Abstract
A major obstacle for effective utilization of therapeutic oligonucleotides such as siRNA, antisense, antimiRs etc. is to deliver them specifically to the target tissues. Toward this goal, nucleic acid aptamers are re-emerging as a prominent class of biomolecules capable of delivering target specific therapy and therapeutic monitoring by various molecular imaging modalities. This class of short oligonucleotide ligands with high affinity and specificity are selected from a large nucleic acid pool against a molecular target of choice. Poor cellular uptake of therapeutic oligonucleotides impedes gene-targeting efficacy in vitro and in vivo. In contrast, aptamer-oligonucleotide chimeras have shown the capacity to deliver siRNA, antimiRs, small molecule drugs etc. toward various targets and showed very promising results in various studies on different diseases models. However, to further improve the bio-stability of such chimeric conjugates, it is important to introduce chemically-modified nucleic acid analogs. In this review, we highlight the applications of nucleic acid aptamers for target specific delivery of therapeutic oligonucleotides.
Keywords: aptamers, siRNA delivery, Aptamer targeted delivery, aptamer chimera, modified nucleotides, miRNA delivery
Introduction
Technological advancement in targeting and delivery of therapies to the site of action within a patient could greatly improve both the standard of living for a patient, as well as decrease costs associated with wasted therapeutics. Toward this goal, nucleic acid aptamers, often termed as chemical antibodies, are an emerging class of synthetic ligands, recently attracted significant attention in various fields.1,2 This class of short single-stranded functional nucleic acids can fold into complex 3-dimensional shapes that can adopt binding pockets and clefts for specific high-affinity recognition of defined molecular targets ranging from small molecules to large proteins and even whole cells. These characteristics make aptamers an attractive platform for applications relating to drug delivery, biosensing and theranostics. During the first decade after the discovery, aptamers gained their foothold in therapeutic development.1,2 In 2004, vascular endothelial growth factor (VEGF) targeting RNA aptamer (Mucagen or Pegaptanib sodium) was approved by the Food and Drug Administration (FDA) for age related macular degeneration.3
Aptamers are typically generated from a large oligonucleotide pool (∼1015 members) via an in vitro reiterative combinatorial selection process called Systematic Evolution of Ligands by EXponential enrichments (SELEX, Fig. 1).4-9 Although this process generally takes around 2–6 months, there are few reports of single or limited step aptamer selection protocols.10-13 It is noteworthy mentioning that aptamer selection procedure may sound simple enough, however, it may not be as straightforward. In some cases, often there may not be any aptamers depending on the diversity of the starting nucleic acid pool, or sometimes the developed aptamers may not be as specific as necessary even with proper negative control selections. Aptamers may possess several advantages over conventional antibody-based therapeutic approaches. First of all, aptamers do not require live animals for production as these can easily be synthesized in a synthetic laboratory setting in very large scale.14 Aptamer synthesis is not vulnerable to bacterial or viral contaminations. They generally have longer shelf-lives and are non-immunogenic, because aptamers are small in size, can easily access protein epitopes and also show better internalization, which is more difficult for large molecules such as antibodies.15,16 Additionally, aptamers offer freedom to introduce chemical modifications for conjugating additional chemical functionalities and also for systematic truncations of the parent aptamer itself.
Figure 1.
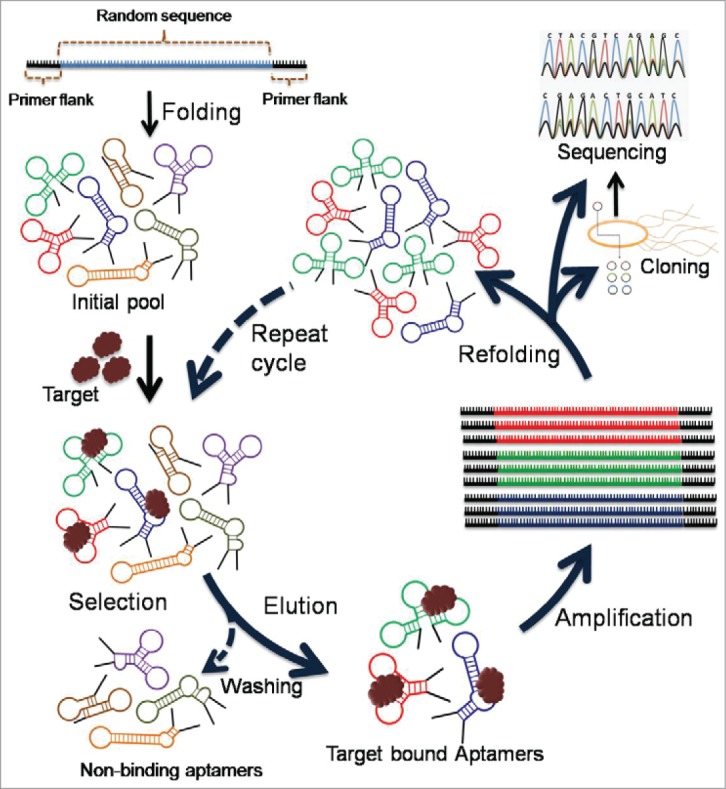
Schematic illustration of aptamer selection procedures by SELEX.
Extremely promising approaches that has evolved during the last decade are the use of RNA interference (RNAi)17,18 using short interfering RNA (siRNA),19 antisense oligo (ASO)20 for silencing gene expression, and targeting microRNAs (miRNA)19-21 that are responsible for several diseases including tumor development. However, while siRNA, antisense and miRNA targeting therapies provide alternatives to conventional chemotherapies, significant hurdles related to the delivery and efficacy of treatment must still be overcome before this technology can be fully utilized. Indeed, in an in vivo setting, the application of nucleic acid-based technologies have been complicated by poor serum stability (due to the presence of nucleases), off-target effects and inability to gain sufficient concentration at the required target site. Thus, it is clear that innovative methods of both packaging, delivery and targeting oligonucleotide therapies are required to advance this technology that has shown such huge promise in vitro. One promising strategy would be to develop and use aptamers targeting cell-surface receptors for effective cellular uptake via receptor-mediated endocytosis.22 In this regard aptamer selection against particular cells in vitro (Cell–SELEX)22-26 and against particular tissues in vivo (in vivo Selection,27 Fig. 2) would be very advantageous.
Figure 2.
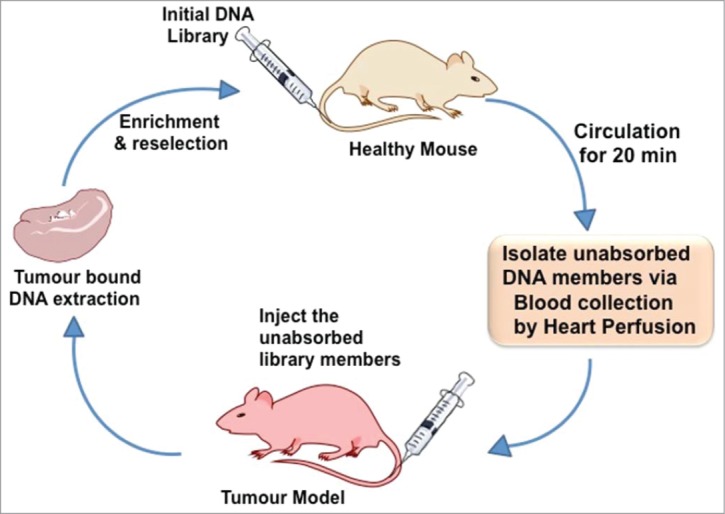
Principles of in vivo aptamer selection.
Aptamers as Tools for siRNA Delivery
RNA interference (RNAi) is a biological process that occurs at the molecular level and mediates gene silencing among the post-transcriptional modification process.18 RNAi has been harnessed for several years to cease the function of several genes for therapeutic purposes toward various diseases.17,28,29 A major obstacle for developing siRNA as therapeutic agents is to deliver them specifically to particular tissues.30 Many scientists aimed to solve this problem by investigating different guidance systems for siRNA, ranging from small molecules, lipids, peptides and synthetic nanostructures.31-34 Aptamers, chemical (non-protein) antibodies, are emerging as a promising tool for delivering siRNA.35
With the dawn of new millennium, the application of aptamers was further extended to target specific delivery of therapeutic compounds.36 Due to their low immunogenicity, ease of production, freedom for chemical alteration and high target specificity, the scientific community quickly accepted this concept. Since then, the application of aptamers for delivering siRNA has been widely explored. For example, in cancer therapy, aptamers have shown great potential to deliver siRNA specifically to tumor cells, minimizing the cytotoxicity to normal cells and harsh side effects of chemotherapeutic drugs.37 Functional aptamer-siRNA chimera toward a wide range of diseases have been developed in recent years, making aptamer-siRNA chimeras one of the most rapidly growing class of therapeutics (Fig. 3 describes a possible mechanism of aptamer-siRNA chimera mediated gene silencing).
Figure 3.
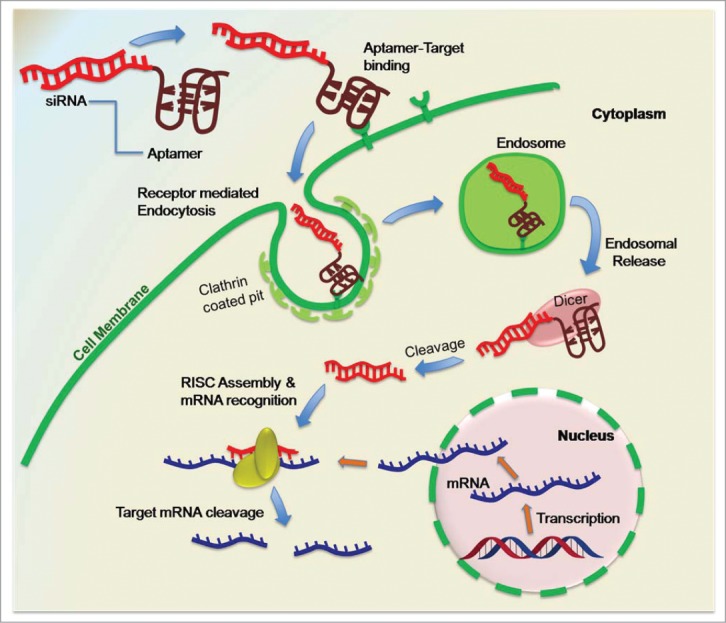
Aptamer-siRNA mediated gene silencing approach.
Chu and colleagues were among the first to perform a functional delivery of siRNA using an aptamer in 2006.38 In this work, they used aptamers against prostate-specific membrane antigen (PSMA). The aptamers A9 and A10 were reported to be capable of transporting nanoparticle into the cells expressing PSMA.39 Streptavidin–biotin interaction was utilized to construct an aptamer-siRNA chimera in which 2 biotinylated anti-PSMA aptamers were connected to 2 biotinylated siRNAs. These conjugates were not only able to deliver the siRNA efficiently to PSMA-expressing LNCaP cells in vitro but also decreased the amount of target mRNA expression level. In the same year, McNamara and colleagues reported the delivery of siRNA targeting polo-like kinase 1 (PLK1) and BCL2 to PSMA-positive LNCaP prostate cancer cells by using PSMA binding RNA aptamer A10.40 This remarkable work clearly demonstrated that the aptamer-guided siRNA delivery system efficiently decreased the proliferation of prostate cancer cells and apoptosis.
In 2008, Zhou and colleagues developed an aptamer-siRNA delivery system with dual inhibitory function for HIV-1 therapy.41 The dual inhibitory function means that both the aptamer and the siRNA components have potent anti-HIV activities, making this capable of targeting the disease at 2 different levels. In this work, they used an anti-gp120 RNA aptamer, targeting the gp120 glycoprotein, a surface protein on the virion that largely determines the entry of HIV into cells, its cellular tropism as well as elements of its pathogenesis.41-44 The aptamer itself is able to bind this protein and neutralize the strains.45 The other part of the chimera contains an anti-tat/rev siRNA that inhibits HIV replication. Zhou et al. showed that the aptamer-siRNA chimera was able to utilize gp120 expressed on HIV infected cells for the delivery of its anti-HIV siRNA. This study demonstrates vast potential of aptamer-siRNA chimeras, because it uses the full capacity of an aptamer and leading the technology from just a target specific ligand to a full therapeutic tool to significantly increase the therapeutic efficacy.
For efficient endocytosis, it has been suggested that multiple ligands to receptor binding may be needed to meet the required free energy for complete wrapping of the membrane.46,47 In regard to this theory, Yoo et al. reported a rod-shaped comb-type aptamer-siRNA chimera.48 In this study, a mucin 1 (MUC1) targeting DNA aptamer was conjugated to the siRNA. MUC1 is a cell surface associated protein, highly over-expressed in malignant adenocarcinomas.49,50 The anti-MUC1-aptamer carrying sense strands of siRNA was hybridized complementary to the multimeric antisense strand to fabricate comb-like-aptamer-siRNA conjugate (Comb-Apt-siR). The intracellular uptake of Comb-Apt-siR in MUC1-positive MCF-7 cells was visually compared to conventional aptamer-siRNA and dimeric aptamer-siRNA conjugates using a red fluorescent dye, POPO-3. Comb-Apt-siR exhibiting the strongest fluorescence, and showed enhanced internalization compared to di- and monomeric aptamer-siRNA conjugates. The enhanced internalization of Comb-Apt-siRNA was explained by its ability to bind multiple receptors on the cell membrane initiating cluster formation leading to efficient endocytosis. The siRNA was designed to target the green fluorescent protein (GFP) gene expression. Despite an enhanced cell uptake, only Comp-Apt-siR inhibited the expression of the GFP gene efficiently, suggesting that the multivalent aptamer-siRNA conjugations might have improved the internalization capabilities compared to the monomers. The mechanism involved in the endosomal release of the chimera after cell entry is not fully understood.
To further improve the efficacy of aptamer-siRNA chimeras, endosome rupturing nanocarrier conjugation can be an alternative. However, Walter et al. showed that the positive net charge of nanomaterials could block the correct folding of an aptamer by triggering it to unfold on the surface.51 Such a conformational change will inhibit any interaction between the aptamer and its target, ultimately destroying its siRNA guiding property. To overcome this problem, Bagalkot and Gao developed a 2-step process using aptamer and siRNA separately to build a functional chimera.52 First, they applied siRNA molecules with a thiol-reactive terminal group to a polyethylene imine coated nanoparticle. This non-covalent interaction reduces some of the positive charge on the carrier. Next, the aptamer containing a single thiol–group was added to form a functional chimera with the nanocarrier bound siRNA. Their approach showed significantly increased gene silencing efficacy compared with conventional one-step assemblies. Recently, a new strategy using a simple protein tag was used to improve the endosome disruption.53 In comparison with nanoparticles, this small protein tag consisted of 2 functional domains; a dsRNA binding domain and a polyhistidine. The dsRNA binding domain binds selectively to the siRNA part of the chimera, and depending on the pH, the polyhistidine induces endosomal membrane disruption. Table 1 summarizes recent efforts on aptamer mediated siRNA delivery for enhanced gene silencing efficacy.
Table 1.
Recent studies on aptamer-targeted siRNA delivery
| Aptamer target | Component | siRNA-Target | In vivo/in vitro target | Aptamer-siRNA linkage | Reference | Further Information |
|---|---|---|---|---|---|---|
| prostate-specific membrane antigen (PSMA) | 2'-Fluoro RNA | Lamin A/C or GAPDH | LNCaP cells | Biotinylated siRNA / aptamer linked by streptavidin | Chu et al., 2006 (38) | |
| 2'-Fluoro RNA | Polo-like kinase1 (PLK1) or BCL2 mRNA | LNCaP cells | Conjugated via combined transcription | McNamara et al., 2006 (40) | ||
| 2'-Fluoro RNA | Polo-like kinase1 (PLK1) or BCL2 mRNA | athymic nude mice | Multiple linking methods | Dassie et al., 2009 (54) | ||
| 2'-Fluoro RNA | Eukaryotic Elongation Factor 2 (EEF2) mRNA | LNCaP cells | Wullner et al., 2008 (55) | |||
| 2'-Fluoro RNA | shRNA: Bcl-xL (anti-apoptotic gene) | LNCAP & PC3 cells | Branched polyethyleneimine (PEI) and polyethylene glycol bridge | Kim et al., 2010 (56) | PSMA aptamer-conjugated PEIePEG (PEIePEGeAPT) | |
| DNA | Smg1 and Upf2 (factors of nonsense-mediated mRNA) | B16/F10 & CT26 tumor cells & Balb/c or Nude mice | Conjugated via combined transcription | Pastor et al., 2010 (57) | ||
| 2'-Fluoro RNA | shRNA: DNA-activated protein kinase (DNAPK); mitotic spindle assembly checkpoint protein MAD2B (MAD2L2); and breast cancer type 2 susceptibility protein (BRCA2) | LNCaP cells | A10-3 aptamer inserted in the loop region of shRNA | Ni et al., 2011 (58) | radiosensitization | |
| RNA | Enhanced green fluorescent protein (EGFP) | Human prostate cancer cell line C4-2B | SPDP crosslinker | Bagalkot et al., 2011 (52) | siRNA-Aptamer Chimeras on QD Nanoparticles | |
| Human epidermal growth factor receptor 2 (HER2) | 2'-Fluoro RNA | Anti-apoptotic gene Bcl-2 | N202.1A cells | Conjugated via combined transcription | Thiel et al., 2012 (59) | |
| CD4 | 2'-Fluoro RNA | Firefly luciferase mRNA | CD4 overexpressing T-cells | Dimerization using phi29 Motor pRNA | Guo et al., 2005 (60) | |
| 2'-Fluoro RNA | Survivin &firefly luciferase mRNA | CD4 overexpressing T-cells | Dimerization using phi29 Motor pRNA | Khaled et al., 2005 (61) | ||
| 2'-Fluoro RNA | gag and vif or host CCR5 | CD4 overexpressing T-cells | Conjugated via combined transcription | Wheeler et al., 2011 (62) | ||
| 2'-Fluoro RNA | gag and vif or host CCR5 | NOD/SCID/IL2Rγ−/− (NSG) mice | Conjugated via combined transcription | Wheeler et al., 2013 (63) | ||
| DNA | HIV-PR | CD4 overexpressing T-cells | Commercial synthesis | Zhu et al., 2012 (64) | ||
| RNA | Asthma STAT5b gen | CD4 overexpressing T-cells | Dimerization using phi29 Motor pRNA | Qiu et al., 2012 (65) | ||
| HIV-1 gp120 | 2' -Fluoro RNA | HIV-1 tat/rev common exon sequence | HIV-1-infected CEM cells & HIV-1 infected Rag-Hu mouse | 4-nucleotide linker (CUCU) | Zhou et al., 2008 (41) | |
| 2′-OMe modified A and G and 2′-F modified U and C | HIV-1 tat/rev common exon sequence | CEM T-cells & primary blood mononuclear cells (PBMCs) | Non-covalent via sticky bridge | Zhou et al., 2009 (66) | ||
| 2'-Fluoro RNA | HIV-1 tat/rev common exon sequence | CD4+ T & Humanized BALB/c-Rag2−/−γc−/− mice | 2-nucleotide linker (UU) | Neff et al., 2011 (67) | ||
| 2'-Fluoro RNA | HIV-1 tat/rev common exon sequence | CHO-WT and CHO-EE cells & PBMCs | Dimerization using phi29 Motor pRNA | Zhou et al., 2011 (68) | ||
| RNA with 2′-OMe–modified A and G and 2′-F–modified U and C sticky end | HIV-1 tat/rev common exon sequence & CD4 & TNPO3 | Humanized BALB/c-Rag2−/−γc−/− mice | Non covalent via 2′-OMe/2′-F GC-rich bridge | Zhou et al., 2013a (69) | Aptamer with siRNA multiplex | |
| CD8 | DNA | GNLY mRNA | CD8 overexpressing T-cells | Non-covalent via sticky bridge | Wang et al., 2013 (70) | |
| CD30 | 2′-O-methyl modified RNA | Anaplastic lymphoma kinase | human anaplastic large cell lymphoma | Non-covalent charge forces to carrier | Zhao et al., 2011 (71) | ALK siRNA and a RNA-based CD30 aptamer probe onto nano-sized polyethyleneimine-citrate carriers |
| Theophylline | RNA | shRNA: albumin mRNA | hepatic (HepG2) cells | Theophylline aptamer inserted in the loop region of shRNA | Tuleuova et al., 2008 (72) | |
| 5′-radiolabeled RNA | shRNA: enhanced green fluorescent protein (EGFP) | HEK293T cells | Theophylline aptamer inserted in the loop region of shRNA | Beisel et al., 2008 (73) | ligand-regulated RNAi | |
| RNA | shRNA: enhanced green fluorescent protein (EGFP) | HEK293 cells | Theophylline aptamer inserted in the loop region of shRNA | An et al., 2006 (74) | ligand-regulated RNAi | |
| RNA | shRNA: firefly luciferase mRNA | HEK293T cells | Theophylline aptamer inserted in the loop region of shRNA | Noguchi et al., 2011 (75) | ||
| Xanthine | 5′-radiolabeled RNA | shRNA: enhanced green fluorescent protein (EGFP) | HEK293T cells | Xanthine aptamer inserted in the loop region of shRNA | Beisel et al., 2008 (73) | ligand-regulated RNAi |
|
Aptamer target |
Component |
siRNA-Target |
in vivo/in vitro target |
Aptamer-siRNA linkage methode |
Reference |
Further Information |
| Malachite green (MG) | 2'-Fluoro RNA | Firefly luciferase mRNA | Human nasopharyngeal carcinoma KB cells | phi29 packaging RNA (pRNA) 3-way junction | Reif et al., 2012 (76) | Fluorogenic RNA NP for Monitoring RNA Folding & Degradation in Real Time |
| Transferrin receptor, CD71 (TfR) | 2'-Fluoro RNA | Enhanced green fluorescent protein (EGFP) | HeLa-EGFP cells | Aptamers conjugated to liposomes | Wilner et al., 2012 (77) | aptamer-targeted siRNA-laden liposomes |
| murine 4-1BB | 2'-Fluoro RNA | Diverse | HEK293T & HEPA1-6 cells | Conjugated via combined transcription | Berezhnoy et al., 2012 (78) | Paper focuses on thermal stability effects on inhibition |
| RNA | raptor mRNA | CD8 overexpressing T-cells | Conjugated via combined transcription | Berezhnoy et al., 2014 (79) | ||
| B-cell–activating factor receptor (BAFF-R) | 2'-Fluoro RNA | STAT3 mRNA | Jeko-1, Z138, Rec-1 & Granta-519 cells | 4-nucleotide linker (CUCU) | Zhou et al., 2013b (80) | |
| 2'-Fluoro RNA | STAT3 mRNA | Jeko-1, Z138, Rec-1 & Granta-519 cells | Non-covalent via sticky bridge | Zhou et al., 2013b (80) | ||
| ανβ3 integrin | RNA | Eukaryotic Elongation Factor 2 (EEF2) mRNA | U-87 MG, SiHa & PC-3 cells | Conjugated via combined transcription | Hussain et al., 2013 (81) | |
| Nucleolin | Oligodeoxy-nucleotides | snail family zinc finger 2 (SLUG) | CL1-5 cells | Hetero-bifunctional crosslinker, sulfo-SMPB | Lai et al., 2014 (82) | |
| Oligodeoxy-nucleotides | neuropilin 1 (NRP1) | CL1-5 cells | Hetero-bifunctional crosslinker, sulfo-SMPB | Lai et al., 2014 (82) | ||
| Oligodeoxy-nucleotides | BRAF gene | A375 cells & Balb/c or Nude mice | Aptamers conjugated to liposomes by PEG-linker | Li et al., 2014 (83) | Nucleolin-targeting liposomes guided by aptamer AS1411 | |
| MUC-1 | DNA | Green fluorescent protein (GFP) gene | MCF-7& A549 cells | siRNA linear linked via crosslinker dithio- bis-maleimidoethane; aptamer to siRNA linking non-covalent via complementary base paring | Yoo et al., 2014 (48) | Multivalent comb-type aptamer–siRNA conjugates |
| Cytotoxic T lymphocyte–associated antigen 4 (CTLA4) | RNA | STAT3 mRNA | CD8 overexpressing T-cells & immunodeficient mice bearing human T cell lymphomas | Unspecified linker | Herrmann et al., 2014 (84) | |
| U87-EGFRvIII cells | DNA | c-Met mRNA | U87MG cells | Biotinylated siRNA / aptamer linked by strepavidin connector | Zhang et al., 2014 (85) |
Aptamer Targeted Delivery of shRNA
Similar to siRNA approach, shRNA (short hairpin RNA) can be used to initiate target gene silencing. shRNAs consist of 2 complementary RNA sequences linked by a short loop region and mimics the naturally-occurring miRNA precursor in miRNA biogenesis. A ribonuclease III family member called Dicer cleaves the shRNAs into small interfering RNA duplexes with symmetric 2–3 nucleotides 3′-overhangs for creating conventional siRNAs.86 In order to trigger high gene silencing efficiency, shRNAs, like conventional siRNAs, are designed to match their target perfectly.
Aptamers can be utilized to further improve the target gene silencing efficacy and the major benefit of using shRNAs-aptamer chimeras is that the whole complex can be synthesized in one step, avoiding the annealing of 2 separated sense and antisense RNA strands, usually required for siRNA. Recently, Ni and colleagues58 used shRNA-aptamer chimeras to target the catalytic subunit of DNA-activated protein kinase, catalytic polypeptide (DNAPK). The aptamer-shRNA conjugate was designed as a single intact nuclease-stabilized 2′-fluoro-modified pyrimidine transcript. The treatments with the chimera lead to significant reductions in DNAPK mRNA levels. This report not only showed the enhanced RNAi capabilities of aptamer-shRNA chimera, but also the simplicity of the chimera synthesis.
Aptamers as Tools for Delivering microRNAs
The discovery of microRNA (miRNA), short endogenous-initiated non-coding RNA molecules, is considered an important breakthrough in the molecular genetics field.21 It was initially revealed as regulator of the larval developmental stages of Caenorhabditis elegans.87 Studies on miRNA received great attention and this area is growing rapidly. The reason for that is the involvement of miRNAs in the regulation of various important gene networks that play a role in the development of various diseases.88-90 miRNAs function as gene modulators inducing either degradation or translational repression of a target mRNA (mRNA). Depending on the degree of complementarity of the miRNA to the target mRNA, negative regulation occurs via the cleavage or by translational biogenesis and regulated repression of the target mRNA. The perfect or almost perfect binding of the miRNA to the target site induces the cleavage of mRNA. This way of interfering is most common in plants, but it was also reported for animals.91 The major regulation pathway in animals as well as in humans, is the translational repression induced by imperfect binding of the miRNA to complementary sites within the 3′ untranslated regions of mRNA blocking the translation into a protein.92,93 As imperfect target binding (compromised Watson-Crick base pairing rules) can block translation, one miRNA is able to regulate multiple mRNAs, making miRNAs an interesting tool for multi-target inhibition.
In comparison with normal cells, tumor cell lines often show a broad deregulation of miRNA expression.94 In most cancer type, miRNA down-regulation correlates with a lack of tumor suppressing functions, indicating their role as tumor suppressors. On the other hand some cancer types exhibit an increased expression of specific miRNAs that target tumor suppressor genes. Therefore, manipulating miRNAs would be a rational therapy considering their diverse roles in tumorigenesis and inducing tumor formation. An increasing number of studies have revealed that depending on the cellular context, one miRNA can act as tumor suppressor as well as an oncomir. One example for this 2-faced activity is miR-221. While being up regulated in most cases of epithelial tumors, miR-221 also play tumor suppressor role in erythroleukemic cells.95 Such examples will further complicate the use of miRNAs as therapeutic agents and demonstrates the requirement for cell specific delivery, further justifying the use of aptamers as a delivery tool.
The miRNAs miR-15a and miR-16-1 are known to act as tumor suppressors in prostate cancer.96-98 In 2011, Wu and colleagues99 used this tumor suppressing character to create a polyamidoamine (PAMAM)-based aptamer conjugation as a target-specific intracellular delivery carrier of miR-15a and miR-16-1 to treat prostate cancer. PAMAM was conjugated to the aptamer using a polyethylenglycol (PEG) linker. ATP-PEG-PAMAM-miRNA complexes were created by an electrostatic interaction between miRNA and PAMAM. By utilizing the aptamer A10-3.2 targeting prostate-specific membrane antigen (PSMA), they were able to deliver the miRNAs specifically to PSMA expressing LNCaP cells and induce cancer cell death.
Another example of utilizing aptamers to deliver miRNA was performed by Dai and colleagues. They conjugated MUC1-aptamers (anti-MUC1 protein) to miRNA-29b to generate the chi-29b chimera for the purpose of re-expressing the tumor-suppressor gene, PTEN. The chi-29b chimera was delivered specifically to OVCAR-3 cells, which express MUC1 protein guided by the aptamer and up-regulated the mRNA of the PTEN gene in the OVCAR-3 cells.100 chi-29b chimera successfully showed anti-tumor effects in ovarian cancer xeno-graft mice models. In another study, MUC1 aptamer was used for target specific delivery of let-7i miRNAs to reverse the paclitaxel-induced chemo-resistance of OVCAR-3-cells in the ovarian carcinomas. The paclitaxel-induced chemo-resistance has been successfully reversed by the MUC1/let-7i chimera, which has down-regulated the expressions of Dicer1, cyclin D1, cyclin D2 and PGRMC.101
Aptamers as Tools for Delivering antimiRs
AntimiRs, short piece of single-stranded nucleic acids targeting miRNA are a recent tool for inhibiting miRNA activity. AntimiRs are mostly modified oligonucleotides binding complementary to the target miRNA preventing from binding to its biological target. For example, Elmen et al. demonstrated the function of LNA-modified antimiRs in vivo, demonstrating antimiRs as an important therapeutic tool.102
In 2012, Kim et al. have developed an AS1411 aptamer-targeted theranostic platform composed of miRNA-221 targeting molecular beacon fused to a magnetic fluorescent nanoparticle.103 The beacon consisted of a perfect reverse compliment sequence to mature miRNA-221. Aptamer and the miRNA beacon were covalently linked to the nanoparticle using the coupling reagent, N-(3-dimethylaminopropyl)-N’-ethyl-carbodiimide hydrochloride. While the aptamer conducts cell specific delivery of the antimiR beacon, the nanoparticle enables tracking and visualization of the complex. They successfully demonstrated a functional system for simultaneous targeting of cancer cells, imaging and oncomir inhibition.
Very recently, Pofahl et al. reported the first successful aptamer based antimiR delivery to the deregulated miRNA target miR-21 in breast cancer cells.104 The antimiR sequence should in principle be specifically delivered to the cancer cells and strongly bind to the target miRNA sequence to inhibit its function. In their study, nucleolin targeting aptamer AS1411105 was used to deliver the antimiR sequence. The antimiR sequence was chemically modified by using phosphorothioate linkages and also by incorporating locked nucleic acid (LNA) nucleotides to enhance the antmiR-miR-21 interaction and to improve the stability. To test antimiR interference, an enhanced green fluorescent protein (EGFP)-expressing MCF-7 cell line was generated. In those cells, the EGFP expression was inhibited by miR-21. The study revealed that the chimera was successfully internalized in MCF-7 cells and exhibited antiproliferative properties while preventing miR-21 dependent EGFP inhibition. They coined the term AptamiR for this type of chimeras for combining aptamer and antimiR.
Aptamer-Oligonucleotide Chimeric Construction Using Oligonucleotide Synthesizer
To link therapeutic oligonucleotides like siRNA, antimiRs, antisense to nucleic acid aptamers, many different approaches can be used (see Table 1). Most procedures adopt appropriate post-oligo conjugation chemistries or interactions including biotin-streptavidin linkages. These approaches often involve time consuming multiple synthesis steps, purification steps and often result in low yields. Some of these chimeras can also be generated by enzymatic methods like ligation, in vitro transcription (recommended for long RNA aptamer siRNA conjugation, e.g. 40mer) and polymerase chain reaction for all DNA constructs. Ideally, it would be convenient to generate the aptamer-oligonucleotide chimera in one step using an oligonucleotide synthesizer via standard phophoramidite chemistry (Fig. 4). There are various methods one can think of; however, the appropriate ones could be to link the 2 functional regions via a disulphide linkage (SS), triethyleneglycol (TEG)/poly carbon (for e.g., C6 linkage or by using polynucleotide linkage (for e.g. –dTdTdT-). All of these amidites are commercially available from different sources, and these synthetic methods do not use large biological molecules like streptavidin, and thus can be less immunogenic.
Figure 4.
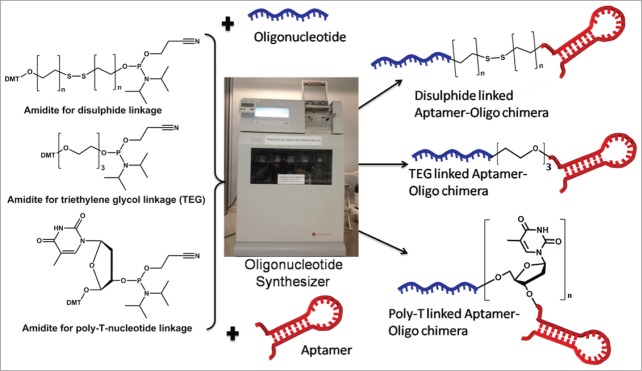
Aptamer-oligonucleotide chimera in one step using an oligonucleotide synthesizer.
Polynucleotide linkage might be the easiest way to link aptamer and therapeutic oligonucleotides. In this case, a special phosphoramidite that may affect the total synthetic yield is not required. It is noteworthy mentioning that polynucleotide linkers are able to engage in base paring with other nucleotides within the sequence or other sequences. Therefore, the linker has to be chosen carefully and also to avoid its influence on the secondary structure of the aptamer. In addition, the polynucleotide linker can make the chimera less flexible compared to other chemistries.
A polyethylene glycol (PEG) based phosphoramidite can be used to establish a PEG linkage between aptamer and oligonucleotides. PEG is hydrophilic, which decreases aggregation and increases solubility of the complex, non-toxic, non-immunogenic and a usual approach for increasing the bioavailability in vivo. Furthermore, a PEG linkage is highly flexible and thus it could be a useful method for conjugation. Disulphide linkages are commonly found in bacterial protein toxins.106 These toxins utilize the cleavage of covalently linked disulphide bond by reducing it to thiol groups. The disulphide bond is mostly stable in serum, due to the oxidizing character of the extracellular space, but if exposed to the reducing intracellular space, the disulphide bond is cleaved. This will facilitate the cleavage of the aptamer-oligonucleotide complex and release of the interfering oligonucleotide upon cell entry. Using this approach, coagulation of aptamer and siRNA/miRNA can be avoided and the efficacy of the interfering oligonucleotide can be improved.
Chemically Modified Aptamer-Oligonucleotide Chimera
Stability of oligonucleotides is key for successful therapeutic efficacy in vivo. Virtually every organism possesses various enzymes to synthesize, modify or hydrolyze nucleic acids. Nucleases are important for nucleic acid turn over and as a defense mechanism against pathogens, such as bacteria and viruses. Consequently, aptamer- therapeutic oligonucleotide chimera composed of naturally occurring DNA or RNA nucleotide monomers have serious limitations toward therapeutic development, as they exhibit shorter half-life in vivo because of their poor nuclease resistance and bio-availability. To tackle these limitations, a number of modified nucleotides have been developed in recent years (Fig. 5).
Figure 5.
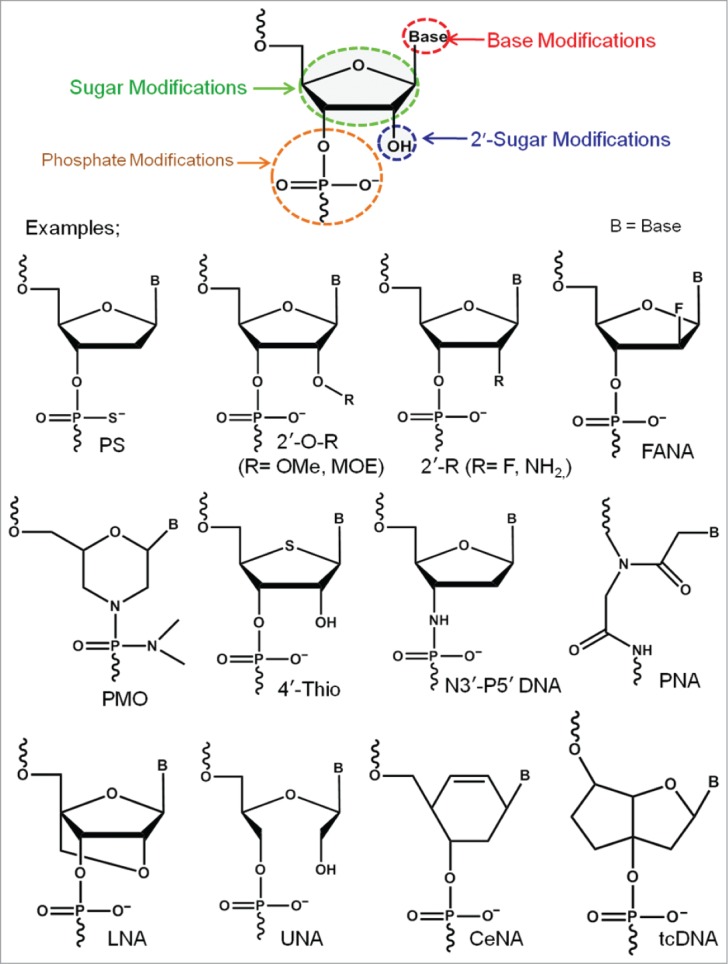
Examples of successful chemically-modified nucleotide analogs.
Some of the most prominent examples are 2′-fluoro (2′-F),107,108 2′-O-methyl (2′-OMe),109 2′-methoxyethyl (2′-MOE),110,111 2′-fluoroarabino (2′-FANA),112 locked nucleic acid (LNA),113,114 unlocked nucleic acid (UNA),115,116 cyclohexenyl (CeNA) nucleic acid,117,118 peptide nucleic acid (PNA),119 phosphoramidate morpholino (PMO)120 etc. Although many of the modified nucleotides have been successfully utilized in various nucleic acid-based therapeutic technologies, their relatively poor or no enzymatic recognition properties often pose a major challenge toward the development of biostable aptamers.
In principle 2 different approaches are used to incorporate modified nucleotides into aptamers. First, fabrication of a pre-selected aptamer introducing modified nucleotides at various positions during solid phase oligonucleotide chemical synthesis (‘post-SELEX’). In this approach, incorporation of a modified nucleotide can result in unfavorable shift or even in total loss of the binding affinity which highlight the importance of a systematic incorporation and analysis. A post-SELEX approach has been used during the development of the first aptamer drug Macugen® (Pegaptanib).3 Pegaptanib is a human vascular endothelial growth factor (VEGF)-binding RNA aptamer containing 2′-F pyrimidine and 2′-OMe purine nucleotides. While the aptamer origins from a 2′-F pyrimidine-containing library via conventional SELEX, the 2′-OMe modifications were introduced post-SELEX by substituting purines to enhance nuclease resistance and serum stability. Kuwahara et al. recently reviewed various successful post-SELEX modified aptamers.121
The second approach is by conventional aptamer selection via SELEX approaches whereby a new aptamer is developed from an oligonucleotide library containing modified nucleotides (in-SELEX approach). The 2′-OH group is a suitable location for introducing chemical modifications, since the modification can be introduced equally in purines and pyrimidines. Furthermore, 2′-modifications is known to increase the stability against chemical and enzymatic degradation.122-125 Very recently, Lauridsen et al. reported a review article describing the enzymatic recognition capabilities of various 2′-modified nucleotides.126 Stemming from their initial enzymatic recognition studies, 2′-amino pyrimidines, 2′-fluoro pyrimidines and 2′-O-Methyl nucleotides have been successfully applied in aptamer development by conventional SELEX-based methodologies.127-134 LNA is one of the successful nucleotide analogs extensively utilized in various fields because of their remarkable properties.113,114 In LNA the sugar ring is conformationally locked by a O2′-C4′methylene linkage to adopt N-type sugar puckering.135-137 Toward developing LNA-modified aptamers, Veedu et al. reported the enzymatic recognition capabilities of LNA nucleotides using DNA and RNA polymerases.138-144 In 2013, Kuwahara and co-workers reported an LNA (BNA) aptamer against thrombin using capillary electrophoresis-based SELEX (CE-SELEX) method.145,146
Summary and Outlook
Since their invention, aptamers have been applied to various applications including therapy, diagnosis, imaging and delivery. Aptamer selection is normally performed with a goal of generating a candidate sequence with very high target binding affinity (low nanomolar level) and specificity to a given molecular target. High affinity would be desirable for most applications, however for aptamers targeting proteins that are overexpressed in a particular disease condition (both intra-cellular and extra-cellular including cell-surface receptors), highest target binding affinity might not be necessary as it could increase the probability of binding to the same proteins needed for normal cellular functions. Aptamers are conventionally selected with a nucleic acid library with primer binding regions flanked to the randomized region. Secondary structures responsible for target binding may usually be expected from the random region; however, it is important to use the full-length oligonucleotide aptamer sequences (with primer flanks) for initial target binding analysis. Systematic truncation of the successful binding aptamer can then be performed using secondary structure prediction algorithms (e.g., mfold).147
In recent years, a number of studies showed the potential of aptamers to improve the efficacy of therapeutic oligonucleotide candidates for target specific gene silencing and generate a better clinical outcome. Endosomal release of aptamer-therapeutic oligonucleotide chimeras could be another problem in addition to cellular uptake, with high amounts of chimeras required to produce relevant changes in gene expression. Attaching endosome disrupting molecules such as a nanoparticle or a protein/peptide tag to the aptamer-oligonucleotide chimera may prove useful to circumvent this limitation. In previous years, the main focus was on aptamer-targeted delivery of siRNA. But, the scope of miRNA targeting and antisense therapy continues to rise and this will surely broaden the applications of aptamers based delivery systems.
A classical approach for targeting mRNA is to use antisense oligonucleotides (ASOs),148 short pieces of single-stranded DNA sequence that anneal to the target mRNA. This RNA:DNA hetero-duplex then recruits the enzyme RNAse H, which specifically cleaves the target mRNA and block translation. Chemically-modified nucleotide-based ASOs are also widely applied for enhanced targeting efficacy and stability, and in this case a steric-block mechanism is also applied for preventing translation. Most importantly, the first therapeutic oligonucleotide entered the clinic is Vitravene (Formivirsen), an ASO for the treatment of cytomagaloviral (CMV) retinititis in patients with HIV infection.149 This approach has been widely explored for its applicability as therapeutics in various disease conditions both in vitro and in vivo. Target specific delivery is very important for high therapeutic efficacy and aptamers can be a vital tool for more efficient delivery of ASOs. However, to the best of our knowledge so far, there are no reports on aptamer-mediated delivery of ASOs.
To summarize, the relatively new field of aptamer-therapeutic oligonucleotide chimera is currently advancing its potential for various therapeutic applications. Aptamer-guided delivery of therapeutic oligonucleotides could be one of the most exciting approaches toward the treatment of diseases and its broad applicability is limited by our knowledge and imagination.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1. Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov 2010; 9:537-50; PMID:20592747; http://dx.doi.org/ 10.1038/nrd3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev 2007; 107:3715-43; PMID:17715981; http://dx.doi.org/ 10.1021/cr0306743 [DOI] [PubMed] [Google Scholar]
- 3. Ng EW, Shima DT, Calias P, Cunningham ET, Jr., Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 2006; 5:123-32; PMID:16518379; http://dx.doi.org/ 10.1038/nrd1955 [DOI] [PubMed] [Google Scholar]
- 4. Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990; 249:505-10; PMID:2200121; http://dx.doi.org/ 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- 5. Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature 1990; 346:818-22; PMID:1697402; http://dx.doi.org/ 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- 6. Brown D, Gold L. Template recognition by an RNA-dependent RNA polymerase: identification and characterization of two RNA binding sites on Q beta replicase. Biochemistry 1995; 34:14765-74; PMID:7578085; http://dx.doi.org/ 10.1021/bi00045a018 [DOI] [PubMed] [Google Scholar]
- 7. Klug SJ, Famulok M. All you wanted to know about SELEX. Mol Biol Rep 1994; 20:97-107; PMID:7536299; http://dx.doi.org/ 10.1007/BF00996358 [DOI] [PubMed] [Google Scholar]
- 8. Gopinath SC. Methods developed for SELEX. Anal Bioanal Chem 2007; 387:171-82; PMID:17072603 [DOI] [PubMed] [Google Scholar]
- 9. Stoltenburg R, Reinemann C, Strehlitz B. SELEX–a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng 2007; 24:381-403; PMID:17627883; http://dx.doi.org/ 10.1016/j.bioeng.2007.06.001 [DOI] [PubMed] [Google Scholar]
- 10. Lauridsen LH, Shamaileh HA, Edwards SL, Taran E, Veedu RN. Rapid one-step selection method for generating nucleic acid aptamers: development of a DNA aptamer against alpha-bungarotoxin. Plos One 2012; 7:e41702; PMID:22860007; http://dx.doi.org/ 10.1371/journal.pone.0041702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nitsche A, Kurth A, Dunkhorst A, Panke O, Sielaff H, Junge W, Muth D, Scheller F, Stöcklein W, Dahmen C, et al. One-step selection of Vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotechnol 2007; 7:48; PMID:17697378; http://dx.doi.org/ 10.1186/1472-6750-7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peng L, Stephens BJ, Bonin K, Cubicciotti R, Guthold M. A combined atomic force/fluorescence microscopy technique to select aptamers in a single cycle from a small pool of random oligonucleotides. Microsc Res Tech 2007; 70:372-81; PMID:17262788; http://dx.doi.org/ 10.1002/jemt.20421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fan M, McBurnett SR, Andrews CJ, Allman AM, Bruno JG, Kiel JL. Aptamer selection express: a novel method for rapid single-step selection and sensing of aptamers. J Biomol Tech 2008; 19:311-9; PMID:19183794 [PMC free article] [PubMed] [Google Scholar]
- 14. Dua P, Kim S, Lee DK. Nucleic acid aptamers targeting cell-surface proteins. Methods 2011; 54:215-25; PMID:21300154; http://dx.doi.org/ 10.1016/j.ymeth.2011.02.002 [DOI] [PubMed] [Google Scholar]
- 15. Liu K, Lin B, Lan X. Aptamers: a promising tool for cancer imaging, diagnosis, and therapy. J Cell Biochem 2013; 114:250-5; PMID:22949372; http://dx.doi.org/ 10.1002/jcb.24373 [DOI] [PubMed] [Google Scholar]
- 16. Liss M, Petersen B, Wolf H, Prohaska E. An aptamer-based quartz crystal protein biosensor. Anal Chem 2002; 74:4488-95.D; PMID:12236360; http://dx.doi.org/ 10.1021/ac011294p [DOI] [PubMed] [Google Scholar]
- 17. Bumcrot D, Manoharan M, Koteliansky V, Sah DW. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol 2006; 2:711-9; PMID:17108989; http://dx.doi.org/ 10.1038/nchembio839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys 2013; 42:217-39; PMID:23654304; http://dx.doi.org/ 10.1146/annurev-biophys-083012-130404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009; 136:642-55; PMID:19239886; http://dx.doi.org/ 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dias N, Stein CA. Antisense oligonucleotides: basic concepts and mechanisms. Mol Cancer Ther 2002; 1:347-55; PMID:12489851; http://dx.doi.org/ 10.4161/cbt.1.4.4 [DOI] [PubMed] [Google Scholar]
- 21. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5:522-31; PMID:15211354; http://dx.doi.org/ 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 22. Ray P, White RR. Aptamers for targeted drug delivery. Pharmaceuticals 2010; 3:1761-1778; http://dx.doi.org/ 10.3390/ph3061761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Blank M, Weinschenk T, Priemer M, Schluesener H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels. selective targeting of endothelial regulatory protein pigpen. J Biol Chem 2001; 276:16464-8; PMID:11279054 [DOI] [PubMed] [Google Scholar]
- 24. Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc Natl Acad Sci U S A 2003; 100:15416-21; PMID:14676325; http://dx.doi.org/ 10.1073/pnas.2136683100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kunii T, Ogura S, Mie M, Kobatake E. Selection of DNA aptamers recognizing small cell lung cancer using living cell-SELEX. Analyst 2011; 136:1310-2; PMID:21321690 [DOI] [PubMed] [Google Scholar]
- 26. Ohuchi S. Cell-SELEX Technology. Biores Open Access 2012; 1:265-72; PMID:23515081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, Clary BM. In vivo selection of tumor-targeting RNA motifs. Nat Chem Biol 2010; 6:22-4; PMID:19946274; http://dx.doi.org/ 10.1038/nchembio.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xie FY, Woodle MC, Lu PY. Harnessing in vivo siRNA delivery for drug discovery and therapeutic development. Drug Discov Today 2006; 11:67-73; PMID:16478693; http://dx.doi.org/ 10.1016/S1359-6446(05)03668-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004; 432:173-8; PMID:15538359; http://dx.doi.org/ 10.1038/nature03121 [DOI] [PubMed] [Google Scholar]
- 30. Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov 2009; 8:129-38; PMID:19180106; http://dx.doi.org/ 10.1038/nrd2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat Biotechnol 2007; 25:1149-57; PMID:17873866; http://dx.doi.org/ 10.1038/nbt1339 [DOI] [PubMed] [Google Scholar]
- 32. Simeoni F, Morris MC, Heitz F, Divita G. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res 2003; 31:2717-24; PMID:12771197; http://dx.doi.org/ 10.1093/nar/gkg385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kam NW, Liu Z, Dai H. Functionalization of carbon nanotubes via cleavable disulfide bonds for efficient intracellular delivery of siRNA and potent gene silencing. J Am Chem Soc 2005; 127:12492-3; PMID:16144388; http://dx.doi.org/ 10.1021/ja053962k [DOI] [PubMed] [Google Scholar]
- 34. Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res 2004; 32:e149; PMID:15520458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou J, Rossi JJ. Cell-specific aptamer-mediated targeted drug delivery. Oligonucleotides 2011; 21:1-10; PMID:21182455; http://dx.doi.org/ 10.1089/oli.2010.0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hicke BJ, Stephens AW. Escort aptamers: a delivery service for diagnosis and therapy. J Clin Invest 2000; 106:923-8; PMID:11032850; http://dx.doi.org/ 10.1172/JCI11324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou J, Rossi JJ. Aptamer-targeted cell-specific RNA interference. Silence 2010; 1:4; PMID:20226078; http://dx.doi.org/ 10.1186/1758-907X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res 2006; 34:e73; PMID:16740739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Farokhzad OC, Jon S, Khademhosseini A, Tran TN, Lavan DA, Langer R. Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells. Cancer Res 2004; 64:7668-72; PMID:15520166; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-2550 [DOI] [PubMed] [Google Scholar]
- 40. McNamara JO, 2nd, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 2006; 24:1005-15; PMID:16823371; http://dx.doi.org/ 10.1038/nbt1223 [DOI] [PubMed] [Google Scholar]
- 41. Zhou J, Li H, Li S, Zaia J, Rossi JJ. Novel dual inhibitory function aptamer-siRNA delivery system for HIV-1 therapy. Mol Ther 2008; 16:1481-9; PMID:18461053; http://dx.doi.org/ 10.1038/mt.2008.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chavda SC, Griffin P, Han-Liu Z, Keys B, Vekony MA, Cann AJ. Molecular determinants of the V3 loop of human immunodeficiency virus type 1 glycoprotein gp120 responsible for controlling cell tropism. J Gen Virol 1994; 75: 3249-53; PMID:7964635; http://dx.doi.org/ 10.1099/0022-1317-75-11-3249 [DOI] [PubMed] [Google Scholar]
- 43. Chesebro B, Nishio J, Perryman S, Cann A, O'Brien W, Chen IS, Wehrly K. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J Virol 1991; 65:5782-9; PMID:1920616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mondor I, Moulard M, Ugolini S, Klasse PJ, Hoxie J, Amara A, Delaunay T, Wyatt R, Sodroski J, Sattentau QJ. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 loops, and sensitivity to neutralizing antibodies. Virology 1998; 248:394-405; PMID:9721247; http://dx.doi.org/ 10.1006/viro.1998.9282 [DOI] [PubMed] [Google Scholar]
- 45. Khati M, Schuman M, Ibrahim J, Sattentau Q, Gordon S, James W. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2'F-RNA aptamers. J Virol 2003; 77:12692-8; PMID:14610191; http://dx.doi.org/ 10.1128/JVI.77.23.12692-12698.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chithrani BD, Chan WC. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett 2007; 7:1542-50; PMID:17465586; http://dx.doi.org/ 10.1021/nl070363y [DOI] [PubMed] [Google Scholar]
- 47. Gao H, Shi W, Freund LB. Mechanics of receptor-mediated endocytosis. Proc Natl Acad Sci U S A 2005; 102:9469-74; PMID:15972807; http://dx.doi.org/ 10.1073/pnas.0503879102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yoo H, Jung H, Kim SA, Mok H. Multivalent comb-type aptamer-siRNA conjugates for efficient and selective intracellular delivery. Chem Commun 2014; 50:6765-7; PMID:24830507; http://dx.doi.org/ 10.1039/c4cc01620c [DOI] [PubMed] [Google Scholar]
- 49. Yu C, Hu Y, Duan J, Yuan W, Wang C, Xu H, Yang XD. Novel aptamer-nanoparticle bioconjugates enhances delivery of anticancer drug to MUC1-positive cancer cells in vitro. Plos One 2011; 6:e24077; PMID:21912664; http://dx.doi.org/ 10.1371/journal.pone.0024077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheever MA, Allison JP, Ferris AS, Finn OJ, Hastings BM, Hecht TT, Mellman I, Prindiville A, Viner JL, Weiner LM, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res 2009; 15:5323-37; PMID:19723653; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-0737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walter JG, Kokpinar O, Friehs K, Stahl F, Scheper T. Systematic investigation of optimal aptamer immobilization for protein-microarray applications. Anal Chem 2008; 80:7372-8; PMID:18729475; http://dx.doi.org/ 10.1021/ac801081v [DOI] [PubMed] [Google Scholar]
- 52. Bagalkot V, Gao X. siRNA-aptamer chimeras on nanoparticles: preserving targeting functionality for effective gene silencing. ACS Nano 2011; 5:8131-9; PMID:21936502; http://dx.doi.org/ 10.1021/nn202772p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu HY, Gao X. A universal protein tag for delivery of SiRNA-aptamer chimeras. Sci Rep 2013; 3:3129; PMID:24196104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, 2nd, Giangrande PH. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat Biotechnol 2009; 27:839-49; PMID:19701187; http://dx.doi.org/ 10.1038/nbt.1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wullner U, Neef I, Eller A, Kleines M, Tur MK, Barth S. Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr Cancer Drug Targets 2008; 8:554-65; PMID:18991566 [DOI] [PubMed] [Google Scholar]
- 56. Kim E, Jung Y, Choi H, Yang J, Suh JS, Huh YM, Kim K, Haam S. Prostate cancer cell death produced by the co-delivery of Bcl-xL shRNA and doxorubicin using an aptamer-conjugated polyplex. Biomaterials 2010; 31:4592-9; PMID:20206379; http://dx.doi.org/ 10.1016/j.biomaterials.2010.02.030 [DOI] [PubMed] [Google Scholar]
- 57. Pastor F, Kolonias D, Giangrande PH, Gilboa E. Induction of tumour immunity by targeted inhibition of nonsense-mediated mRNA decay. Nature 2010; 465:227-30; PMID:20463739; http://dx.doi.org/ 10.1038/nature08999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ni X, Zhang Y, Ribas J, Chowdhury WH, Castanares M, Zhang Z, Laiho M, DeWeese TL, Lupold SE. Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts. J Clin Invest 2011; 121:2383-90; PMID:21555850; http://dx.doi.org/ 10.1172/JCI45109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, McNamara JO, 2nd, Giangrande PH. Delivery of chemo-sensitizing siRNAs to HER2+-breast cancer cells using RNA aptamers. Nucleic Acids Res 2012; 40:6319-37; PMID:22467215; http://dx.doi.org/ 10.1093/nar/gks294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum Gene Ther 2005; 16:1097-109; PMID:16149908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Khaled A, Guo S, Li F, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett 2005; 5:1797-808; PMID:16159227; http://dx.doi.org/ 10.1021/nl051264s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wheeler LA, Trifonova R, Vrbanac V, Basar E, McKernan S, Xu Z, Seung E, Deruaz M, Einarsson JI, Yang L, et al. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J Clin Invest 2011; 121:2401-12; PMID:21576818; http://dx.doi.org/ 10.1172/JCI45876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wheeler LA, Vrbanac V, Trifonova R, Brehm MA, Gilboa-Geffen A, Tanno S, Greiner DL, Luster AD, Tager AM, Lieberman J. Durable knockdown and protection from HIV transmission in humanized mice treated with gel-formulated CD4 aptamer-siRNA chimeras. Mol Ther 2013; 21:1378-89; PMID:23629001; http://dx.doi.org/ 10.1038/mt.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhu Q, Shibata T, Kabashima T, Kai M. Inhibition of HIV-1 protease expression in T cells owing to DNA aptamer-mediated specific delivery of siRNA. Eur J Med Chem 2012; 56:396-9; PMID:22907035 [DOI] [PubMed] [Google Scholar]
- 65. Qiu C, Peng WK, Shi F, Zhang T. Bottom-up assembly of RNA nanoparticles containing phi29 motor pRNA to silence the asthma STAT5b gene. Genet Mol Res 2012; 11:3236-45; PMID:23079817 [DOI] [PubMed] [Google Scholar]
- 66. Zhou J, Swiderski P, Li H, Zhang J, Neff CP, Akkina R, Rossi JJ. Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells. Nucleic Acids Res 2009; 37:3094-109; PMID:19304999; http://dx.doi.org/ 10.1093/nar/gkp185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H, Smith DD, Swiderski P, Rossi JJ, Akkina R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci Transl Med 2011; 3:66ra6; PMID:21248316; http://dx.doi.org/ 10.1126/scitranslmed.3001581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhou J, Shu Y, Guo P, Smith DD, Rossi JJ. Dual functional RNA nanoparticles containing phi29 motor pRNA and anti-gp120 aptamer for cell-type specific delivery and HIV-1 inhibition. Methods 2011; 54:284-94; PMID:21256218; http://dx.doi.org/ 10.1016/j.ymeth.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou J, Neff CP, Swiderski P, Li H, Smith DD, Aboellail T, Remling-Mulder L, Akkina R, Rossi JJ. Functional in vivo delivery of multiplexed anti-HIV-1 siRNAs via a chemically synthesized aptamer with a sticky bridge. Mol Ther 2013; 21:192-200; PMID:23164935; http://dx.doi.org/ 10.1038/mt.2012.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang CW, Chung WH, Cheng YF, Ying NW, Peck K, Chen YT, Hung SI. A new nucleic acid-based agent inhibits cytotoxic T lymphocyte-mediated immune disorders. J Allergy Clin Immunol 2013; 132:713-22.e11; PMID:23791505; http://dx.doi.org/ 10.1016/j.jaci.2013.04.036 [DOI] [PubMed] [Google Scholar]
- 71. Zhao N, Bagaria HG, Wong MS, Zu Y. A nanocomplex that is both tumor cell-selective and cancer gene-specific for anaplastic large cell lymphoma. J Nanobiotech 2011; 9:2; PMID:21281497; http://dx.doi.org/ 10.1186/1477-3155-9-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tuleuova N, An CI, Ramanculov E, Revzin A, Yokobayashi Y. Modulating endogenous gene expression of mammalian cells via RNA-small molecule interaction. Biochem Biophys Res Commun 2008; 376:169-73; PMID:18765226; http://dx.doi.org/ 10.1016/j.bbrc.2008.08.112 [DOI] [PubMed] [Google Scholar]
- 73. Beisel CL, Bayer TS, Hoff KG, Smolke CD. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol Syst Biol 2008; 4:224; PMID:18956013; http://dx.doi.org/ 10.1038/msb.2008.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. An CI, Trinh VB, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA 2006; 12:710-6; PMID:16606868; http://dx.doi.org/ 10.1261/rna.2299306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Noguchi K, Ishitu Y, Takaku H. Evaluating target silencing by short hairpin RNA mediated by the group I intron in cultured mammalian cells. BMC Biotechnol 2011; 11:79; PMID:21781346; http://dx.doi.org/ 10.1186/1472-6750-11-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Reif R, Haque F, Guo P. Fluorogenic RNA nanoparticles for monitoring RNA folding and degradation in real time in living cells. Nucleic Acid Ther 2012; 22:428-37; PMID:23113765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wilner SE, Wengerter B, Maier K, de Lourdes Borba Magalhaes M, Del Amo DS, Pai S, Opazo F, Rizzoli SO, Yan A, Levy M. An RNA alternative to human transferrin: a new tool for targeting human cells. Mol Ther Nucleic Acids 2012; 1:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Berezhnoy A, Brenneman R, Bajgelman M, Seales D, Gilboa E. Thermal Stability of siRNA Modulates Aptamer- conjugated siRNA Inhibition. Mol Ther Nucleic Acids 2012; 1:e51; PMID:23344651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Berezhnoy A, Castro I, Levay A, Malek TR, Gilboa E. Aptamer-targeted inhibition of mTOR in T cells enhances antitumor immunity. J Clin Invest 2014; 124:188-97; PMID:24292708; http://dx.doi.org/ 10.1172/JCI69856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhou J, Tiemann K, Chomchan P, Alluin J, Swiderski P, Burnett J, Zhang X, Forman S, Chen R, Rossi J. Dual functional BAFF receptor aptamers inhibit ligand-induced proliferation and deliver siRNAs to NHL cells. Nucleic Acids Res 2013; 41:4266-83; PMID:23470998; http://dx.doi.org/ 10.1093/nar/gkt125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hussain AF, Tur MK, Barth S. An aptamer-siRNA chimera silences the eukaryotic elongation factor 2 gene and induces apoptosis in cancers expressing alphavbeta3 integrin. Nucleic Acid Ther 2013; 23:203-12; PMID:23544955 [DOI] [PubMed] [Google Scholar]
- 82. Lai WY, Wang WY, Chang YC, Chang CJ, Yang PC, Peck K. Synergistic inhibition of lung cancer cell invasion, tumor growth and angiogenesis using aptamer-siRNA chimeras. Biomaterials 2014; 35:2905-14; PMID:24397988; http://dx.doi.org/ 10.1016/j.biomaterials.2013.12.054 [DOI] [PubMed] [Google Scholar]
- 83. Li L, Hou J, Liu X, Guo Y, Wu Y, Zhang L, Yang Z. Nucleolin-targeting liposomes guided by aptamer AS1411 for the delivery of siRNA for the treatment of malignant melanomas. Biomaterials 2014; 35:3840-50; PMID:24486214 [DOI] [PubMed] [Google Scholar]
- 84. Herrmann A, Priceman SJ, Kujawski M, Xin H, Cherryholmes GA, Zhang W, Zhang C, Kowolik C, Forman SJ, Kortylewski M, et al. CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells. J Clin Invest 2014; 124:2977-87; PMID:24892807; http://dx.doi.org/ 10.1172/JCI73174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhang X, Liang H, Tan Y, Wu X, Li S, Shi Y. A U87-EGFRvIII cell-specific aptamer mediates small interfering RNA delivery. Biomed Rep 2014; 2:495-9; PMID:24944794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kim VN. MicroRNA biogenesis: corrdinated cropping and dicing. Nat Rev Mol Cell Biol 2005; 6:376-85; PMID:15852042; http://dx.doi.org/ 10.1038/nrm1644 [DOI] [PubMed] [Google Scholar]
- 87. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391:806-11; PMID:9486653; http://dx.doi.org/ 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- 88. Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol 2011; 223:102-15; PMID:21125669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 2012; 4:143-59; PMID:22351564; http://dx.doi.org/ 10.1002/emmm.201100209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Garofalo M, Condorelli GL, Croce CM, Condorelli G. MicroRNAs as regulators of death receptors signaling. Cell Death Differ 2010; 17:200-8; PMID:19644509; http://dx.doi.org/ 10.1038/cdd.2009.105 [DOI] [PubMed] [Google Scholar]
- 91. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 92. Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 2007; 17:118-26; PMID:17197185; http://dx.doi.org/ 10.1016/j.tcb.2006.12.007 [DOI] [PubMed] [Google Scholar]
- 93. Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE 2007; 2007:re1; PMID:17200520 [DOI] [PubMed] [Google Scholar]
- 94. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857-66; PMID:17060945; http://dx.doi.org/ 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- 95. Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med 2012; 12:27-33; PMID:22082479; http://dx.doi.org/ 10.2174/156652412798376170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kawasaki H, Wadhwa R, Taira K. World of small RNAs: from ribozymes to siRNA and miRNA. Differentiation 2004; 72:58-64; PMID:15066185; http://dx.doi.org/ 10.1111/j.1432-0436.2004.07202006.x [DOI] [PubMed] [Google Scholar]
- 97. Pontes O, Pikaard CS. siRNA and miRNA processing: new functions for Cajal bodies. Curr Opin Genet Dev 2008; 18:197-203; PMID:18337083; http://dx.doi.org/ 10.1016/j.gde.2008.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lin SL, Chang D, Ying SY. Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene 2005; 356:32-8; PMID:16005165; http://dx.doi.org/ 10.1016/j.gene.2005.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wu X, Ding B, Gao J, Wang H, Fan W, Wang X, Ye L, Zhang M, Ding X, Liu J, et al. Second-generation aptamer-conjugated PSMA-targeted delivery system for prostate cancer therapy. Int J Nanomed 2011; 6:1747-56; PMID:21980237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dai F, Zhang Y, Zhu X, Shan N, Chen Y. Anticancer role of MUC1 aptamer-miR-29b chimera in epithelial ovarian carcinoma cells through regulation of PTEN methylation. Target Oncol 2012; 7:217-25; PMID:23179556; http://dx.doi.org/ 10.1007/s11523-012-0236-7 [DOI] [PubMed] [Google Scholar]
- 101. Liu N, Zhou C, Zhao J, Chen Y. Reversal of paclitaxel resistance in epithelial ovarian carcinoma cells by a MUC1 aptamer-let-7i chimera. Cancer Invest 2012; 30:577-82; PMID:22812695; http://dx.doi.org/ 10.3109/07357907.2012.707265 [DOI] [PubMed] [Google Scholar]
- 102. Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansten HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008; 452:896-9; PMID:18368051; http://dx.doi.org/ 10.1038/nature06783 [DOI] [PubMed] [Google Scholar]
- 103. Kim JK, Choi KJ, Lee M, Jo MH, Kim S. Molecular imaging of a cancer-targeting theragnostics probe using a nucleolin aptamer- and microRNA-221 molecular beacon-conjugated nanoparticle. Biomaterials 2012; 33:207-17; PMID:21944470; http://dx.doi.org/ 10.1016/j.biomaterials.2011.09.023 [DOI] [PubMed] [Google Scholar]
- 104. Pofahl M, Wengel J, Mayer G. Multifunctional nucleic acids for tumor cell treatment. Nucleic Acid Ther 2014; 24:171-7; PMID:24494617; http://dx.doi.org/ 10.1089/nat.2013.0472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol 2009; 86:151-64; PMID:19454272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Falnes PO, Sandvig K. Penetration of protein toxins into cells. Curr Opin Cell Biol 2000; 12:407-13; PMID:10873820; http://dx.doi.org/ 10.1016/S0955-0674(00)00109-5 [DOI] [PubMed] [Google Scholar]
- 107. Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2'-deoxy-2'-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J Med Chem 1993; 36:831-41; PMID:8464037; http://dx.doi.org/ 10.1021/jm00059a007 [DOI] [PubMed] [Google Scholar]
- 108. Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Kinetic characterization of ribonuclease-resistant 2'-modified hammerhead ribozymes. Science 1991; 253:314-7; PMID:1857967; http://dx.doi.org/ 10.1126/science.1857967 [DOI] [PubMed] [Google Scholar]
- 109. Majlessi M, Nelson NC, Becker MM. Advantages of 2’-O-methyl oligoribonucleotides probes for detecting RNA targets. Nucleic Acids Res 1998; 26:2224-9; PMID:9547284; http://dx.doi.org/ 10.1093/nar/26.9.2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM, Bennett CF. 2’-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation oft he ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J Biol Chem 1997; 272:11994-2000; PMID:9115264; http://dx.doi.org/ 10.1074/jbc.272.18.11994 [DOI] [PubMed] [Google Scholar]
- 111. Geary RS, Watanabe TA, Truong L, Freier S, Lesnik EA, Sioufi NB, Sasmor H, Manoharan M, Levin AA. Pharmacokinetic properties of 2’-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J Pharmacol Exp Ther 2001; 296:890-7; PMID:11181921 [PubMed] [Google Scholar]
- 112. Wilds CJ, Damha MJ. 2’-Deoxy-2’-fluoro-β-D-arabinonucleosides and oligonucleotides (2’F-ANA): synthesis and physicochemical studies. Nucleic Acids Res 2000; 28:3625-35; PMID:10982885; http://dx.doi.org/ 10.1093/nar/28.18.3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Veedu RN, Wengel J. Locked nucleic acid as a novel class of therapeutic agents. RNA Biol 2009; 6: 321-23; PMID:19458498; http://dx.doi.org/ 10.4161/rna.6.3.8807 [DOI] [PubMed] [Google Scholar]
- 114. Veedu RN, Wengel J. Locked nucleic acids: promising nucleic acid analogs for therapeutic applications. Chem Biodivers 2010; 7: 536-42; PMID:20232325; http://dx.doi.org/ 10.1002/cbdv.200900343 [DOI] [PubMed] [Google Scholar]
- 115. Nielson P, Dreiøe LH, Wengel J. Synthesis and evaluation of oligodeoxynucleotides containing acyclic nucleosides. Bioorg Med Chem 1995; 3:19-28; PMID:8612043; http://dx.doi.org/ 10.1016/0968-0896(94)00143-Q [DOI] [PubMed] [Google Scholar]
- 116. Langkjær N, Pasternak A, Wengel J. UNA (unlocked nucleic acid): A flexible RNA mimc that allows engineering of nucleic acid duplex stability. Bioorg Med Chem 2009; 71:5420-5; PMID:19604699; http://dx.doi.org/ 10.1016/j.bmc.2009.06.045 [DOI] [PubMed] [Google Scholar]
- 117. Herdewijn P. Cyclohexene nucleic acids: serum stable oligonucleotides that activate RNase H and increase duplex stability with complementary RNA. Abstr Pap Am Chem Soc 2001; 221:U155 [Google Scholar]
- 118. Herdewijn P, De Clercq E. The cyclohexene ring as bioisostere of a furanose ring: synthesis and antiviral activity of cyclohexenyl nucleosides. Bioorg Med Chem Lett 2001; 11:1591-7; PMID:11412988; http://dx.doi.org/ 10.1016/S0960-894X(01)00270-0 [DOI] [PubMed] [Google Scholar]
- 119. Hyrup B, Nielsen P. Peptide Nucleic Acids (PNA): Synthesis, properties and potential applications. Bioorg Med Chem 1996; 4:5-23; http://dx.doi.org/ 10.1016/0968-0896(95)00171-9 [DOI] [PubMed] [Google Scholar]
- 120. Summerton J, Weller D. Morpholino antisense oligomers: design, perparation, and properties. Antisense Nucleic Acid Drug Dev 1997; 7:187-95; PMID:9212909; http://dx.doi.org/ 10.1089/oli.1.1997.7.187 [DOI] [PubMed] [Google Scholar]
- 121. Kuwahara M, Sugimoto N. Molecular evolution of functional nucleic acids with chemical modifications. Molecules 2010; 15:5423-44; PMID:20714306; http://dx.doi.org/ 10.3390/molecules15085423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. De Mesmaeker A, Häner R, Martin P, Moser HE. Antisense oligonucleotides. Acc Chem Res 1995; 28:366 [Google Scholar]
- 123. Eaton BE, Pieken WA. Ribonucleosides and RNA. Annu Rev Biochem 1995; 64:837-63; PMID:7574502; http://dx.doi.org/ 10.1146/annurev.bi.64.070195.004201 [DOI] [PubMed] [Google Scholar]
- 124. Sioud M, Sorensen DR. A nuclease-resistant protein kinase C alpha ribozyme blocks glioma cell growth. Nat Biotechnol 1998; 16:556-61; PMID:9624687; http://dx.doi.org/ 10.1038/nbt0698-556 [DOI] [PubMed] [Google Scholar]
- 125. Zinnen SP, Domenico K, Wilson M, Dickinson BA, Beaudry A, Mokler V, Daniher AT, Burgin A, Beigelman L. Selection, design, and characterization of a new potentially therapeutic ribozyme. RNA 2002; 8:214-28; PMID:11911367; http://dx.doi.org/ 10.1017/S1355838202014723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Lauridsen LH, Rothnagel JA, Veedu RN. Enzymatic recognition of 2'-modified ribonucleoside 5'-triphosphates: towards the evolution of versatile aptamers. Chembiochem 2012; 13:19-25; PMID:22162282; http://dx.doi.org/ 10.1002/cbic.201100648 [DOI] [PubMed] [Google Scholar]
- 127. Lin Y, Qiu Q, Gill SC, Jayasena SD. Modified RNA sequence pools for in vitro selection. Nucleic Acids Res 1994; 22:5229-34; PMID:7529404; http://dx.doi.org/ 10.1093/nar/22.24.5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Jellinek D, Green LS, Bell C, Lynott CK, Gill N, Vargeese C, Kirschenheuter G, McGee DP, Abesinghe P, Pieken WA, et al. Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 1995; 34:11363-72; PMID:7547864; http://dx.doi.org/ 10.1021/bi00036a009 [DOI] [PubMed] [Google Scholar]
- 129. Lin Y, Nieuwlandt D, Magallanez A, Feistner B, Jayasena SD. High-affinity and specific recognition of human thyroid stimulating hormone (hTSH) by in vitro-selected 2′-amino-modified RNA. Nucleic Acids Res 1996; 24:3407-14; PMID:8811096; http://dx.doi.org/ 10.1093/nar/24.17.3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Proske D, Gilch S, Wopfner F, Schatzl HM, Winnacker EL, Famulok M. Prion-protein-specific aptamer reduces PrPSc formation. Chembiochem 2002; 3:717-25; PMID:12203970; http://dx.doi.org/ 10.1002/1439-7633(20020802)3:8%3c717::AID-CBIC717%3e3.0.CO;2-C [DOI] [PubMed] [Google Scholar]
- 131. Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 2002; 419:90-4; PMID:12214238; http://dx.doi.org/ 10.1038/nature00963 [DOI] [PubMed] [Google Scholar]
- 132. Biesecker G, Dihel L, Enney K, Bendele RA. Derivation of RNA aptamer inhibitors of human complement C5. Immunopharmacology 1999; 42:219-30; PMID:10408383; http://dx.doi.org/ 10.1016/S0162-3109(99)00020-X [DOI] [PubMed] [Google Scholar]
- 133. Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC,Epstein DM, Wilson C, et al. Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem Biol 2005; 12:25-33; PMID:15664512 [DOI] [PubMed] [Google Scholar]
- 134. Burmeister PE, Wang C, Killough JR, Lewis SD, Horwitz LR, Ferguson A, Thompson KM, Pendergrast PS, McCauley TG, Kurz M, et al. 2′-Deoxy purine, 2′-O-methyl pyrimidine (dRmY) aptamers as candidate therapeutics. Oligonucleotides 2006; 16:337-51; PMID:17155909 [DOI] [PubMed] [Google Scholar]
- 135. Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucelic Acid): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 1998, 54:3607-3630 [Google Scholar]
- 136. Singh SK, Koshkin AA, Wengel J, Nielsen P. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem Commun 1998; 39:455-456; http://dx.doi.org/ 10.1039/a708608c [DOI] [Google Scholar]
- 137. Obika S, Nanbu D, Hari Y, Andoh JI, Morio KI, Doi T, Imanishi T. Stability and structural features of the duplex containing nucleoside analogues with a fixed N-type conformation, 2’-O,4’-C-methyleneribonucleotieds. Tetrahedron Lett. 1998; 39:5401; http://dx.doi.org/ 10.1016/S0040-4039(98)01084-3 [DOI] [Google Scholar]
- 138. Veedu RN, Vester B, Wengel J. Polymerase chain reaction and transcription using locked nucleic acid nucleotide triphosphates. J Am Chem Soc 2008; 130:8124-5; PMID:18533656; http://dx.doi.org/ 10.1021/ja801389n [DOI] [PubMed] [Google Scholar]
- 139. Veedu RN, Vester B, Wengel J. Enzymatic incorporation of LNA nucleotides into DNA strands. ChemBioChem 2007; 8:490-2; PMID:17315250; http://dx.doi.org/ 10.1002/cbic.200600501 [DOI] [PubMed] [Google Scholar]
- 140. Veedu RN, Vester B, Wengel J. Efficient Enzymatic Synthesis of LNA-modified DNA duplexes using KOD DNA polymerase. Org Biomol Chem 2009; 7:1404-9; PMID:19300826; http://dx.doi.org/ 10.1039/b819946a [DOI] [PubMed] [Google Scholar]
- 141. Veedu RN, Vester B, Wengel J. In Vitro incorporation of Locked Nucleic Acids. Nucleosides Nucleotides Nucleic Acids 2007; 26:1207; PMID:18058567; http://dx.doi.org/ 10.1080/15257770701527844 [DOI] [PubMed] [Google Scholar]
- 142. Veedu RN, Wengel J. Locked nucleic acid nucleoside triphosphates and polymerases: On the way towards evolution of LNA aptamers. Mol Biosyst 2009; 5: 787-92; PMID:19603111; http://dx.doi.org/ 10.1039/b905513b [DOI] [PubMed] [Google Scholar]
- 143. Doessing H, Hansen L, Veedu RN, Wengel J, Vester B. Amplification and Re-Generation of LNA-Modified Libraries. Molecules 2012; 17: 13087-97; PMID:23128088; http://dx.doi.org/ 10.3390/molecules171113087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Crouzier L, Dubois C, Edwards SL, Lauridsen LH, Wengel J, Veedu RN. Efficient Reverse Transcription Using Locked Nucleic Acid Nucleotides towards the Evolution of Nuclease Resistant RNA Aptamers. Plos One 2012; 7: e35990; PMID:22558297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Kasahara Y, Irisawa Y, Ozaki H, Obika S, Kuwahara M. 2',4'-BNA/LNA aptamers: CE-SELEX using a DNA-based library of full-length 2'-O,4'-C-methylene-bridged/linked bicyclic ribonucleotides. Bioorg Med Chem Lett 2013; 23:1288-92; PMID:23374873; http://dx.doi.org/ 10.1016/j.bmcl.2012.12.093 [DOI] [PubMed] [Google Scholar]
- 146. Kuwahara M, Obika S. In vitro selection of BNA (LNA) aptamers. Artif DNA PNA XNA 2013; 4:39-48; PMID:24044051; http://dx.doi.org/ 10.4161/adna.25786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kushner DM, Silverman RH. Antisense cancer therapy: the state of the science. Curr Oncol Rep 2000; 2:23-30; PMID:11122821; http://dx.doi.org/ 10.1007/s11912-000-0007-y [DOI] [PubMed] [Google Scholar]
- 149. Perry CM, Balfour JA. Fomivirsen. Drugs 1999; 57:375-80; discussion 81; PMID:10193689; http://dx.doi.org/ 10.2165/00003495-199957030-00010 [DOI] [PubMed] [Google Scholar]


