Abstract
Local synthesis of proteins near their activity site has been demonstrated in many biological systems, and has diverse contributions to cellular functions. Studies in recent years have revealed that hundreds of mitochondria-destined proteins are synthesized by cytosolic ribosomes near the mitochondrial outer membrane, indicating that localized translation also occurs at this cellular locus. Furthermore, in the last year central factors that are involved in this process were identified in yeast, Drosophila, and human cells. Herein we review the experimental evidence for localized translation on the cytosolic side of the mitochondrial outer membrane; in addition, we describe the factors that are involved in this process and discuss the conservation of this mechanism among various species. We also describe the relationship between localized translation and import into the mitochondria and suggest avenues of study that look beyond cotranslational import. Finally we discuss future challenges in characterizing the mechanisms for localized translation and its physiological significance.
Keywords: cotranslational import, mRNA localization, mitochondria, NAC, Om14, Puf3, ribosome, translation, Tom20
Introduction
The synthesis of many proteins has been shown to be localized to specific sites within the cell, usually close to where the encoded protein is active. Synthesizing a protein at its action site has significant advantages, which include reducing the probability of malfunctioning at other cellular loci and lowering the energy costs of protein transport. Furthermore, the synthesis of proteins with similar roles in proximity to each other may provide on-site regulation of expression and a faster response to local needs.1-4 Segregating the translation process to distinct loci throughout the cell may also be beneficial in cases of stress, in that some translation sites are more protected from stress than others and maintain some protein synthesis in adverse conditions,5,6 presumably to support cell survival. Thus, localized translation further compartmentalizes cells into small functional units, which provide better response to local needs and improves overall physiological efficiency.
Considering the advantages of localized translation, it is not surprising that it has been found in many systems,7,8 particularly those involved in developmental processes. Notable examples are found in Drosophila melanogaster embryonal development,9 Xenopus development,8 cell division and polarized growth in Saccharomyces cerevisiae,10,11,12 and hyphal growth in Ustilago maydis.13 Localized translation is also very common in neuronal synapses and in the growing edges of fibroblasts.14,15 In these systems, mRNAs are transported in an untranslated manner to their target through a process that requires the activity of various RNA-binding proteins (RBPs) and sometimes motor proteins to shuttle the transcript along the cytoskeleton.16 Once at the destination, and upon a proper cue, translation inhibition is released and the mRNA is translated by local ribosomes and translation factors. As a result, a local concentration of the encoded protein is generated, which exerts local effects (either developmental or physiological).8
Localized translation has also been shown to occur near cellular organelles.17 The most established example of this is found in the translation of many secreted and membrane proteins at the ER, where the synthesis of these proteins is usually coupled to their transport into the ER lumen or membrane (i.e. cotranslational transport). Coupling translation with transport minimizes the chances of aggregation of hydrophobic proteins in the hydrophilic environment of the cytosol. Furthermore, it lowers the energetic costs of transport, as the energy used for protein synthesis serves also to fuel protein insertion into the ER. Translation at the ER also allows fast coordination with the unfolded protein response, as ribosomes quickly dissociate from the ER membrane upon the detection of stress.4 Surprisingly, many mRNAs that encode cytosol-destined proteins have been found to be associated with the ER, and translated by ER-associated ribosomes.18,19 A possible explanation for this phenomenon was suggested following the observation that ER-associated ribosomes remain active under stress conditions (such as viral infection or hypoxia), while cytosolic translation is inhibited.5,6 This suggests that the ER is a sheltered site for protein synthesis under stress, where proteins that are important for the stress response (even if they are destined for the cytosol) can still be synthesized.
In developmental systems, transport of mRNAs to their destination is translation-independent. However, transport into the ER has been shown to require a translational process, which begins in the cytoplasm. Once a signal peptide emerges from the ribosome's exit tunnel, it is recognized by the signal recognition particle (SRP). The SRP-Ribosome-mRNA complex is targeted to the ER, anchored to the membrane by the SRP receptor, and the ribosome then docks onto the translocon. At this stage, translation resumes, yielding localized translation that is coupled to protein transport into the ER. It has been shown, however, that the knock out/down of various components of this pathway is not lethal, suggesting the presence of alternative transport pathways.20 Indeed, SRP-independent mechanisms of transport have also been discovered (reviewed in ref. 21 as well as, more intriguingly, translation-independent mechanisms.22-25 It is possible that these alternative pathways operate on different sets of mRNAs or under different physiological conditions.
The ER is the first organelle near which ribosomes and translation were identified26 and consequently the mechanisms for translation here are well-known (reviewed in ref. 27). Translation near other organelles, however, is less studied. An obvious limitation for such studies is the intracellular abundance of organelles, which obstructs spatial studies of localization. Another limitation is organelles' need for a constant supply of proteins, the continual flow of which can obscure temporal studies of mRNA- or ribosome traffic. Also, organelles usually contain hundreds of different proteins, which are likely to have redundant modes of targeting. For these reasons, our knowledge regarding localized translation near cellular organelles lags behind studies in developmental systems or polarized cells. Nevertheless, studies from recent years have yielded novel findings regarding translation near peroxisomes, chloroplasts, the nucleus, and mitochondria.17,28,29 In the last year few important publications30-32 have provided molecular details of localized translation near the mitochondria. We cover herein data pertained to this organelle.
Experimental Evidence for Localized Translation Near the Mitochondria
The vast majority of the hundreds of mitochondrial proteins are encoded in the nucleus, translated in the cytoplasm, and imported into the mitochondria. Studies in the last 4 decades have shown that proteins can be imported following their complete synthesis by cytosolic ribosomes (i.e., post-translationally).33,34 This suggested that mitochondria-destined proteins are translated throughout the cytoplasm and targeted to the mitochondria with the help of cytosolic chaperones that maintain them in an unfolded state. However, in the last decade, various lines of evidence have revived older models that propose the co-existence of localized translation near the mitochondria. Below we describe these lines of evidence, divided into 3 sections based on the main hallmarks of translation: mRNA, ribosomes, and the synthesized protein.
mRNA localization near the mitochondria
Localization of an mRNA to a specific cellular site does not necessarily imply its translation at this site. For example, many mRNAs are localized into cytoplasmic processing bodies, yet their translation does not occur at these sites.35 However, in cases in which the localization of an mRNA coincides with the activity site of its encoded protein, it is reasonable to deduce that localized translation occurs at this site. Hence, the localization of mRNAs that encode mitochondrial proteins near the mitochondrial outer membrane can be used as a reasonable proxy for the existence of localized translation. As there are diverse methodologies to identify mRNAs and quantify their abundance and localization (detailed below), mRNA analysis has become the main approach in the study of localized translation.
In situ hybridization combined with electron microscopy (EM) was among the first methodologies used to detect mRNAs near the mitochondria. An early study examined 2 nuclear-encoded mRNAs of the oxidative phosphorylation pathway in rat hepatocytes. It found that while the mRNA that encoded the alpha subunit of F1-ATPase was spread throughout the cytosol, the mRNA for the F1beta subunit appeared in clusters in close proximity to the mitochondria 36. Further experiments showed that the F1beta mRNA is translated, either free or attached to mitochondria, yet the pre-protein was not observed in the cytosol. This result suggested immediate translocation through import-sites on the mitochondria.37
A few years later, several seminal studies from the laboratory of C. Jacq demonstrated that many more mRNAs are localized to the vicinity of the mitochondria in the yeast model system. Yeast cells were fractionated by differential centrifugation and the mRNAs that were associated with the mitochondrial fraction were identified. Initial work utilized northern analysis to examine a few selected mRNAs,38 while later studies used unbiased, genome-wide methods such as filter arrays and oligonucleotide microarrays.39,40 Several hundred mRNAs were identified, with most of them encoding mitochondria-destined proteins. The extent of the association with mitochondria appeared to differ among genes, and bioinformatics analyses indicated that transcripts of prokaryotic origin are enriched among those tended to localize to mitochondria.40 Importantly, the association between mRNAs and mitochondria was weakened when EDTA was added to the mitochondrial fractions, supporting the idea that ribosomes (i.e. active translation) are important for association.
These studies have been supported by an alternative, in vivo tagging methodology. Here, the 3′-UTRs of mRNAs of interest were fused to MS2 coat-protein binding sites, and co-expressed with MS2 coat protein that had been fused to GFP (MS2-GFP). As the fusion MS2-GFP bound to the mRNA, fluorescent microscopy was used to detect sites of mRNA localization. Beyond in vivo confirmation of the biochemical fractionation results, this method provided important support for the role of several different 3′-UTRs in localization.38,39,41 Recently, advanced modification of this method has allowed the detection of endogenously expressed mRNAs. MS2-binding sites were introduced into genomic loci by homologous recombination and selection markers were removed. In this way, the transcripts were tagged with only minimal interference to their native functions.42 In vivo imaging of 24 mRNAs that were tagged by this methodology revealed mitochondrial association in most.43
RNA fluorescent in situ hybridization (FISH) is advantageous in its ability to detect the localization of endogenous, unmodified transcripts. Studies that applied this methodology on a single-cell level have provided further support for the association of mRNAs to the mitochondrial outer membrane in both yeast44 and Drosophila muscle tissue.30
Finally, the recently developed proximity-specific ribosome profiling method (see below) allows the isolation and high-throughput characterization of mRNAs that are translated by mitochondria-associated ribosomes.32 This has led to the identification of many mRNAs that are translated near the mitochondria, particularly of those that encode inner-membrane proteins.
Ribosome association to mitochondria
A critical line of evidence supporting localized translation near the mitochondria was found in the presence of cytosolic ribosomes near the mitochondrial outer membrane, described in the pioneering work by Kellems and Butow. These researchers were the first to identify mitochondria-associated ribosomes, which proved to be of cytosolic type (i.e., 80S) in several aspects, including sedimentation in sucrose gradients, sensitivity to cycloheximide, and insensitivity to chloramphenicol inhibition.45 Under electron microscopy analysis, these ribosomes appeared to be attached to the outer membrane of mitochondria from cyclohexamide-treated yeast cells.46 The number of mitochondria-bound ribosomes depended on the metabolic state of the cells, with approximately 4 times more ribosomes bound to log-phase than stationary or starved yeast mitochondria.47
In vitro, only a finite number of ribosomes can bind to isolated mitochondria, suggesting that binding is mediated by a receptor that is present in limited quantities.48 The presence of a ribosome-specific receptor on the outer membrane was further supported by binding assays that showed significant binding inhibition after proteinase-K treatment (which removes cytosolic protrusions of outer membrane proteins) or when binding was performed under high ionic conditions (0.5 M KCl).48 Ribosome binding in vitro was found to be inhibited in the presence of GTP; however, this inhibition was rescued when ribosomes were charged with a mitochondrial nascent chain, while a non-mitochondrial nascent protein (luciferase) could not induce a similar effect.49 Thus, it is apparent that the mitochondrial targeting sequence is also important for ribosome binding. In addition, Kellems and colleagues showed that a high-salt treatment of isolated yeast mitochondria resulted in the release of approximately one-third of the bound ribosomes. The rest of the ribosomes dissociated only after treatment with puromycin, which disrupts ribosome-nascent chain interactions;46 this observation suggested a role for the nascent chain in stabilizing mitochondria-ribosome attachment. Taken together, these data reveal at least 2 modes of association between ribosomes and mitochondria: one dependent on nascent chains and one dependent on ribosome receptors on the mitochondrial outer membrane. Recent studies have revealed the molecular players that are involved in both of these modes (see Section 3).30,31,50,51
Ribosomes' association with mitochondria can be measured by western analysis of mitochondrial fractions with an antibody that recognizes a ribosomal protein.31,52 This technique, however, should be interpreted cautiously, as mitochondrial fractions usually contain ER fragments, which also contain ribosomes.53 A recent method, proximity-specific ribosome profiling,32 nicely overcomes this problem. In this approach, the mitochondrial outer membrane protein Om45 is fused to a biotin ligase (BirA), while the cytosolic ribosomes contain a biotin target sequence (AviTag) fused to one of the ribosomal proteins. A short pulse of biotin to the medium leads to the specific tagging of ribosomes that are in proximity to Om45-BirA, i.e. ribosomes that are near the outer membrane. Biotinylated ribosomes are then isolated using streptavidin beads and their associated mRNAs are identified via deep sequencing of the ribosome-protected fragments. By applying this powerful methodology to yeast cells, Williams and colleagues were able to specifically tag ribosomes associated with mRNAs that encode mitochondrial proteins, with enrichment of those encode proteins that reside in the inner membrane. This study therefore confirmed that ribosomes near the outer membrane are active and translate mitochondrial proteins.32
Translated protein
Localized translation can also be inferred from assays that monitor the synthesized protein. For example, isolating mitochondria and inducing translation in vitro has resulted in the production of mitochondrial proteins, in particular those of the inner membrane.54,55 These proteins appeared resistant to protease treatment, indicating that they were efficiently imported into the mitochondria.
Another approach utilized the mitochondria targeting sequence (MTS) as a handle to identify localized translation. Ni and colleagues56 attached an MTS to the N-terminus of eGFP, and an ER targeting signal to its C-terminus. Analysis of the localization of the dual-signal eGFP in HeLa cells revealed it was present exclusively in mitochondria. A simple interpretation of this result is that the reporter protein was synthesized near the mitochondria and thereby more likely to be inserted into this organelle. A related possibility is that the insertion into the mitochondria started while the proteins were being translated (i.e., cotranslational import), so that the C-terminal ER signal was synthesized only after the proteins were largely inside the mitochondria. The latter possibility was explored by fusing the MTS to dihydrofolate reductase (DHFR), and testing import in the presence of methotrexate.57 Since methotrexate binds folded DHFR at high affinity and prevents its import into mitochondria, the detection of mitochondrial DHFR would probably be due to cotranslational import of the protein in its unfolded state. Western analysis of mitochondrial fractions revealed that ∼70% of the DHFR appeared in the mitochondria. Thus, a significant amount of the protein was imported in a cotranslational manner. Similarly, import of a COX IV-DHFR protein fusion was not affected in vivo by the addition of methotrexate, consistent with cotranslational import.58 Indeed, since cotranslational import is a specific case of localized translation, studies of cotranslational import (either in vitro or in vivo)58,59,60 provide another layer of support for the existence of localized translation.
A group of mitochondrial proteins are known to be present also in other cellular locations.61,62 The most studied example is fumarase, which appears in both the mitochondria and the cytosol. Interestingly, both cytosolic and mitochondrial fumarase lack an MTS, due to cleavage by mitochondrial peptidases. The current model for fumarase maturation posits that fumarase is first imported cotranslationally into mitochondria, has its MTS cleaved off, and then moves back into the cytosol.63-65 This process therefore implies that fumerase is locally translated. Despite a lack of in-depth studies, the large number of dual-targeted proteins (estimated at approximately a third of mitochondrial proteins)62 might indicate the breadth of localized translation.
Factors Controlling Localized Translation
Localization of an mRNA and its translation are mediated by both cis sequences embedded in the transcript and protein factors that utilize these elements in order to transport and anchor the mRNA at the target site. The cis elements that control localized translation near the mitochondria can comprise non-translated regions (usually the 3′-UTR) or coding regions, in particular the mitochondria targeting sequence (MTS). These two elements are not mutually exclusive (Fig. 1). In yeast, either the MTS or 3′-UTR was sufficient to independently target ATM1 mRNA to the vicinity of the mitochondria.38 Similarly, attaching an MTS to GFP induced its mRNA localization to the mitochondria, and adding a 3′-UTR from a mitochondria-targeted mRNA further increased the strength of the association.51 Likewise, in HeLa cells, attaching the MTS from SOD2 to transcripts of ATP6 led to a significant association with the mitochondrial outer membrane, and adding both SOD2 MTS and 3′UTR to the ATP6 coding region strengthened the association.66
Figure 1.
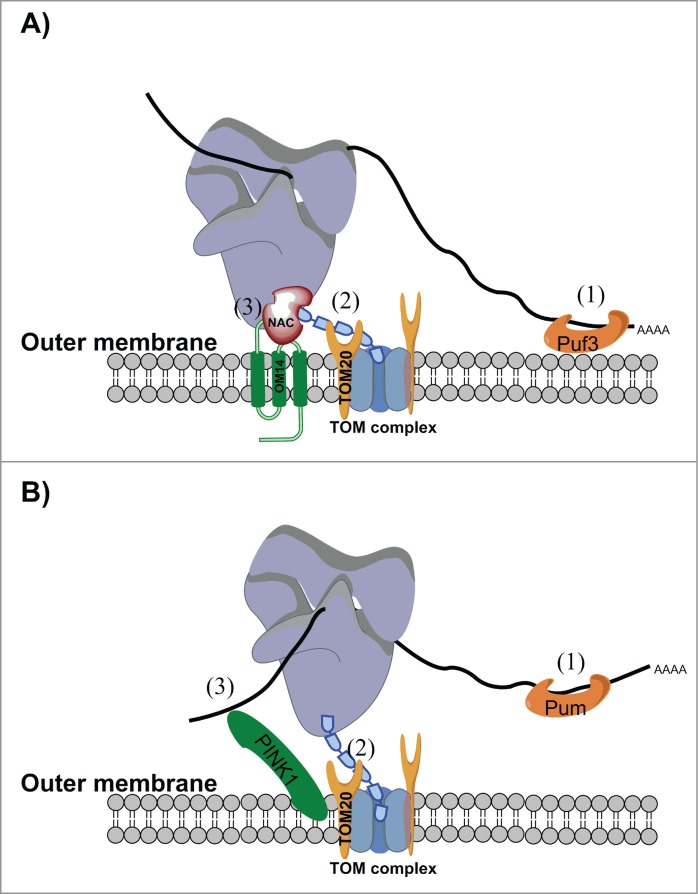
Proteins that mediate translation near mitochondria. A schematic of the working model for localized translation near mitochondria from (A) S. cerevisiae or (B) Drosophila and human cells. Recent studies indicate that multiple contact sites and multiple modes of association coordinate localized translation. A) In S. cerevisiae, mRNAs may form direct contact with Puf3 (1) via sequences in their 3′-UTR.72 Since Puf3 is associated with the mitochondria,71 this interaction anchors the mRNA in proximity to the mitochondria without interfering with translation. Additional nonexclusive contacts have been reported, but all involve a translational process. The interaction between the mitochondria targeting sequence (MTS) undergoing translation and Tom20 (2)51 is likely to be important for specifying which ribosomes should be maintained near the mitochondria. Nevertheless, once the MTS is imported into the mitochondria other anchoring factors are necessary. The interaction between NAC and Om14 (3) might provide such support, as this interaction permits cotranslational import31 and therefore might be maintained during the import process. In higher eukaryotes, the role of NAC in localized translation, as well as a functional Om14 homolog, have yet to be demonstrated. B) In Drosophila and human cell lines, the outer-membrane kinase PINK1 was shown to directly interact with mRNA (3) and to assist in the localization of some mRNAs to the mitochondrial outer membrane.30 This interaction was important in maintaining the mRNAs in a translationally-repressed state. Repression was relieved via interaction with Parkin and the subsequent removal of translation repressors such as Pum and Gro/hnRNP-F (not depicted here). The protein receptor Tom20 was shown to be important for mRNA localization, presumably via interaction with the incoming MTS (2). This mode of interaction therefore seems to be conserved among yeast, flies, and humans.30,51 A PUF protein (Pum) was also shown to interact with mitochondria-associated mRNAs (1), with a role in translation repression. Interestingly, in multicellular organisms Pum is not mitochondria-associated (as in yeast), and therefore does not serve as an anchoring factor. Conversely, a role for Puf3 in translation regulation near yeast mitochondria has yet to be demonstrated.
The impact of the MTS on localization is mediated, at least partly, through its known interaction with the protein receptor Tom20, the deletion of which reduces the association of many mRNAs with the mitochondria.51 Furthermore, localization of an MTS-GFP reporter mRNA was significantly reduced in tom20Δ cells, indicating that localization is mediated through the Tom20:MTS interaction. Interestingly, mRNA localization was not completely abolished in tom20Δ cells, which points to the involvement of additional factors in association with coding regions.51
Another important factor in localized translation near the mitochondria appears to be the heterodimeric, ribosome-associated nascent chain-associated complex (NAC). In vitro studies have shown that NAC stimulates the association of ribosome-bound nascent chains (RNCs) with mitochondria and consequent protein import.67 Additionally, protein analysis of mitochondria isolated from both wild type and NAC-deleted yeast strains revealed weaker signal of a ribosome marker in the deletion strain, indicating that NAC affects ribosomes' association with mitochondria also in vivo.52 NAC is composed of an α subunit (Egd2 in S. cerevisiae) and either a β1 (Egd1) or a β3 (Btt1) subunit, thereby forming 2 different heterodimers.68 Affinity purification of NAC proteins with their associated RNCs revealed enrichment in mRNAs that encode mitochondrial proteins for the β3 subunit.69 In addition, the association of NAC with mitochondria was shown recently to be mediated by the mitochondrial outer membrane protein Om1431(Fig. 1). The deletion of this protein resulted in less NAC being associated with mitochondria as well as a reduction in mitochondria-RNC association. Moreover, Om14 was shown to support, in vitro, the cotranslational import of proteins into isolated mitochondria.31 The interaction between NAC and Om14 may therefore provide a physical mechanism for the association of ribosomes with mitochondria, presumably after the MTS detaches from Tom20 and enters the mitochondria (Fig. 1A).
While the role of the 3′-UTR in localized translation has been established by many studies, the proteins that mediate its function are largely unknown. Studies of the Puf3 protein in S. cerevisiae, which belongs to the PUF family of RNA-binding proteins, have yielded significant results. Puf3 preferentially binds mRNAs that encode mitochondrial proteins70 and is associated with the mitochondrial outer membrane.71 Because of this, it was predicted to assist in localized translation near the mitochondria (Fig. 1A). Indeed, deletion of Puf3 affected the localization of many mRNAs to the mitochondria, and ablation of the Puf3 binding site in the 3′-UTR of BCS1 reduced mitochondrial localization of this mRNA.72 Together, these results point to a direct role for Puf3 in localization and this protein may provide an additional anchor for localized translation near the mitochondria (Fig. 1). Consistent with this idea, while yeast strains that lack either Tom20 or Puf3 are viable when grown on a non-fermentable carbon source, the double deletion is lethal.51 This suggests that MTS:Tom20 and 3′-UTR:Puf3 interactions are redundant to each other under conditions that require mitochondrial activity (Fig. 1, denoted as (1) and (2)). Puf3 was also shown to play a role in regulation of translation and mRNA stability,73-76 functions that might also be significant for mRNA localization near the mitochondria. In particular, differential stabilization of mRNAs is one of the modes through which localization in Drosophila embryos is achieved.8 It is therefore possible that Puf3 mediates mRNA localization by stabilizing mRNAs near the mitochondria, or destabilizing transcripts in the cytosol.
In higher eukaryotes, proteins that mediate the association with mitochondria have been identified only recently, in a comprehensive work that analyzed Drosophila and mammalian cells30 (Fig. 1B). The outer membrane kinase PINK1 was found to regulate the localized translation of some mRNAs that encode members of the oxidative phosphorylation complexes. These mRNAs are maintained in a translationally repressed state near mitochondria. Repression is relieved upon association with Parkin and subsequent release of a PUF family protein (Pum) and Gro/hnRNP-F. Interestingly, as in yeast,51 the protein receptor Tom20 was found to be important for mRNA localization. Furthermore, Tom20 was associated with PINK1 in a manner that was sensitive to RNase A (i.e. through RNA molecules). Depletion of Tom20 further reduced mRNA localization in PINK1-mutated cells, whereas Tom20 overexpression increased mRNA association with mitochondria. These data reveal a genetic and physical interaction between Tom20 and PINK1 in promoting localized translation. Overall, it appears that the combined functions of PINK1, Tom20, and Pum are important for the localization of translationally repressed mRNAs30 (Fig. 1B).
The proteins that transport mRNA, ribosomes, and translation factors to the mitochondria are largely unknown. In yeast, however, investigations of the Hsp70 chaperone Ssa1 provided some insights. An analysis of cells with reduced levels of this protein revealed a decreased association between mitochondria and certain mRNAs, many of which encode proteins with a high hydrophobic score.50 These results were consistent with a role for this Hsp70 protein in the folding of hydrophobic domains. Indeed, Ssa1 was shown to interact with RNCs,77 suggesting that its impact on the localization of mRNAs occurs during their translation. Importantly, Ssa1 is a cytosolic chaperone that is absent from the mitochondrial fraction,50 therefore its effects take place during mRNA transport to the mitochondria.
Conservation Among Species
The results and models depicted above are mainly derived from studies that utilize the yeast S. cerevisiae as a model. Nevertheless, various studies suggest that localized translation also occurs in other organisms. For example, mRNAs that encoded a GFP with an N-terminal MTS were shown to be associated with mitochondria isolated from cells of the social amoeba Dictyostelium discoideum. Intriguingly, a mitochondria-mRNA association was apparent also for GFP fusions with an insufficient targeting sequence, thus suggesting that targeting is independent of translation.78 In fractionated potato (Solanum tuberosum) cells, mRNAs of 6 different genes were identified in the mitochondrial fraction.79 More recently, cellular fractionations and in vivo imaging using the MS2-tagging system revealed that VDAC3 mRNAs are associated with mitochondria in Arabidopsis thaliana.80 Interestingly, VDAC3 transcripts that varied in their 3′-UTR length differed in their association, and a region of 140 nts from the 3′-UTR appeared sufficient to induce localization of a heterologous mRNA to the mitochondria. These results indicate that localization of VDAC3 mRNA can occur in a non-translational manner, via cis elements from the 3′-UTR.
Mitochondria from mammalian cells are also associated with mRNA. As mentioned earlier, samples from rat liver revealed that mRNA of F1β-ATPase is localized near mitochondria, in a manner that involves sequences from both the coding region and the 3′-UTR.81 Similarly, a broader analysis in human cell lines of 27 oxidative phosphorylation-related mRNAs revealed that about half of them were localized to the mitochondria.82 This group was significantly enriched in mRNAs that encode hydrophobic, inner-membrane proteins, consistent with data from yeast.32 Gehrke and colleagues also identified a few mRNAs linked with oxidative phosphorylation that were associated with the outer membrane protein PINK1,30 a protein which, together with several regulators of translation, was shown to have a similar function in both human cells and Drosophila muscle cells.
Functional conservation between yeast and human cells was directly demonstrated by Sylvestre and colleagues, who found that the mRNA encoding the inner membrane protein Oxa1 is localized to the mitochondria in both HeLa cells and yeast. Introduction of the human gene into yeast cells led to mitochondrial localization and to the rescue of a respiration defect in oxa1Δ yeast. Importantly, rescue was dependent on the 3′-UTR of the mRNA. Thus, yeast proteins are able to recognize the human mRNA and transport it to the mitochondria.83
Localization to the mitochondria is not restricted to mRNAs that encode inner-membrane proteins. For example, TMEM126A is an outer-membrane protein that is implicated in optic neuropathies. Its mRNA was found to be localized to mitochondria in mouse retina and in various human cell lines.84,85 Whether this localization is important for mitochondrial dysfunction or pathogenesis has yet to be determined.
Functional Relevance
The functional significance of localized translation near the mitochondria has not been fully established. Localized translation is usually linked to the import process, allowing the import of unfolded proteins with a minimal need for protein chaperones. An early study, which used electron microscopy to examine thin sections of yeast cells, indicated a significantly higher degree of binding between ribosomes and the mitochondrial surface at regions where the inner and outer mitochondrial membranes were in close proximity to each other (contact sites);47 these same regions were later found to be enriched with import machinery.86,87 The proximity between mitochondria-associated ribosomes and mitochondrial import was later supported by many analyses of cotranslational import (reviewed by).88,89 This coupling between translation and import appears to be important to proteins that have dual localization (mitochondria and cytoplasm) and must undergo a maturation step inside the mitochondria.63,65
Localized translation has also been suggested to contribute to the proper assembly of protein complexes. For example, mRNAs that encode factors that are important for the assembly of cytochrome C oxidase and components of ATP-synthase are translated near the mitochondria.30,32,90 Intriguingly, some components of these complexes are translated by mitochondrial ribosomes that are associated with the inner membrane.91 This suggests coordination in the synthesis of components from either side of the mitochondria.30,44
Studies from numerous taxa have demonstrated the importance of localized translation to mitochondrial activity. In Arabadopsis thaliana, interference with the localization of mRNA of the membrane protein VDAC3 affected mitochondrial morphology and size80 and in yeast, the correct localization of ATP2 mRNA was shown to be important for respiration.41 In human fibroblasts, induced localization of ATP6 mRNA to the mitochondrial outer membrane resulted in the rescue of mitochondrial defects.92 Despite the widespread occurrence and significance of localized translation, the underlying molecular mechanisms have yet to be determined.
Future Directions
The large amount of data described above, utilizing various methodologies and diverse organisms, demonstrate the extent of localized translation near the mitochondria. As mentioned above, though, one of the main future challenges is to establish the molecular players that mediate this process. Molecularly, localized translation near the mitochondria has been linked to protein import (i.e., cotranslational import), and a few of the proteins that are involved in this association have been identified.30,31, 50,51,72 However, these proteins probably represent only a fraction of the machinery that is necessary to identify, transport, and anchor translating ribosomes to the mitochondria. Furthermore, much remains obscure about the dynamics of the process (e.g., whether translation precedes transport), whether the cytoskeleton and motor proteins are necessary for transport, and whether there is a vectorial insertion of the synthesized polypeptide that utilizes the energy from peptide bond formation. All these questions remain unanswered.
Another important future challenge is to establish the physiological significance of this phenomenon. To date, there have been very few studies that demonstrate the significance of localized translation to cellular physiology. As described above, many studies have linked localized translation with protein import. However, an impact on cellular physiology following a defect in cotranslational import has not been shown. For example, both NAC and Om14 were shown to be important for cotranslational import31,67 yet single-deletion of NAC or om14 did not show severe growth defects.31 This indicates that tools of high resolution are necessary to measure changes in mitochondrial function and to differentiate the effects of cotranslational import from the redundant post-translational import process.
It is likely that localized translation has other roles in organellar physiology, beyond cotranslational import. A plausible possibility is faster and localized regulation of translation. Localized translation machinery can utilize local factors to provide a quick response to mitochondrial needs and on-site regulation of expression. A potential example of such localized regulation was provided recently in,30 in which the association of Parkin with PINK1 led to the removal of translation repressors from mRNAs that were attached to the mitochondrial outer membrane. In addition, localized translation might serve to provide a secluded site that maintains translation when other cellular compartments are affected. This has been shown previously for translation at the ER upon viral infection or hypoxic stress,5,6 and might occur near the mitochondria as well. Finally, the association of mRNAs with yeast ER was suggested to be important for the inheritance of mRNAs and translation factors to the daughter cell (reviewed in).93,94 This might also be the case for the mRNAs that are associated with mitochondria. Moreover, association of the translation machinery with the mitochondria might facilitate its concordant distribution with mitochondria during fission and fusion events, and during cellular divisions. All these possibilities provide advantages to the cell and may improve its physiological efficiency. None of these, however, has been established for localized translation near the mitochondria. Therefore, future studies should look beyond cotranslational import to explore other facets of this phenomenon.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Work in our lab is supported by grants from the Israel Science Foundation (1096/13), Binational Science Foundation (2011013) and the Russell Berrie Nanotechnology Institute (RBNI) and Life Science and Engineering program.
References
- 1.Choi SB, Wang C, Muench DG, Ozawa K, Franceschi VR, Wu Y, Okita TW. Messenger RNA targeting of rice seed storage proteins to specific ER subdomains. Nature 2000. 407(6805):765-7; PMID:11048726; http://dx.doi.org/ 10.1038/35037633 [DOI] [PubMed] [Google Scholar]
- 2.Washida H, Sugino A, Doroshenk KA, Satoh-Cruz M, Nagamine A, Katsube-Tanaka T, Ogawa M, Kumamaru T, Satoh H, Okita TW. RNA targeting to a specific ER sub-domain is required for efficient transport and packaging of α-globulins to the protein storage vacuole in developing rice endosperm. Plant J 2012. 70(3):471-9; PMID:22168839; http://dx.doi.org/ 10.1111/j.1365-313X.2011.04880.x [DOI] [PubMed] [Google Scholar]
- 3.Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol 2008. 9(12):971-80; PMID:19023284; http://dx.doi.org/ 10.1038/nrm2548 [DOI] [PubMed] [Google Scholar]
- 4.Reid DW, Chen Q, Tay AS, Shenolikar S, Nicchitta CV. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell 2014. 158(6):1362-74; PMID:25215492; http://dx.doi.org/ 10.1016/j.cell.2014.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lerner RS, Nicchitta CV. mRNA translation is compartmentalized to the endoplasmic reticulum following physiological inhibition of cap-dependent translation. RNA 2006. 12(5):775-89; PMID:16540694; http://dx.doi.org/ 10.1261/rna.2318906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Staudacher JJ, Naarmann-de Vries IS, Ujvari SJ, Klinger B, Kasim M, Benko E, Ostareck-Lederer A, Ostareck DH, Bondke Persson A, Lorenzen S, et al.. Hypoxia-induced gene expression results from selective mRNA partitioning to the endoplasmic reticulum. Nucleic Acids Res 2015. 43(6):3219-36; PMID:25753659; http://dx.doi.org/ 10.1093/nar/gkv167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science 2009. 326(5957):1212-6; PMID:19965463; http://dx.doi.org/ 10.1126/science.1176488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medioni C, Mowry K, Besse F. Principles and roles of mRNA localization in animal development. Development 2012. 139(18):3263-76; PMID:22912410; http://dx.doi.org/ 10.1242/dev.078626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007. 131(1):174-87; PMID:17923096; http://dx.doi.org/ 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 10.Aronov S, Gelin-Licht R, Zipor G, Haim L, Safran E, Gerst JE. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol 2007. 27(9):3441-55; PMID:17339339; http://dx.doi.org/ 10.1128/MCB.01643-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heym RG, Niessing D. Principles of mRNA transport in yeast. Cell Mol Life Sci 2012. 69(11):1843-53; PMID:22159587; http://dx.doi.org/ 10.1007/s00018-011-0902-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singer-Kruger B, Jansen RP. Here, there, everywhere. mRNA localization in budding yeast. RNA Biol 2014. 11(8):1031-9; PMID:25482891; http://dx.doi.org/ 10.4161/rna.29945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jansen RP, Niessing D, Baumann S, Feldbrügge M. mRNA transport meets membrane traffic. Trends Genet 2014. 30(9):408-17; PMID:25110341; http://dx.doi.org/ 10.1016/j.tig.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 14.Holt CE, Schuman EM. The central dogma decentralized: new perspectives on RNA function and local translation in neurons. Neuron 2013. 80(3):648-57; PMID:24183017; http://dx.doi.org/ 10.1016/j.neuron.2013.10.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Condeelis J, Singer RH. How and why does β-actin mRNA target? Biol Cell 2005. 97(1):97-110; PMID:15601261; http://dx.doi.org/ 10.1042/BC20040063 [DOI] [PubMed] [Google Scholar]
- 16.Bullock SL. Messengers, motors and mysteries: sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem Soc Trans 2011. 39(5):1161-5; PMID:21936782; http://dx.doi.org/ 10.1042/BST0391161 [DOI] [PubMed] [Google Scholar]
- 17.Weis BL, Schleiff E, Zerges W. Protein targeting to subcellular organelles via MRNA localization. Biochim Biophys Acta 2013. 1833(2):260-73; PMID:23457718; http://dx.doi.org/ 10.1016/j.bbamcr.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 18.Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA 2003. 9(9):1123-37; PMID:12923260; http://dx.doi.org/ 10.1261/rna.5610403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephens SB, Dodd RD, Brewer JW, Lager PJ, Keene JD, Nicchitta CV. Stable ribosome binding to the endoplasmic reticulum enables compartment-specific regulation of mRNA translation. Mol Biol Cell 2005. 16(12):5819-31; PMID:16221886; http://dx.doi.org/ 10.1091/mbc.E05-07-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hann BC, Walter The signal recognition particle in S. cerevisiae. Cell 1991. 67(1):131-44; PMID:1655273; http://dx.doi.org/ 10.1016/0092-8674(91)90577-L [DOI] [PubMed] [Google Scholar]
- 21.Cui XA, Palazzo AF. Localization of mRNAs to the endoplasmic reticulum. Wiley Interdiscip Rev RNA 2014. 5(4):481-92; PMID:24644132; http://dx.doi.org/ 10.1002/wrna.1225 [DOI] [PubMed] [Google Scholar]
- 22.Loya A, Pnueli L, Yosefzon Y, Wexler Y, Ziv-Ukelson M, Arava Y. The 3′-UTR mediates the cellular localization of an mRNA encoding a short plasma membrane protein. RNA 2008. 14(7):1352-65; PMID:18492794; http://dx.doi.org/ 10.1261/rna.867208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pyhtila B, Zheng T, Lager PJ, Keene JD, Reedy MC, Nicchitta CV. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA 2008. 14(3):445-53; PMID:18192611; http://dx.doi.org/ 10.1261/rna.721108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui XA, Zhang H, Palazzo AF. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol 2012. 10(5):e1001336; PMID:22679391; http://dx.doi.org/ 10.1371/journal.pbio.1001336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kraut-Cohen J, Afanasieva E, Haim-Vilmovsky L, Slobodin B, Yosef I, Bibi E, Gerst JE. Translation- and SRP-independent mRNA targeting to the endoplasmic reticulum in the yeast Saccharomyces cerevisiae. Mol Biol Cell 2013. 24(19):3069-84; PMID:23904265; http://dx.doi.org/ 10.1091/mbc.E13-01-0038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palade GE. A small particulate component of the cytoplasm. J Biophys Biochem Cytol 1955. 1(1):59-68; PMID:14381428; http://dx.doi.org/ 10.1083/jcb.1.1.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reid DW, Nicchitta CV. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat Rev Neurosci 2015; 16(4):221-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zipor G, Haim-Vilmovsky L, Gelin-Licht R, Gadir N, Brocard C, Gerst JE. Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2009. 106(47):19848-53; PMID:19903887; http://dx.doi.org/ 10.1073/pnas.0910754106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian L, Okita TW. mRNA-based protein targeting to the endoplasmic reticulum and chloroplasts in plant cells. Curr Opin Plant Biol 2014. 22:77-85; PMID:25282588; http://dx.doi.org/ 10.1016/j.pbi.2014.09.007 [DOI] [PubMed] [Google Scholar]
- 30.Gehrke S, Wu Z, Klinkenberg M, Sun Y, Auburger G, Guo S, Lu B. PINK1 and Parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab 2015. 21(1):95-108; PMID:25565208; http://dx.doi.org/ 10.1016/j.cmet.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lesnik C, Cohen Y, Atir-Lande A, Schuldiner M, Arava Y. OM14 is a mitochondrial receptor for cytosolic ribosomes that supports co-translational import into mitochondria. Nat Commun 2014. 5:5711; PMID:25487825; http://dx.doi.org/ 10.1038/ncomms6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science 2014. 346(6210):748-51; PMID:25378625; http://dx.doi.org/ 10.1126/science.1257522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt O, Pfanner N, Meisinger C. Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol 2010. 11(9):655-67; PMID:20729931; http://dx.doi.org/ 10.1038/nrm2959 [DOI] [PubMed] [Google Scholar]
- 34.Wenz LS, Opaliński Ł, Wiedemann N, Becker T. Cooperation of protein machineries in mitochondrial protein sorting. Biochim Biophys Acta 2015. 1853(5):1119-1129; PMID:25633533; http://dx.doi.org/ 10.1016/j.bbamcr.2015.01.012 [DOI] [PubMed] [Google Scholar]
- 35.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol 2012. 4(9):a012286; PMID:22763747; http://dx.doi.org/ 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Egea G, Izquierdo JM, Ricart J, San Martín C, Cuezva JM. mRNA encoding the β-subunit of the mitochondrial F1-ATPase complex is a localized mRNA in rat hepatocytes. Biochem J 1997. 322 ( Pt 2):557-65; PMID:9065777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricart J, Egea G, Izquierdo JM, San Martín C, Cuezva JM. Subcellular structure containing mRNA for β subunit of mitochondrial H+-ATP synthase in rat hepatocytes is translationally active. Biochem J 1997. 324 ( Pt 2):635-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corral-Debrinski M, Blugeon C, Jacq C. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol Cell Biol 2000. 20(21):7881-92; PMID:11027259; http://dx.doi.org/ 10.1128/MCB.20.21.7881-7892.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marc P, Margeot A, Devaux F, Blugeon C, Corral-Debrinski M, Jacq C. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep 2002. 3(2):159-64; PMID:11818335; http://dx.doi.org/ 10.1093/embo-reports/kvf025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sylvestre J, Vialette S, Corral Debrinski M, Jacq C. Long mRNAs coding for yeast mitochondrial proteins of prokaryotic origin preferentially localize to the vicinity of mitochondria. Genome Biol 2003. 4(7):R44; PMID:12844360; http://dx.doi.org/ 10.1186/gb-2003-4-7-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margeot A, Blugeon C, Sylvestre J, Vialette S, Jacq C, Corral-Debrinski M. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J 2002. 21(24):6893-904; PMID:12486010; http://dx.doi.org/ 10.1093/emboj/cdf690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haim L, Zipor G, Aronov S, Gerst JE. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods 2007. 4(5):409-12; PMID:17417645 [DOI] [PubMed] [Google Scholar]
- 43.Gadir N, Haim-Vilmovsky L, Kraut-Cohen J, Gerst JE. Localization of mRNAs coding for mitochondrial proteins in the yeast Saccharomyces cerevisiae. RNA 2011. 17(8):1551-65; PMID:21705432; http://dx.doi.org/ 10.1261/rna.2621111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia M, Darzacq X, Delaveau T, Jourdren L, Singer RH, Jacq C. Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes. Mol Biol Cell 2007. 18(2):362-8; PMID:17108321; http://dx.doi.org/ 10.1091/mbc.E06-09-0827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kellems RE, Butow RA. Cytoplasmic-type 80 S ribosomes associated with yeast mitochondria. I. Evidence for ribosome binding sites on yeast mitochondria. J Biol Chem 1972. 247(24):8043-50; PMID:4629740 [PubMed] [Google Scholar]
- 46.Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J Biol Chem 1974. 249(10):3297-303; PMID:4598123 [PubMed] [Google Scholar]
- 47.Kellems RE, Allison VF, Butow RA. Cytoplasmic type 80S ribosomes associated with yeast mitochondria. IV. Attachment of ribosomes to the outer membrane of isolated mitochondria. J Cell Biol 1975. 65(1):1-14; PMID:1092698; http://dx.doi.org/ 10.1083/jcb.65.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacKenzie JA, Payne RM. Ribosomes specifically bind to mammalian mitochondria via protease-sensitive proteins on the outer membrane. J Biol Chem 2004. 279(11):9803-10; PMID:14668341; http://dx.doi.org/ 10.1074/jbc.M307167200 [DOI] [PubMed] [Google Scholar]
- 49.Crowley KS, Payne RM. Ribosome binding to mitochondria is regulated by GTP and the transit peptide. J Biol Chem 1998. 273(27):17278-85; PMID:9642299; http://dx.doi.org/ 10.1074/jbc.273.27.17278 [DOI] [PubMed] [Google Scholar]
- 50.Eliyahu E, Lesnik C, Arava Y. The protein chaperone Ssa1 affects mRNA localization to the mitochondria. FEBS Lett 2012. 586(1):64-9; PMID:22138184; http://dx.doi.org/ 10.1016/j.febslet.2011.11.025 [DOI] [PubMed] [Google Scholar]
- 51.Eliyahu E, Pnueli L, Melamed D, Scherrer T, Gerber AP, Pines O, Rapaport D, Arava Y. Tom20 mediates localization of mRNAs to mitochondria in a translation-dependent manner. Mol Cell Biol 2010. 30(1):284-94; PMID:19858288; http://dx.doi.org/ 10.1128/MCB.00651-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.George R, Walsh P, Beddoe T, Lithgow T. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett 2002. 516(1–3):213-6; PMID:11959135; http://dx.doi.org/ 10.1016/S0014-5793(02)02528-0 [DOI] [PubMed] [Google Scholar]
- 53.Lesnik C, Arava Y. Isolation of mRNAs associated with yeast mitochondria to study mechanisms of localized translation. J Vis Exp 2014; 85; PMID:24686138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ades IZ, Butow RA. The products of mitochondria-bound cytoplasmic polysomes in yeast. J Biol Chem 1980; 255(20):9918-24; PMID:6448841 [PubMed] [Google Scholar]
- 55.Suissa M, Schatz G. Import of proteins into mitochondria. Translatable mRNAs for imported mitochondrial proteins are present in free as well as mitochondria-bound cytoplasmic polysomes. J Biol Chem 1982. 257(21):13048-55; PMID:6752146 [PubMed] [Google Scholar]
- 56.Ni L, Heard TS, Weiner H. In vivo mitochondrial import. A comparison of leader sequence charge and structural relationships with the in vitro model resulting in evidence for co-translational import. J Biol Chem 1999. 274(18):12685-91; PMID:10212250; http://dx.doi.org/ 10.1074/jbc.274.18.12685 [DOI] [PubMed] [Google Scholar]
- 57.Mukhopadhyay A, Ni L, Weiner H. A co-translational model to explain the in vivo import of proteins into HeLa cell mitochondria. Biochem J 2004. 382(Pt 1):385-92; PMID:15153070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujiki M, Verner K. Coupling of cytosolic protein synthesis and mitochondrial protein import in yeast. Evidence for cotranslational import in vivo. J Biol Chem 1993. 268(3):1914-20; PMID:8380582 [PubMed] [Google Scholar]
- 59.Fujiki M, Verner K. Coupling of protein synthesis and mitochondrial import in a homologous yeast in vitro system. J Biol Chem 1991. 266(11):6841-7; PMID:1849892 [PubMed] [Google Scholar]
- 60.Nobumoto M, Yamada M, Song S, Inouye S, Nakazawa A. Mechanism of mitochondrial import of adenylate kinase isozymes. J Biochem 1998. 123(1):128-35; PMID:9504419; http://dx.doi.org/ 10.1093/oxfordjournals.jbchem.a021899 [DOI] [PubMed] [Google Scholar]
- 61.Regev-Rudzki N, Pines O. Eclipsed distribution: a phenomenon of dual targeting of protein and its significance. Bioessays 2007. 29(8):772-82; PMID:17621655; http://dx.doi.org/ 10.1002/bies.20609 [DOI] [PubMed] [Google Scholar]
- 62.Ben-Menachem R, Tal M, Shadur T, Pines O. A third of the yeast mitochondrial proteome is dual localized: a question of evolution. Proteomics 2011. 11(23):4468-76; PMID:21910249; http://dx.doi.org/ 10.1002/pmic.201100199 [DOI] [PubMed] [Google Scholar]
- 63.Stein I, Peleg Y, Even-Ram S, Pines O. The single translation product of the FUM1 gene (fumarase) is processed in mitochondria before being distributed between the cytosol and mitochondria in Saccharomyces cerevisiae. Mol Cell Biol 1994. 14(7):4770-8; PMID:8007976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knox C, Sass E, Neupert W, Pines O. Import into mitochondria, folding and retrograde movement of fumarase in yeast. J Biol Chem 1998. 273(40):25587-93; PMID:9748223; http://dx.doi.org/ 10.1074/jbc.273.40.25587 [DOI] [PubMed] [Google Scholar]
- 65.Yogev O, Karniely S, Pines O. Translation-coupled translocation of yeast fumarase into mitochondria in vivo. J Biol Chem 2007. 282(40):29222-29229; PMID:17666392; http://dx.doi.org/ 10.1074/jbc.M704201200 [DOI] [PubMed] [Google Scholar]
- 66.Kaltimbacher V, Bonnet C, Lecoeuvre G, Forster V, Sahel JA, Corral-Debrinski M. mRNA localization to the mitochondrial surface allows the efficient translocation inside the organelle of a nuclear recoded ATP6 protein. RNA 2006. 12(7):1408-17; PMID:16751614; http://dx.doi.org/ 10.1261/rna.18206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Funfschilling U, Rospert S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol Biol Cell 1999. 10(10):3289-99; PMID:10512867; http://dx.doi.org/ 10.1091/mbc.10.10.3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Preissler S, Deuerling E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci 2012. 37(7):274-83; PMID:22503700; http://dx.doi.org/ 10.1016/j.tibs.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 69.del Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol 2011. 9(7):e1001100; PMID:21765803; http://dx.doi.org/ 10.1371/journal.pbio.1001100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2004. 2(3):E79; PMID:15024427; http://dx.doi.org/ 10.1371/journal.pbio.0020079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Rodriguez LJ, Gay AC, Pon LA. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J Cell Biol 2007. 176(2):197-207; PMID:17210948; http://dx.doi.org/ 10.1083/jcb.200606054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS One 2008. 3(6):e2293; PMID:18523582; http://dx.doi.org/ 10.1371/journal.pone.0002293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rowe W, Kershaw CJ, Castelli LM, Costello JL, Ashe MP, Grant CM, Sims PF, Pavitt GD, Hubbard SJ. Puf3p induces translational repression of genes linked to oxidative stress. Nucleic Acids Res 2014. 42(2):1026-41; PMID:24163252; http://dx.doi.org/ 10.1093/nar/gkt948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller MA, Russo J, Fischer AD, Lopez Leban FA, Olivas WM. Carbon source-dependent alteration of Puf3p activity mediates rapid changes in the stabilities of mRNAs involved in mitochondrial function. Nucleic Acids Res 2014. 42(6):3954-70; PMID:24371272; http://dx.doi.org/ 10.1093/nar/gkt1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA 2011. 2(4):471-92; PMID:21957038; http://dx.doi.org/ 10.1002/wrna.69 [DOI] [PubMed] [Google Scholar]
- 76.Jiang H, Xu L, Wang Z, Keene J, Gu Z. Coordinating expression of RNA binding proteins with their mRNA targets. Sci Rep 2014. 4:7175; PMID:25417751; http://dx.doi.org/ 10.1038/srep07175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 2006. 124(1):75-88; PMID:16413483; http://dx.doi.org/ 10.1016/j.cell.2005.11.039 [DOI] [PubMed] [Google Scholar]
- 78.Ahmed AU, Beech PL, Lay ST, Gilson PR, Fisher PR. Import-associated translational inhibition: novel in vivo evidence for cotranslational protein import into Dictyostelium discoideum mitochondria. Eukaryot Cell 2006. 5(8):1314-27; PMID:16896215; http://dx.doi.org/ 10.1128/EC.00386-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Michaud M, Marechal-Drouard L, Duchene AM. RNA trafficking in plant cells: targeting of cytosolic mRNAs to the mitochondrial surface. Plant Mol Biol 2010. 73(6):697-704; PMID:20506035; http://dx.doi.org/ 10.1007/s11103-010-9650-3 [DOI] [PubMed] [Google Scholar]
- 80.Michaud M, Ubrig E, Filleur S, Erhardt M, Ephritikhine G, Maréchal-Drouard L, Duchêne AM. Differential targeting of VDAC3 mRNA isoforms influences mitochondria morphology. Proc Natl Acad Sci U S A 2014. 111(24):8991-6; PMID:24889622; http://dx.doi.org/ 10.1073/pnas.1402588111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ricart J, Izquierdo JM, Di Liegro CM, Cuezva JM. Assembly of the ribonucleoprotein complex containing the mRNA of the β-subunit of the mitochondrial H+-ATP synthase requires the participation of two distal cis-acting elements and a complex set of cellular trans-acting proteins. Biochem J 2002. 365(Pt 2):417-28; PMID:11952427; http://dx.doi.org/ 10.1042/BJ20011726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Matsumoto S, Uchiumi T, Saito T, Yagi M, Takazaki S, Kanki T, Kang D. Localization of mRNAs encoding human mitochondrial oxidative phosphorylation proteins. Mitochondrion 2012. 12(3):391-8; PMID:22406259; http://dx.doi.org/ 10.1016/j.mito.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 83.Sylvestre J, Margeot A, Jacq C, Dujardin G, Corral-Debrinski M. The role of the 3′ untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells. Mol Biol Cell 2003. 14(9):3848-56; PMID:12972568; http://dx.doi.org/ 10.1091/mbc.E03-02-0074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hanein S, Garcia M, Fares-Taie L, Serre V, De Keyzer Y, Delaveau T, Perrault I, Delphin N, Gerber S, Schmitt A. TMEM126A is a mitochondrial located mRNA (MLR) protein of the mitochondrial inner membrane. Biochim Biophys Acta 2013. 1830(6):3719-33; PMID:23500070; http://dx.doi.org/ 10.1016/j.bbagen.2013.02.025 [DOI] [PubMed] [Google Scholar]
- 85.Hanein S, Perrault I, Roche O, Gerber S, Khadom N, Rio M, Boddaert N, Jean-Pierre M, Brahimi N, Serre V. TMEM126A, encoding a mitochondrial protein, is mutated in autosomal-recessive nonsyndromic optic atrophy. Am J Hum Genet 2009. 84(4):493-8; PMID:19327736; http://dx.doi.org/ 10.1016/j.ajhg.2009.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schleyer M, Neupert W. Transport of proteins into mitochondria: translocational intermediates spanning contact sites between outer and inner membranes. Cell 1985. 43(1):339-50; PMID:2866845; http://dx.doi.org/ 10.1016/0092-8674(85)90039-X [DOI] [PubMed] [Google Scholar]
- 87.Pon L, Moll T, Vestweber D, Marshallsay B, Schatz G. Protein import into mitochondria: ATP-dependent protein translocation activity in a submitochondrial fraction enriched in membrane contact sites and specific proteins. J Cell Biol 1989. 109(6 Pt 1):2603-16; PMID:2556402; http://dx.doi.org/ 10.1083/jcb.109.6.2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ahmed AU, Fisher PR. Import of nuclear-encoded mitochondrial proteins: a cotranslational perspective. Int Rev Cell Mol Biol 2009. 273:49-68; PMID:19215902 [DOI] [PubMed] [Google Scholar]
- 89.Verner K. Co-translational protein import into mitochondria: an alternative view. Trends Biochem Sci 1993. 18(10):366-71; PMID:8256283; http://dx.doi.org/ 10.1016/0968-0004(93)90090-A [DOI] [PubMed] [Google Scholar]
- 90.Margeot A, Garcia M, Wang W, Tetaud E, di Rago JP, Jacq C. Why are many mRNAs translated to the vicinity of mitochondria: a role in protein complex assembly? Gene 2005. 354:64-71; PMID:15979254; http://dx.doi.org/ 10.1016/j.gene.2005.04.022 [DOI] [PubMed] [Google Scholar]
- 91.Mick DU, Fox TD, Rehling Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat Rev Mol Cell Biol 2011. 12(1):14-20; PMID:21179059; http://dx.doi.org/ 10.1038/nrm3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bonnet C, Kaltimbacher V, Ellouze S, Augustin S, Bénit P, Forster V, Rustin P, Sahel JA, Corral-Debrinski M. Allotopic mRNA localization to the mitochondrial surface rescues respiratory chain defects in fibroblasts harboring mitochondrial DNA mutations affecting complex I or v subunits. Rejuvenation Res 2007. 10(2):127-44; PMID:17518546; http://dx.doi.org/ 10.1089/rej.2006.0526 [DOI] [PubMed] [Google Scholar]
- 93.Hermesh O, Jansen RP. Take the (RN)A-train: localization of mRNA to the endoplasmic reticulum. Biochim Biophys Acta 2013. 1833(11):2519-25; PMID:23353632; http://dx.doi.org/ 10.1016/j.bbamcr.2013.01.013 [DOI] [PubMed] [Google Scholar]
- 94.Gerst JE. Message on the web: mRNA and ER co-trafficking. Trends Cell Biol 2008. 18(2):68-76; PMID:18215524; http://dx.doi.org/ 10.1016/j.tcb.2007.11.005 [DOI] [PubMed] [Google Scholar]


