable until advanced age when it appears to decline.9,10
This research was done to determine the safety of strawberry-flavored yogurt supplemented with the strain BB-12 and to determine the effect of probiotic treatment on whole blood cell expression of inflammatory response-associated genes. This trial serves as the first in a two-stage process to establish the safety profile of a BB-12-supplemented yogurt drink in adults and children. The long-term objective is to obtain the necessary FDA approval to proceed to an efficacy study to determine how BB-12-supplemented yogurt may impact human health.
Results
Recruitment, enrollment and participant flow
Two hundred and 11 individuals were screened for eligibility and 40 participants were enrolled in the study (Fig. 1). Nineteen participants were in the BB-12 group and 21 participants were in the control group (Fig. 1). There were 8 participants who self-withdrew or were withdrawn early by the investigator from the study. All forty participants were included in the analyses as per the intention-to-treat principle, and all participants were included for assessment of stool samples. Fifteen of the 40 participants consented to participate in the sub-study to evaluate gene expression in whole blood cells: 6 participants in the BB-12 group and 9 participants in the control group.
Figure 1.
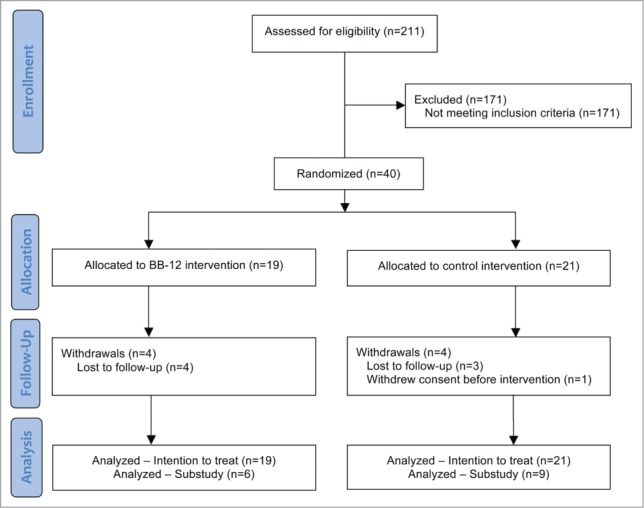
Flow diagram of study participation.
Baseline health and demographics
There were no significant differences in any of the demographic or baseline health characteristics between the BB-12 and control groups (Table 1). All participants were on antibiotics. The most common diagnosis resulting in the antibiotic prescription was strep throat (42% in the BB-12 group and 38% in the control group). The other diagnoses were sinusitis, otitis media, cold, bacterial infection, throat infection and pneumonia.
Table 1.
Participant demographics and baseline health characteristics
| BB-12 N (%) | Control N (%) | p-value | |
|---|---|---|---|
| Number of participants | 19 | 21 | |
| Gender (Female) | 12 (63) | 12 (57) | 0.70 |
| Gender (Male) | 7 (37) | 9 (43) | |
| Age, Mean (SD) | 33 (19) | 29 (12) | 0.43 |
| Does anyone in the household smoke (Yes) | 4 (21) | 4 (19) | 0.59 |
| Marital Status | 0.78 | ||
| Divorced | 1 (5) | 3 (15) | |
| Married | 7 (33) | 5 (26) | |
| Partner | 3 (14) | 3 (16) | |
| Single | 10 (48) | 8 (42) | |
| Race | 1.00 | ||
| Asian | 1 (5) | 0 (0) | |
| Black or African American | 1 (5) | 1 (5) | |
| Other | 6 (29) | 5 (26) | |
| White | 13 (62) | 13 (69) | |
| Hispanic (Yes) | 9 (47) | 6 (29) | 0.18 |
| Education | 0.62 | ||
| High School or less | 7 (37) | 5 (24) | |
| Some College or Associate | 7 (37) | 8 (38) | |
| College or more | 5 (26) | 8 (38) | |
| Health Insurance (Yes)a | 17 (90) | 19 (91) | 0.66 |
| Income Level (N = 23) | 1.00 | ||
| Less than $30K | 4 (33) | 3 (27) | |
| $75K-$150K | 6 (50) | 7 (64) | |
| Above $150K | 2 (17) | 1 (9) | |
| Have you ever heard of probiotics or active cultures (Yes) | 11 (58) | 13 (62) | 0.53 |
| Interview conducted in | 0.29 | ||
| English | 13 (68) | 17 (81) | |
| Spanish | 6 (32) | 4 (19) | |
| Diagnosis/Reason for antibiotic prescription | 0.31 | ||
| Strep throat | 8 (42) | 8 (38) | |
| Otitis media (ear infection) | 4 (21) | 1 (5) | |
| Sinusitis (sinus infection) | 4 (21) | 10 (48) | |
| Cold | 1 (5) | 1 (5) | |
| Other (bacterial infection, throat infection, pneumonia) | 2 (11) | 1 (5) | |
| Antibiotic prescribed (N = 38) | 0.63 | ||
| Amoxicillin | 12 (67) | 11 (55) | |
| Augmentin | 1 (6) | 3 (15) | |
| Pen-Vee K | 4 (22) | 6 (30) | |
| Zinacef (Cefuroxime) | 1 (5) | 0 (0) | |
| Height (inches) Mean (SD) | 66.6 (3.4) (N = 17) | 67.4 (4.5) (N = 18) | 0.56* |
| Weight (pounds) Mean (SD) | 162 (36) | 161 (38) | 0.93* |
| Pulse Oxygen Mean (SD) | 98.4 (0.8) (N = 18) | 98.4 (1.0) (N = 19) | 1.00* |
2-sample t-test.
Bivariate analyses for this variable was conducted using Fisher's exact test due to small number of frequencies in the cells. P-values reflect one-sided significance levels.
Interventions
Viable counts of BB-12 and pH of the probiotic containing and control yogurt-based drinkable products were evaluated over the 30 d the products were stored at 4°C. All product information has been previously described elsewhere.11 Data demonstrated a stable, viable BB-12 population in the test product and less than 100 colony forming units/gram of BB-12 in the control product. The pH of the drinkable products remained constant over time. The maintenance of viability and constant pH of the drinkable products indicated that the product was stable throughout the experiment.11
Compliance
The self-reported number of yogurt beverages consumed over the 10-day intervention was statistically similar between the groups: 9.4 total yogurt drinks in BB-12 group and 9.8 total yogurt drinks in control group. The PCR results for day 7 fecal samples show that overall, 66.7% of the participants were compliant (out of 39 participants who initiated the intervention); of the participants in the treated group, 63.2% tested positive for B. lactis and of the participants in the control group, 70.0% tested negative for B. lactis. Blinding worked appropriately as, when surveyed at the end of the intervention period as to which yogurt beverage the participant believed he or she consumed, 50% of participants (out of 28 respondents) correctly guessed their assignment.
Primary outcome
A total of 165 adverse events were reported in this study (Table 2). There were 98 adverse events reported in the control group and 67 adverse events reported in the BB-12 intervention group. There were also no reported allergic reactions or hypersensitivity to the yogurts. No serious adverse events were reported and no participant deaths occurred. There were no participant withdrawals from the study due to adverse events.
Table 2.
Adverse events1
| Events/Outcomes | BB-12 group N = 19 | Control group N = 21 | ||
|---|---|---|---|---|
| N | % | N | % | |
| Abdominal pain | 0 | 0 | 1 | 5 |
| Acid reflux | 3 | 16 | 0 | 0 |
| Allergies (seasonal, allergic rhinitis) | 0 | 0 | 1 | 5 |
| Back pain | 1 | 5 | 0 | 0 |
| Bloating | 0 | 0 | 3 | 14 |
| Bowel sounds | 0 | 0 | 1 | 5 |
| Breathing problems | 0 | 0 | 3 | 14 |
| Constipation | 2 | 11 | 3 | 14 |
| Cough | 6 | 32 | 8 | 38 |
| Decreased appetite | 5 | 26 | 7 | 33 |
| Diarrhea | 2 | 11 | 2 | 10 |
| Dizziness | 1 | 5 | 0 | 0 |
| Drug hypersensitivity | 0 | 0 | 1 | 5 |
| Ear aches | 3 | 16 | 5 | 24 |
| Fever | 2 | 11 | 2 | 10 |
| Gas | 1 | 5 | 6 | 29 |
| Headache | 5 | 26 | 7 | 33 |
| Irritability | 1 | 5 | 1 | 5 |
| Lethargy | 4 | 21 | 8 | 38 |
| Loose stool | 4 | 21 | 9 | 43 |
| Muscle pain | 1 | 5 | 0 | 0 |
| Nasal congestion | 9 | 47 | 7 | 33 |
| Nausea | 0 | 0 | 1 | 5 |
| Runny nose | 5 | 26 | 7 | 33 |
| Sore throat | 6 | 32 | 5 | 24 |
| Stomach pain | 3 | 16 | 8 | 38 |
| Tonsil swelling | 0 | 0 | 1 | 5 |
| Vaginal discomfort | 0 | 0 | 1 | 5 |
| Vomiting | 1 | 5 | 0 | 0 |
| Yeast infection | 2 | 11 | 0 | 0 |
| Total events reported | 67 | 98 | ||
All adverse events occurred during the intervention or closely after; none were reported at day 180.
Secondary outcomes
There was a near significant difference in the number of total stools over the intervention period between the groups: 12.7 stools in the BB-12 group (n = 10) and 19.2 stools in the control group (n = 12), P = 0.06.
Microbiota composition among treatment groups
DNA from stool samples collected at baseline, days 3, 7, 10 and 28 was isolated and used as a template to determine bacterial species abundance. Copy numbers for total bacteria (Eubacteria) were not affected by probiotic treatment throughout the experiment. Similarly B. fragilis spp., Bifidobacterium spp and Lactobacillus spp. abundance did not change after consumption of yogurt with or without B. lactis BB-12. However, relative abundance of B. lactis in the stool was marginally increased in stools from participants consuming the yogurt with B. lactis BB-12 with a higher abundance at day 10 (P = 0.071) and at day 28 (P = 0.1) when compared with the control group (Fig. 2). Pairwise comparisons within treatment groups indicated a significant increase in B. lactis abundance at day 3 when compared to day 0 (P < 0.05), with non-significant changes detected at other times or within the non-supplemented yogurt group (Table 3).
Figure 2.
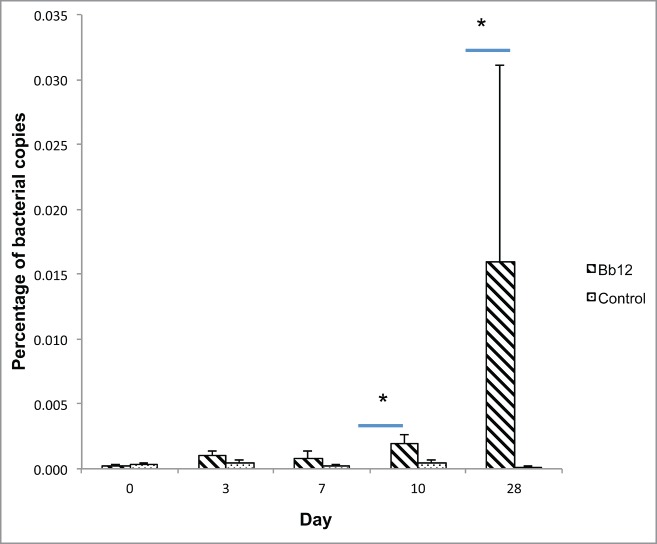
Relative abundance of Bifidobacterium lactis in patients after intervention. Abundance of B. lactis is expressed as percentage of total bacteria. Stripped bars represent copies from B. lactis treated patients. Dotted filled bars represent copies from control patients. * Denotes significant differences among groups (P ≤ 0.1).
Table 3.
Bacterial abundance in stools of participants consuming yogurts with or without BB-12 supplementation
| p-value Pairwise Comparison |
p-value Repeated Measures |
|||||||
|---|---|---|---|---|---|---|---|---|
| Bacteria | Day | BB-12 | Control | Treatment | Day | Treatment *Day | ||
| Eubacteria | 0 | 1.28 × 1012 ± 0.54* | 0.92 × 1012 ± 0.49 | 0.9063 | 0.5609 | 0.1942 | 0.7368 | |
| 3 | 1.18 × 1012 ± 0.45 | 1.36 × 1012 ± 0.70 | 0.6373 | |||||
| 7 | 1.12 × 1012 ± 0.40 | 1.49 × 1012 ± 0.51 | 0.2053 | |||||
| 10 | 0.53 × 1012 ± 0.16 | 1.49 × 1012 ± 0.52 | 0.6804 | |||||
| 28 | 1.07 × 1012 ± 0.60 | 1.17 × 1012 ± 0.66 | 0.8975 | |||||
| Bacteroides fragilis spp | 0 | 6.58 × 1011 ± 3.73 (51.4 ± 29.1) | 1.47 × 1011 ± 0.59 (16.0 ± 6.4) | 0.8206 | 0.572 | 0.1986 | 0.7776 | |
| 3 | 5.08 × 1011 ± 2.84 (43.1 ± 24.1) | 5.19 × 1011 ± 2.07 (38.2 ± 15.2) | 0.8898 | |||||
| 7 | 3.78 × 1011 ± 1.61 (33.8 ± 14.4) | 8.39 × 1011 ± 5.10 (56.3 ± 34.2) | 0.3895 | |||||
| 10 | 1.50 × 1011 ± 0.63 (28.3 ± 11.9) | 4.26 × 1011 ± 1.54 (28.6 ± 10.3) | 0.3441 | |||||
| 28 | 2.16 × 1011 ± 1.31 (20.2 ± 12.2) | 2.28 × 1011 ± 0.85 (19.5 ± 7.3) | 0.3967 | |||||
| Lactobacillus spp. | 0 | 2.00 × 106 ± 1.32 (0.0002 ± 0.0001) | 1.63 × 106 ± 0.86 (0.0002 ± 0.0001) | 0.63 | 0.9213 | 0.5583 | 0.9801 | |
| 3 | 1.20 × 106 ± 0.36 (0.0001 ± 0.0000) | 1.28 × 106 ± 0.46 (0.0001 ± 0.0000) | 0.9396 | |||||
| 7 | 1.26 × 106 ± 0.44 (0.0001 ± 0.0000) | 4.16 × 106 ± 2.41 (0.0003 ± 0.0002) | 0.7249 | |||||
| 10 | 0.99 × 106 ± 0.45 (0.0002 ± 0.0001) | 0.93 × 106 ± 0.45 (0.0001 ± 0.0000) | 0.9877 | |||||
| 28 | 0.65 × 106 ± 0.27 (0.0001 ± 0.0000) | 4.88 × 106 ± 3.97 (0.0004 ± 0.0003) | 0.9002 | |||||
| Bifidobacterium spp | 0 | 0.94 × 109 ± 0.49 (0.0734 ± 0 .0383) | 0.58 × 109 ± 0.27 (0.0630 ± 0.0293) | 0.9401 | 0.7327 | 0.247 | 0.5926 | |
| 3 | 0.59 × 109 ± 0.49 (0.0500 ± 0.0415) | 0.38 × 109 ± 0.15 (0.0279 ± 0.0110) | 0.3428 | |||||
| 7 | 0.51 × 109 ± 0.20 (0.0455 ± 0.0179) | 0.85 × 109 ± 0.32 (0.0570 ± 0.0215) | 0.6979 | |||||
| 10 | 1.19 × 109 ± 0.97 (0.2245 ± 0.1830) | 1.65 × 109 ± 0.73 (0.1107 ± 0.0490) | 0.6869 | |||||
| 28 | 4.54 × 109 ± 4.42 (0.4243 ± 0.4131) | 0.70 × 109 ± 0.31 (0.0598 ± 0.0265) | 0.2872 | |||||
| Bifidobacterium lactis | 0 | 0.30 × 107 ± 0.13 (0.0002 ± 0.0001)a | 0.28 × 107 ± 0.12 (0.0003 ± 0.0001)a | 0.5339 | 0.122 | 0.0422 | 0.5525 | |
| 3 | 1.21 × 107 ± 0.43 (0.0010 ± 0.0004)b | 0.56 × 107 ± 0.30 (0.0004 ± 0.0002)a | 0.1584 | |||||
| 7 | 0.93 × 107 ± 0.54 (0.0008 ± 0.0005)a | 0.35 × 107 ± 0.12 (0.0002 ± 0.0001)a | 0.6132 | |||||
| 10 | 1.00 × 107 ± 0.39 (0.0019 ± 0.0007)ab | 0.67 × 107 ± 0.38 (0.0004 ± 0.0003)a | 0.0706 | |||||
| 28 | 1.71 × 108 ± 1.62 (0.0160 ± 0.0151)ab | 0.13 × 107 ± 0.07 (0.0001 ± 0.0001)a | 0.1017 | |||||
Data represents bacterial abundance expressed as copies per gram of feces or as percentage for each bacterial species in parenthesis. P-values represent treatment pairwise comparison at each separate day or the overall treatment, day or treatment*day interaction modeling bacterial abundance counts with lognormal distribution in a repeated measures ANOVA. Pairwise comparison were also done within treatment groups for B. lactis species, any non-identical letters indicate significant differences among group means from different times (p < 0 .05).
Gene expression in whole blood cells among treatment groups
RNA from whole blood samples collected at baseline, day 7 and 14 were isolated and converted to cDNA to be used as a template for measuring gene expression of a selected group of genes associated with cell activation and inflammatory response. Consumption of yogurt supplemented with B. lactis BB-12 for 10 d induced an increase in transcription factor, Interferon Regulatory Factor 8 (IRF-8) which regulates expression of genes stimulated by type I interferons (IFNs), Toll like receptor-2 (TLR2) involved in antigen recognition, and tumor necrosis factor receptor superfamily member 14 (TNFSF14), a gene which encodes a protein from the TNF-receptor superfamily involved in mediating signal transduction pathways that activate the immune response (P < 0.05). Treatment X day interaction effects in response to BB-12 treatment were detected for GATA3 expression (P = 0.0134) a transcription factor involved in the regulation of T-cell development, and mildly for CXCL10 (P = 0.09) a chemokine involved in early stimulation and cell recruitment.
Most of the BB-12 induced gene expression changes compared to the control group were seen at day 14. Levels of transcription factor GATA3; CD80, an early inducer of T-cell proliferation; CXCL10, and pro-inflammatory cytokine tumor necrosis factor alpha (TNFA) were upregulated at least five-fold in blood cells isolated from participants receiving B. lactis when compared to participants receiving the control (P < 0.05). Changes in transcription levels for other genes associated with early cell activation (CD40), immune regulation (forkhead box P3 (FOXP3), tumor growth factor beta-2 (TGFB2)) and co-stimulatory signal (TNFSF14) were also observed at day 14 (P < 0.1); but with a higher variability as these changes were detected at α = 0.10. With the exception of a mild BB-12-induced threefold increase in chemokine CCL4 expression at day 7 (P = 0.09), no other changes in gene transcription levels were seen at baseline or day 7 (Table 4).
Table 4.
Gene expression levels in whole blood after 10 d post-antibiotic treatment with probiotic BB-12
| Gene | Treatment | Baseline | Collection time Day 7 | Day 14 | Treatment | P-value*** Day | Treatment X day interaction | |
|---|---|---|---|---|---|---|---|---|
| Transcription factors | ||||||||
| TBX21 | Control | ΔCT* | 7.40 ± 0.42 a | 7.38 ± 0.81 a | 7.63 ± 0.55 a | 0.9519 | 0.4135 | 0.1698 |
| BB-12 | ΔCT | 7.61 ± 0.46a | 8.08 ± 0.83 a | 6.57 ± 0.56 a | ||||
| FC** | −1.16 | −1.62 | 2.08 | |||||
| GATA3 | Control | ΔCT | 9.85 ± 0.59 a | 10.44. ± 0.59 a | 10.83 ± 0.59 a | 0.2427 | 0.7478 | 0.0134 |
| BB-12 | ΔCT | 10.00 ± 0.66 a | 9.57 ± 0.69 a | 8.63 ± 0.66 b | ||||
| FC | −1.11 | 1.83 | 4.59 | |||||
| FOXP3 | Control | ΔCT | 9.64 ± 0.73 a | 9.04 ± 0.60 a | 11.14 ± 0.84 a | 0.1432 | 0.3712 | 0.296 |
| BB-12 | ΔCT | 8.82 ± 0.73 a | 9.26 ± 0.73 a | 9.07 ± 0.65 c | ||||
| FC | 1.77 | −1.16 | 4.2 | |||||
| IRF8 | Control | ΔCT | 8.15 ± 1.16 a | 6.83 ± 1.06 a | 7.81 ± 1.40 a | 0.044 | 0.4523 | 0.7055 |
| BB-12 | ΔCT | 7.26 ± 1.30 a | 5.32 ± 1.28 a | 4.88 ± 1.39 a | ||||
| FC | 1.85 | 2.85 | 7.62 | |||||
| Toll like receptors | ||||||||
| TLR2 | Control | ΔCT | 8.00 ± 1.06 a | 6.79 ± 0.98 a | 7.63 ± 1.30 a | 0.0406 | 0.7529 | 0.7483 |
| BB-12 | ΔCT | 5.88 ± 1.16 a | 5.67 ± 1.16 a | 4.71 ± 1.30 a | ||||
| FC | 4.35 | 2.17 | 7.57 | |||||
| TLR4 | Control | ΔCT | 3.72 ± 1.16 a | 3.42 ± 1.49 a | 6.79 ± 1.83 a | 0.632 | 0.505 | 0.1559 |
| BB-12 | ΔCT | 2.85 ± 1.83 a | 6.00 ± 1.16 a | 3.26 ± 1.49 a | ||||
| FC | 1.83 | −5.98 | 11.55 | |||||
| TLR9 | Control | ΔCT | 9.06 ± 1.14 a | 7.59 ± 1.06 a | 8.72 ± 1.40 a | 0.1093 | 0.6304 | 0.7326 |
| BB-12 | ΔCT | 7.50 ± 1.25 a | 6.83 ± 1.25 a | 5.93 ± 1.40 a | ||||
| FC | 2.95 | 1.69 | 6.92 | |||||
| Inflammation | ||||||||
| TNFA | Control | ΔCT | 10.10 ± 0.51 a | 10.91 ± 0.94 a | 11.34 ± 0.63 a | 0.1645 | 0.5652 | 0.0987 |
| BB-12 | ΔCT | 9.62 ± 0.55 a | 9.90 ± 1.03 a | 9.04 ± 0.65 b | ||||
| FC | 1.39 | 2.01 | 4.92 | |||||
| IFNG | Control | ΔCT | 10.52 ± 0.98 a | 10.57 ± 1.27 a | 11.23 ± 1.55 a | 0.2753 | 0.9791 | 0.7392 |
| BB-12 | ΔCT | 12.44 ± 1.55 a | 12.31 ± 1.10 a | 11.21 ± 1.27 a | ||||
| FC | −3.78 | −3.34 | 1.01 | |||||
| IL1B | Control | ΔCT | 6.87 ± 0.72 a | 8.24 ± 0.76 a | 8.39 ± 0.72 a | 0.4639 | 0.0614 | 0.4332 |
| BB-12 | ΔCT | 6.60 ± 0.77 a | 8.05 ± 0.78 a | 6.92 ± 0.78 a | ||||
| FC | 1.2 | 1.14 | 2.77 | |||||
| IL6 | Control | ΔCT | 11.94 ± 1.12 a | 12.47 ± 1.12 a | 13.25 ± 1.29 a | 0.4076 | 0.9101 | 0.763 |
| BB-12 | ΔCT | 11.77 ± 1.00 a | 12.02 ± 1.29 a | 11.38 ± 1.29 a | ||||
| FC | 1.13 | 1.37 | 3.65 | |||||
| Regulatory | ||||||||
| TGFB2 | Control | ΔCT | 4.93 ± 0.78 a | 4.71 ± 0.85 a | 6.04 ± 0.78 a | 0.4531 | 0.7489 | 0.1981 |
| BB-12 | ΔCT | 4.32 ± 0.85 a | 5.76 ± 0.85 a | 4.08 ± 0.78 c | ||||
| FC | 1.53 | −2.07 | 3.89 | |||||
| IL10 | Control | ΔCT | 10.76 ± 0.88 a | 10.53 ± 0.88 a | 12.07 ± 1.46 a | 0.3251 | 0.5466 | 0.4826 |
| BB-12 | ΔCT | 10.53 ± 0.88 a | 10.94 ± 1.44 a | 7.43 ± 2.51a | ||||
| FC | 1.17 | 2.19 | 26.72 | |||||
| Cell Activation | ||||||||
| CD40 | Control | ΔCT | 7.05 ± 0.88a | 8.37 ± 1.96a | 8.72 ± 0.92a | 0.4578 | 0.504 | 0.4227 |
| BB-12 | ΔCT | 6.60 ± 0.87a | 8.44 ± 1.94a | 6.13 ± 0.79c | ||||
| FC | 1.37 | −1.05 | 6.02 | |||||
| HLA-DRA | Control | ΔCT | 7.01 ± 0.88a | 6.24 ± 0.82a | 7.29 ± 1.08a | 0.0792 | 0.7071 | 0.8219 |
| BB-12 | ΔCT | 5.92 ± 0.97a | 5.18 ± 0.97a | 5.10 ± 1.08a | ||||
| FC | 2.13 | 2.08 | 4.56 | |||||
| CD80 | Control | ΔCT | 9.51 ± 0.79a | 10.07 ± 0.64a | 13.35 ± 0.91a | 0.0553 | 0.038 | 0.0806 |
| BB-12 | ΔCT | 9.20 ± 0.79a | 10.03 ± 0.71a | 9.84 ± 0.79b | ||||
| FC | 1.24 | 1.03 | 11.39 | |||||
| CD86 | Control | ΔCT | 5.35 ± 0.74a | 5.43 ± 0.22a | 5.93 ± 0.53a | 0.2198 | 0.7521 | 0.8516 |
| BB-12 | ΔCT | 5.09 ± 0.80a | 4.88 ± 0.29a | 4.98 ± 0.47a | ||||
| FC | 1.19 | 1.46 | 1.93 | |||||
| CD274 | Control | ΔCT | 9.13 ± 1.04a | 9.40 ± 0.95a | 9.93 ± 1.16a | 0.1305 | 0.7977 | 0.9526 |
| BB-12 | ΔCT | 7.69 ± 1.04a | 8.40 ± 1.16a | 8.27 ± 1.04a | ||||
| FC | 2.71 | 2 | 3.16 | |||||
| TNFSF14 | Control | ΔCT | 8.39 ± 1.18a | 7.21 ± 1.08a | 8.35 ± 1.42a | 0.0065 | 0.5753 | 0.6246 |
| BB-12 | ΔCT | 6.66 ± 1.33a | 5.38 ± 1.31a | 4.38 ± 1.40c | ||||
| FC | 3.32 | 3.56 | 15.67 | |||||
| Chemokines | ||||||||
| CXCL10 | Control | ΔCT | 10.02 ± 0.60a | 10.69 ± 0.59a | 10.88 ± 0.71a | 0.132 | 0.7968 | 0.0952 |
| BB-12 | ΔCT | 10.33 ± 0.73a | 9.22 ± 0.73a | 8.72 ± 0.65b | ||||
| FC | −1.24 | 2.77 | 4.47 | |||||
| CCL4 | Control | ΔCT | 8.88 ± 0.65a | 8.90 ± 0.60a | 9.37 ± 0.80a | 0.3993 | 0.1553 | 0.1427 |
| BB-12 | ΔCT | 9.94 ± 0.71a | 7.17 ± 0.80c | 8.54 ± 0.71a | ||||
| FC | −2.08 | 3.32 | 1.78 | |||||
| CCL3 | Control | ΔCT | 9.49 ± 0.70a | 8.82 ± 0.65a | 9.81 ± 0.85a | 0.3554 | 0.4666 | 0.5122 |
| BB-12 | ΔCT | 9.66 ± 0.76a | 8.54 ± 0.76a | 8.16 ± 0.85a | ||||
| FC | −1.13 | 1.21 | 3.14 |
ΔCt Mean ± SE gene expression values inversely proportional to signal abundance are normalized to housekeeping gene RPL32.
**FC, pairwise comparison were done by one-way ANOVA significant n-fold changes relative to results from control patients are designated with different superscripts letters b (P < 0.05), c (P < 0.1).
***Repeated measures in a treatment * day ANOVA with covariance structure among days for BB-12 and for control subjects identified by log likelihood ratio test as described in Patients and Methods.
Discussion
The aim of this randomized, controlled study was to assess the effect of a probiotic-supplemented yogurt containing the probiotic strain B. lactis BB-12 on the safety of generally healthy adults consuming antibiotics for upper respiratory infections. The baseline and demographic characteristics of the study participants, the duration of the study and the compliance of the product consumption were similar for the BB-12-supplemented and control groups. There was no difference in adverse events between the 2 groups. Additionally, there were no withdrawals from the study for adverse events related to product consumption.
The present study is novel in that it was conducted under FDA oversight, with the BB-12 probiotic yogurt considered an investigational new drug. While infant formula and yogurts containing BB-12 are marketed as foods in the United States, the intended use of a product to impact human health is considered as a drug purpose by the FDA. As such, this BB-12-supplemented probiotic yogurt was regarded as a drug; for the study to proceed in the United States, it was necessary to conduct the protocol under IND guidelines. Few studies on probiotics as INDs have been conducted in the United States. This is the first study on BB-12 to adhere to FDA regulatory policies for drug products and thus, is on track for continued research as a drug in the United States.
Molecular-based detection methods for enumeration of B. lactis in the stools indicated that not all participants were free of B. lactis at the beginning of the intervention, as low abundance levels for B. lactis species were detected in some participants’ baseline samples. This finding suggests that participants had some inherent level of this bacterial species in their intestinal tract as a result of previous colonization, and/or due to consumption of a product containing B. lactis species shortly before study enrollment. This later possibility should have been precluded, as adjunctive probiotic yogurt consumption was an exclusion criterion at the time of study entry interview. Despite this low initial detection level, participants were found to be in compliance with the intervention, as BB-12 signals were detected by PCR (66.7%) collected from subjects in the BB-12 intervention group after 7 d of supplementation and with no detection in 70% of participants from the control group. As in previous studies, BB-12 remained viable throughout the shelf life of the product and was absent in the control product throughout the study.11 No changes in bacterial abundance were detected for total bacteria (Eubacteria), or other beneficial bacterial species such as Bacteroides fragilis, Bifidobacterium and Lactobacillus species in fecal samples from both intervention groups after 10-day supplementation with yogurt containing B. lactis, indicating that major changes in composition of intestinal microbiota are not seen after short-term interventions with BB-12 at the current dose or may not be affected by a short term consumption of antibiotics (Table 3). Alternatively, high variability in bacterial abundance within a reduced group of subjects may have contributed to the lack of detectable change at a species level. However, the absolute (adjusted BB-12 copies per gram of feces) or relative (% of total bacteria) determined by species-specific real-time PCR indicated that the intervention had an overall significant day effect (P = 0.0422). When compared to baseline levels a mild but significant increase of B. lactis in feces was observed in BB-12 supplemented group at day 3 of intervention (Table 3). When the BB-12 supplemented group was compared to the non-supplemented group, B. lactis levels remained modestly higher at day 10 at a time BB-12 and antibiotic consumption was stopped and at day 28 after 18 d of treatment, suggesting that concomitant antibiotic treatment does not affect B. lactis levels. Our results are in agreement with another study where subjects who consumed B. lactis with amoxicillin-clavulanate antibiotic treatment had an initial transient reduction of fecal B. lactis but with a mild recovery to baseline or higher levels at the end of antibiotic intervention and follow-up period when compared to a placebo treated group.2,12 Intervention studies with other probiotic strains also coincided in reporting discrete changes in detectable probiotic in fecal samples after short-term interventions.13
This double-blinded, randomized, controlled intervention study was not designed to assess the effect of B. lactis on systemic immune response as patients’ immune systems were also naturally stimulated with bacterial or viral pathogens associated with upper respiratory infections. However, since the capacity of B. lactis to affect local cytokine production and regulatory cell populations has been demonstrated in ex vivo experiments with human derived cells,14-16 it is reasonable to assume that by increasing B. lactis abundance in vivo, the immune response may also be affected. Therefore, with the intent of investigating possible changes in immune function, RNA from peripheral whole blood voluntarily contributed from participants from both intervention groups was isolated to study a limited number of transcriptomic changes associated with antigen recognition, cell activation and inflammatory response. Gene expression levels of selected markers at baseline and 7 d after starting intervention, did not show changes with the exception of CCL4. The expression for CCL4, a monokine with inflammatory and chemokinetic properties, temporarily showed a mild increase in blood cells of individuals who received the B. lactis-supplemented yogurt for 7 d (P < 0.1). Significant changes in gene expression were detected at day 14 after the intervention was interrupted for 3 d. Notably, GATA3, a transcription factor involved in the early differentiation and T-cell lineage commitment,17 CD80 a transmembrane receptor associated with antigen recognition,18 and CXCL10, a chemokine involved in early cell stimulation, were upregulated at least fivefold only in blood of participants from the B. lactis-supplemented yogurt group (P < 0.05). A fivefold induction of tumor necrosis factor α (TNFA) expression in response to BB-12 supplementation is suggestive of a Th1 polarization effect as previously shown in in vitro culture experiments with isolated human peripheral blood mononuclear cells (PBMC),14,19 indicating a direct activation of blood monocytes, as well as T lymphocytes toward a mild inflammatory Th1 response (Table 4).
The impact of controlled administration of Bifidobacterium animalis subsp. lactis BB-12 on expression of Toll-like receptors, a major pattern in the induction of innate immunity through the recognition of exogenous microbial-associated molecular patterns,20 was also evaluated. However, high expression levels detected in the B. lactis-supplemented intervention group at day 14 for all 3 Toll receptors (TLR2, TLR4, TLR9) evaluated, did not reach statistical significance. This result may be due to a high variability in the response. The contribution of antigenic load associated with upper respiratory infection may be a reflection of these findings.21
Our results are also in agreement with previously described ex vivo studies with whole blood cells14 or enriched cell populations where dendritic cells exposed to different Bifidobacterium ssp were also able to upregulate the expression of HLA-DR and the co-stimulatory molecules CD86 and CD80, which can help trigger antigen specific T-cell responses indicating a higher immune stimulating ability.25,30 In this study, we wanted to determine the potency of B. lactis to regulate the co-stimulatory molecules CD80, CD86, CD40 and HLA-DR, which are required for an effective activation of T-cells. B. lactis induced upregulation of CD40 and CD80 expression. It is particularly interesting that short-term supplementation with B. lactis–containing yogurt was found to increase expression of these immune activation markers. Although the effect of Bifidobacterium cannot be independently evaluated under our experimental conditions, our data suggest that a short-term supplementation with BB-12 was able to activate whole blood cells, presumably those involved in the initiation of the immune response. We were able to identify changes in expression of transcription factors and genes associated with cell activation and migration, which presumably constitutes an advantage to the host by improving immune recognition function.
This randomized, controlled trial demonstrated that BB-12 is safe and well tolerated in healthy adults concurrently receiving antibiotic treatment, and provided the evidence for safety needed for the next staging of the IND process to include a healthy pediatric population. Future trials are necessary to understand the effects of BB-12 on improving health outcomes and further investigate the immunological response BB-12 demonstrates.
Patients and Methods
Study design
A phase I, double-blinded, randomized controlled pilot study was conducted with 2 parallel arms. The study protocol was approved by the Georgetown University Institutional Review Board (IRB #2008-588, Washington, DC) and registered at ClinicalTrials.gov (NCT00848003). An independent Data Safety Monitoring Board reviewed the protocol prior to study initiation and adverse event data at approximately 25%, 50%, and 75% data completion. Monitoring was also conducted by the FDA/CBER, under IND#13691 and the National Institutes of Health (NIH), National Center for Complementary and Alternative Medicine (NCCAM), including its Office of Clinical and Regulatory Affairs.
Study participants who passed the initial screenings had baseline physical examinations conducted by licensed and trained Medical Doctors or Nurse Practitioners for vital signs (pulse rate, respiratory rate, blood pressure and oxygen saturation) and to ensure general health. Eligible participants who provided written informed consent were enrolled and randomized to either the BB-12 or control yogurt drink. Participants consumed 4 ounces of the assigned yogurt beverage daily for 10 consecutive days. The participants agreed to refrain from consuming any other probiotic foods or supplements during the study period and were supplied with a list of excluded products.
To assess the safety of the interventions, research assistants conducted follow-up interviews at days 6, 11, 15, and 180, and a second physical examination was performed on day 14. A 14-day daily assessment diary was completed by the participants, which captured data on compliance, symptoms and adverse events. Fecal specimens were collected by the participants prior to (day 0) and 3, 7, 10, and 28 d after the initiation of the yogurt intervention and antibiotic treatment. At enrollment, participants were also informed of optional participation in an additional sub-study intended to monitor gene expression changes in whole blood cells in response to BB-12 exposure. Interested participants completed a separate consent form and were asked to provide 9 mL (mls) whole blood per visit at day 0, 7, and 14. Blood samples were collected in PAXgene tubes (BD, New Jersey, USA) by research personnel trained in phlebotomy.
Participants
The participants in the study were healthy individuals between the ages of 18–65 y and were prescribed treatment with a penicillin-class antibiotic regimen for a respiratory infection. A respiratory infection was classified as any infection the physician designates as Streptococcus or non-Streptococcus pharyngitis, otitis media, pneumonia, sinusitis or bronchitis that results in a 10-day prescription of antibiotics.
Inclusion and exclusion criteria
The inclusion criteria for participants were: ability to speak and write English or Spanish; access to a refrigerator for proper storage of the yogurt drink; access to a telephone; enrollment must have taken place within 24 hours of starting the antibiotic regimen; prescribed treatment with a penicillin-class antibiotic regimen for 10 d for an upper respiratory infection; antibiotic dose prescribed at least twice a day; and were outpatients. Participants were ineligible if they had any of the following: any chronic condition regardless of the requirement for medication; allergy to strawberry, active diarrhea, allergy to penicillin-class antibiotics; use of any other medicines except prescribed antibiotic and anti-pyretic medicines; allergy to tetracycline, erythromycin, trimethoprim or ciprofloxacin; lactose intolerance; during baseline physical exam, had vital signs outside the normal range: systolic blood pressure >140, systolic blood pressure <90, diastolic >90, oxygen saturation <98%, pulse rate >100, pulse rate <55 and respiratory rate >17; history of heart disease, including valvulopathies or cardiac surgery, any implantable device or prosthetic; history of gastrointestinal surgery or disease; milk-protein allergy; allergy to any component of the product or the yogurt vehicle; or females who were pregnant at the time of enrollment or were planning to become pregnant during the study.
Setting
The participants were recruited from ambulatory care clinics through the Capital Area Primary Care Research Network, a practice based research network, and from the greater local community through print and web-based advertising.
Interventions
The control and treatment interventions were strawberry-flavored yogurt drinks developed at the Pennsylvania State University using YFL-702, a commercial blend of the active cultures Streptococcus thermophilus and Lactobacillus delbrueckii subsp bulgaricus (methods described elsewhere).11 All yogurts used starter cultures Streptococcus thermophilus and Lactobacillus delbrueckii subsp Bulgaricus. The BB-12 product was supplemented additionally with the investigational agent Bifidobacterium animalis subsp. lactis strain BB-12 by thoroughly mixing frozen concentrated culture into the yogurt drink after fermentation. The microbiological composition of the active yogurt drink at the end of its 30-day shelf life met targets of at least 1 × 1010 colony forming units per 100 mL serving of BB-12.11 Both the YFL-702 starter culture and the BB-12 probiotic were supplied as frozen concentrated cultures by the manufacturer, Chr. Hansen (Milwaukee, WI).
To verify the viable count of B. lactis in the yogurt drinks, both control and treatment products were analyzed immediately after manufacture and the treatment was measured weekly by pour plating suitable dilutions on selective MRS agar followed by anaerobic incubation at 37°C for 48 hours. Random colonies counted as B. lactis were picked and confirmed to be B. lactis by PCR using subspecies specific primers.22
Primary outcome measures
The primary outcome was to determine the frequency and severity of adverse events reported during the study. Data on adverse events were collected via participant daily assessment diaries, spontaneous calls to the 24-hour study phone line and regularly scheduled phone interviews with the research personnel. An adverse event refers to any untoward event experienced by a participant during a clinical trial, whether or not it is associated with the use of the study products. This includes symptoms that were not present at the start of the study, as well as those symptoms that were present at baseline but worsened in severity during the course of the study. Adverse events were tabulated by type, intensity/severity, solicited or unsolicited and charted over time. The events were graded for severity using the National Cancer Institute Common Terminology Criteria for Adverse Events v3.0.23 Serious adverse events were defined as any incidences of death, life-threatening event, hospitalization, prolongation of a hospital stay or an event resulting in permanent disability.
Secondary outcome measures
The secondary aim was to assess the ability of BB-12 to affect the expression of whole blood immune markers associated with cell activation and inflammatory response. Other outcomes included total yogurt beverages consumed over the intervention period and total number of stools.
Fecal sample processing
DNA from stool samples provided by participants on days 0, 3, 7, 10, and 28 was isolated using the QIAamp DNA Stool isolation mini-kit (Qiagen, Valencia, CA).24 Briefly, one gram of homogenized contents was weighed and immediately re-suspended with lysis buffer. After heating the suspension at 95°C to increase DNA yield, removal of inhibitors and proteinase K digestion was done before DNA was bound to a column, washed, and eluted in TE buffer. DNA concentration was determined by NanoDrop (Thermo Fisher Scientific, Waltham, MA). The amount of bacterial copy numbers in fecal DNA was determined by real-time PCR using previously validated primers and probe sets to identify a common 16S rRNA (rRNA) sequence fragment of Domain bacteria for determination of total bacterial load,25 Lactobacillus and Bifidobacterium species,26-28 Bacteroides fragilis group (B. fragilis group)29 or using a specific primer-probe set for identification of a genomic tuf gene fraction used for identification and relative quantification of Bifidobacterium animalis subsp lactis.24 A bacterial strain representative for each bacterial species was used as a positive amplification control. The Ct value expressing the target gene copy numbers for different bacterial species were compared to a standard curve generated from a series of dilutions of a purified target gene fragment to determine specificity and efficiency of the real-time PCR assay. The size of the fragment was verified and molecular mass was quantified by automated electrophoresis system (DNAchip, Experion, Biorad, Hercules, CA). A linear relationship was established between the Ct value and number of target gene copies ranging between 101 to 1010 copies/mL and this relationship was subsequently used to estimate values of log10 target gene copy numbers in fecal samples. In all assays used, the amplification efficiency was >90% and the standard curve showed a linear range across at least 5 logs of DNA gene concentrations with a correlation coefficient >0.9. The lowest detection limit of all assays was between 10–100 copies of specific bacterial gene copy per reaction.30 All molecular assays were performed in the 7700-ABI PRISM (PerkinElmer, Waltham, MA) using a 25 uL PCR amplification mixture containing 1X Thermo-start QPCR master mix with ROX (Abgene, Rochester, NY), forward, reverse, probe and an equivalent of 20 ng of DNA. The amplification conditions were 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute. Mean copy numbers (expressed as log10 target gene copies) per gram of feces were compared among treatment groups.
Blood sample processing
Whole blood collected at baseline, day 7 and day 14 in PAXgene Blood RNA tubes (BD, New Jersey, USA) was processed for isolation of RNA with on-column DNase digestion following the manufacturer's manual. The mRNA expression level of selected genes associated with transcription factors: T-box transcription factor (TBX21), GATA binding protein 3 (GATA 3), Forkhead box P3 (FOXP3), Interferon regulatory factor (IRF8); Toll like receptors TLR2, TLR4 and TLR9; pro-inflammatory and regulatory cytokines: Tumor necrosis factor α (TNFA), Interleukin 6 (IL6), Interleukin 1b (IL1B), Tumor necrosis factor ligand superfamily member 14 (TNFSF14), interferon gamma (IFNg), Tumor growth factor β-2 (TGFB2), Interleukin 10 (IL10); cell activation markers: CD40, HLA-DRA, CD80, CD86, CD274; and chemokines CXCL10, CCL3, CCL4 was measured on cDNA synthesized from each sample using 1 μg of total RNA.31 DNAse-treated RNA was quantified using the Agilent Bioanalyzer 2100 and RNA 6000 Labchip Kit (Agilent Technologies, Palo Alto, CA).32 Briefly, cDNA was synthesized with Superscript RT (Invitrogen) and oligo (dT), and 50 ng of this cDNA was used for real-time PCR amplifications using a Thermo-Start DNA Polymerase master mix (Abgene, Rochester, NY) and the ABI PRISM 7700 Sequence detector system (Applied Biosystems, Foster City, CA). Amplification conditions were: 50°C for 2 minutes; 95°C for 10 minutes; 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute. All probes and primers selected for real-time PCR were designed using the Primer Express (Applied Biosystems, Foster City, CA) software package and nucleotide sequences were obtained from Genbank. The sequence information for human genes assayed can be found in the Porcine Translational Research Database from the Diet, Genomics, and Immunology Laboratory, Beltsville Human Nutrition Research Center, Agricultural Research Service-United States Department of Agriculture (http://www.ars.usda.gov/Services/docs.htm?docid = 6065). Fluorescence signals were processed after amplification and were considered positive if the fluorescence intensity was 20-fold more or greater that the standard deviation of the baseline fluorescence. Gene expression was normalized based upon a constant amount of RNA and cDNA amplified.32 Relative quantification of target gene expression was evaluated by comparing Ct values from cDNA processed from patients at different times after normalization with the housekeeping gene RPL32. Up- or downregulation in gene expression is denoted by fold changes in Ct values.33
Randomization
Participants were allocated to either the control or BB-12 intervention arm in a 1:1 ratio using permuted block randomization of block size 4 using SAS® version 9.1 (SAS Institute Inc., Cary, NC). Once eligibility was determined, the participant was randomly assigned a number between 1 and 10, corresponding to a control or BB-12 yogurt drink. True allocation concealment was ensured, as research personnel had no methods to alter randomization or enrollment.
Compliance
Compliance was measured by self-report on the daily assessment diary, follow-up phone interviews and by PCR analysis of the fecal samples collected at Day 7 of the intervention. Results showing the presence of B. lactis in the feces of the BB-12 group or the absence of B. lactis in the feces of the control group were considered to be compliant. Results showing the presence of B. lactis in the control group or the absence of B. lactis in the treated group, and missing fecal samples, were considered to be noncompliant.
Data analysis
Sample size calculations are not applicable for a phase I safety study. Statistical analyses were conducted using Stata® 10 (StataCorp LP, College Station, TX) statistical software. Continuous variables were summarized using means, medians and standard deviations, and frequency percentages were calculated for categorical variables. The frequency and severity of the adverse events were described using frequencies and percentages. Baseline demographics and health characteristics were compared between the treated and control groups using Chi-square or Fisher's exact test for categorical variables and t-tests and Wilcoxon rank test for continuous variables.
Blood gene expression data and bacterial counts data were analyzed modeling days as repeated measures in a treatment X day ANOVA with covariance structure among days for BB-12 and for control groups identified by log-likelihood ratio test (relative to independence) from among the possible covariance structures: compound symmetric, first-order auto-regressive, heterogeneous compound symmetric, or heterogeneous first-order auto-regressive. Data for blood gene expression, CT values were collected at baseline, day 7 and day 14; normalized to the housekeeping gene RPL32 (ΔCT) and expressed as fold change compared to a basal group which was designated as 1 fold change. Absolute bacterial counts, expressed as copies per gram values, or relative counts, expressed as percentage of total bacteria, were generated from real-time PCR analysis for samples of stool contents collected at baseline, day 3, 7, 10, and 28; and were modeled using a lognormal distribution (after adding one to each observed bacterial count to allow observed zero counts to be represented as zero on the log scale) in a generalized linear mixed effects ANOVA model with between – subject treatment X within –subject day and covariance structure among days. At each time, pairwise comparisons among treatment groups were conducted for all blood gene expression and bacterial data. Any non-identical letters (abc) indicate significant differences among treatment groups means (P < 0.05 or P < 0.1). All analyses were performed using SAS® version 9.3, Proc GLIMMIX (SAS Institute Inc., Cary, NC).
Disclosure of Potential Conflicts of Interest
DJM previously served as a paid expert witness for General Mills, Inc., Nestlé Nutrition, Bayer, and the Proctor & Gamble Company. MES consults for numerous probiotic manufacturers. The remaining authors have no conflicts of interest to declare.
Acknowledgments
We thank all the individuals who participated in this study, Emily Furumoto, the Berkey Creamery at Penn State University, Pilar Hamilton, Haewon Park, R. Scott Flinn, student research assistants, and the Capital Area Primary Care Research Network. We would also like to thank Data Safety Monitoring Board Chair Marie Diener-West and members Felice Roggen, Marguerite Duane and Tamar Ringel-Kulka, and Bryan Vinyard, Director of Biometrics Consulting Service, ARS-USDA, for statistical analysis of transcriptomic data. We would also like to express our gratitude to Linda C. Duffy, National Institutes of Health program officer for the NIH Division of Extramural Research at NCCAM, and the Office of Clinical and Regulatory Affairs for their help and support.
Funding
The project described was supported by Award Number U01AT003600 from the NIH National Center for Complementary and Alternative Medicine and partial support from in-house ARS-USDA project 1235-51000-051-00D. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Complementary and Alternative Medicine or the National Institutes of Health.
Authorship
DJM, RFR, and GSA designed research; DJM, KHS, and TPT conducted research and acquired data; DJM, TPT, GSA, AM, MM, NMS, and RFR analyzed and interpreted data; MM, NMS, and DJM performed the statistical analyses; DJM, TPT, and GSA drafted the initial manuscript; MM, NMS, AM, RFR, and MES provided critical revision for important intellectual content; DJM had primary responsibility for the final content. All authors have seen and approved the final manuscript.
References
- 1. Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. Cordoba, Argentina: World Health Organization. [Google Scholar]
- 2. Guillemard E, Tondu F, Lacoin F, Schrezenmeir J. Consumption of a fermented dairy product containing the probiotic Lactobacillus casei DN-114001 reduces the duration of respiratory infections in the elderly in a randomised controlled trial. Br J Nutr 2010; 103:58-68; PMID:19747410; http://dx.doi.org/ 10.1017/S0007114509991395 [DOI] [PubMed] [Google Scholar]
- 3. Lehtoranta L, Pitkaranta A, Korpela R. Probiotics in respiratory virus infections. Eur J Clin Microbiol Infect Dis: Off Pub Eur Soc Clin Microbiol 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rutten NB, Besseling-Van der Vaart I, Klein M, De Roock S, Vlieger AM, Rijkers GT. In vitro assessment of the immunomodulatory effects of multispecies probiotic formulations for management of allergic diseases. Benef Microbes 2011; 2:183-92; PMID:21986357; http://dx.doi.org/ 10.3920/BM2011.0012 [DOI] [PubMed] [Google Scholar]
- 5. Rodriguez B, Prioult G, Hacini-Rachinel F, Moine D, Bruttin A, Ngom-Bru C, Labellie C, Nicolis I, Berger B, Mercenier A, et al. Infant gut microbiota is protective against cow's milk allergy in mice despite immature ileal T-cell response. FEMS Microbiol Ecol 2012; 79:192-202; PMID:22029421; http://dx.doi.org/ 10.1111/j.1574-6941.2011.01207.x [DOI] [PubMed] [Google Scholar]
- 6. Engelbrektson AL, Korzenik JR, Sanders ME, Clement BG, Leyer G, Klaenhammer TR, Kitts CL. Analysis of treatment effects on the microbial ecology of the human intestine. FEMS Microbiol Ecol 2006; 57:239-50; PMID:16867142; http://dx.doi.org/ 10.1111/j.1574-6941.2006.00112.x [DOI] [PubMed] [Google Scholar]
- 7. Clausen MR, Bonnen H, Tvede M, Mortensen PB. Colonic fermentation to short-chain fatty-acids is decreased in antibiotic-associated diarrhea. Gastroenterology 1991; 101:1497-504; PMID:1955116 [DOI] [PubMed] [Google Scholar]
- 8. Johnston BC, Goldenberg JZ, Vandvik PO, Sun X, Guyatt GH. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev 2011:CD004827; PMID:22071814; http://dx.doi.org/ 10.1002/14651858.CD004827 [DOI] [PubMed] [Google Scholar]
- 9. Klijn A, Mercenier A, Arigoni F. Lessons from the genomes of bifidobacteria. FEMS Microbiol Rev 2005; 29:491-509; PMID:15939502; http://dx.doi.org/ 10.1016/j.fmrre.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 10. Langhendries JP, Detry J, Van Hees J, Lamboray JM, Darimont J, Mozin MJ, Secretin MC, Senterre J. Effect of a fermented infant formula containing viable bifidobacteria on the fecal flora composition and pH of healthy full-term infants. J Pediatr Gastroenterol Nutr 1995; 21:177-81; PMID:7472904; http://dx.doi.org/ 10.1097/00005176-199508000-00009 [DOI] [PubMed] [Google Scholar]
- 11. Merenstein D, Gonzalez J, Young AG, Roberts RF, Sanders ME, Petterson S. Study to investigate the potential of probiotics in children attending school. Eur J Clin Nutr 2011; 65:447-53; PMID:21326270; http://dx.doi.org/ 10.1038/ejcn.2010.290 [DOI] [PubMed] [Google Scholar]
- 12. Forssten S, Evans M, Wilson D, Ouwehand AC. Influence of a probiotic mixture on antibiotic induced microbiota disturbances. World J Gastroenterol 2014; 20:11878-85; PMID:25206295; http://dx.doi.org/ 10.3748/wjg.v20.i33.11878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mangalat N, Liu Y, Fatheree NY, Ferris MJ, Van Arsdall MR, Chen Z, Rahbar MH, Gleason WA, Norori J, Tran DQ, et al. Safety and tolerability of Lactobacillus reuteri DSM 17938 and effects on biomarkers in healthy adults: results from a randomized masked trial. PLoS One 2012; 7:e43910; PMID:22970150; http://dx.doi.org/ 10.1371/journal.pone.0043910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lopez P, Gueimonde M, Margolles A, Suarez A. Distinct Bifidobacterium strains drive different immune responses in vitro. Int J Food Microbiol 2010; 138:157-65; PMID:20071048; http://dx.doi.org/ 10.1016/j.ijfoodmicro.2009.12.023 [DOI] [PubMed] [Google Scholar]
- 15. Donkor ON, Ravikumar M, Proudfoot O, Day SL, Apostolopoulos V, Paukovics G, Vasiljevic T, Nutt SL, Gill H. Cytokine profile and induction of T helper type 17 and regulatory T cells by human peripheral mononuclear cells after microbial exposure. Clin Exp Immunol 2012; 167:282-95; PMID:22236005; http://dx.doi.org/ 10.1111/j.1365-2249.2011.04496.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weiss G, Christensen HR, Zeuthen LH, Vogensen FK, Jakobsen M, Frokiaer H. Lactobacilli and bifidobacteria induce differential interferon-beta profiles in dendritic cells. Cytokine 2011; 56:520-30; PMID:21889358; http://dx.doi.org/ 10.1016/j.cyto.2011.07.024 [DOI] [PubMed] [Google Scholar]
- 17. Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 2011; 35:299-311; PMID:21867929; http://dx.doi.org/ 10.1016/j.immuni.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol 2013; 13:227-42; PMID:23470321; http://dx.doi.org/ 10.1038/nri3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopez P, Gonzalez-Rodriguez I, Gueimonde M, Margolles A, Suarez A. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS One 2011; 6:e24776; PMID:21966367; http://dx.doi.org/ 10.1371/journal.pone.0024776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lavelle EC, Murphy C, O'Neill LA, Creagh EM. The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol 2010; 3:17-28; PMID:19890268; http://dx.doi.org/ 10.1038/mi.2009.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagalingam NA, Cope EK, Lynch SV. Probiotic strategies for treatment of respiratory diseases. Trends Microbiol 2013; 21:485-92; PMID:23707554; http://dx.doi.org/ 10.1016/j.tim.2013.04.008 [DOI] [PubMed] [Google Scholar]
- 22. Ventura M, Reniero R, Zink R. Specific identification and targeted characterization of bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl Env Microbiol 2001; 67:2760-5; PMID:11375192; http://dx.doi.org/ 10.1128/AEM.67.6.2760-2765.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Therapy Evaluation Program Common Terminology Criteria for Adverse Events, Version 3.0. Division of Cancer Treatment and Diagnosis, National Cancer Institute, National Institutes of Health, United States Department of Health and Human Services. August 9, 2006. http://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/ctcaev3.pdf [Google Scholar]
- 24. Solano-Aguilar G, Dawson H, Restrepo M, Andrews K, Vinyard B, Urban JF, Jr. Detection of Bifidobacterium animalis subsp. lactis (Bb12) in the intestine after feeding of sows and their piglets. Appl Env Microbiol 2008; 74:6338-47; PMID:18689506; http://dx.doi.org/ 10.1128/AEM.00309-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002; 148:257-66; PMID:11782518 [DOI] [PubMed] [Google Scholar]
- 26. Delroisse JM, Boulvin AL, Parmentier I, Dauphin RD, Vandenbol M, Portetelle D. Quantification of Bifidobacterium spp. and Lactobacillus spp. in rat fecal samples by real-time PCR. Microbiol Res 2008; 163:663-70; PMID:19216105; http://dx.doi.org/ 10.1016/j.micres.2006.09.004 [DOI] [PubMed] [Google Scholar]
- 27. Haarman M, Knol J. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl Env Microbiol 2005; 71:2318-24; PMID:15870317; http://dx.doi.org/ 10.1128/AEM.71.5.2318-2324.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haarman M, Knol J. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl Env Microbiol 2006; 72:2359-65; PMID:16597930; http://dx.doi.org/ 10.1128/AEM.72.4.2359-2365.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 2006; 118:511-21; PMID:16882802; http://dx.doi.org/ 10.1542/peds.2005-2824 [DOI] [PubMed] [Google Scholar]
- 30. Solano-Aguilar G, Fernandez KP, Ets H, Molokin A, Vinyard B, Urban JF, Gutierrez MF. Characterization of fecal microbiota of children with diarrhea in 2 locations in Colombia. J Pediatr Gastroenterol Nutr 2013; 56:503-11; PMID:23254448; http://dx.doi.org/ 10.1097/MPG.0b013e318282aa12 [DOI] [PubMed] [Google Scholar]
- 31. Solano-Aguilar GI, Vengroski KG, Beshah E, Douglass LW, Lunney JK. Characterization of lymphocyte subsets from mucosal tissues in neonatal swine. Dev Comp Immunol 2001; 25:245-63; PMID:11164889; http://dx.doi.org/ 10.1016/S0145-305X(00)00053-7 [DOI] [PubMed] [Google Scholar]
- 32. Dawson HD, Beshah E, Nishi S, Solano-Aguilar G, Morimoto M, Zhao A, Madden KB, Ledbetter TK, Dubey JP, Shea-Donohue T, et al. Localized multigene expression patterns support an evolving Th1/Th2-like paradigm in response to infections with Toxoplasma gondii and Ascaris suum. Infect Immun 2005; 73:1116-28; PMID:15664955; http://dx.doi.org/ 10.1128/IAI.73.2.1116-1128.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protocols 2008; 3:1101-8; PMID:18546601; http://dx.doi.org/ 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]


