Abstract
Long noncoding RNAs (lncRNAs) are dysregulated in many cancer types and are believed to play crucial roles in regulating several hallmarks of cancer biology. Currently, most studies support the concept that lncRNAs are involved in either transcriptional or post-transcriptional processes via binding/targeting epigenetic modifiers or hRNP complexes. The discovery of new biological functions of lncRNA and novel RNA binding proteins suggests that lncRNAs may be implicated in a broad spectrum of biological processes such as signal transduction, allosteric regulation of cytoplasmic enzymatic activities, among other potential processes. In a recent report that we have made, based on open-ended lncRNA pulldown technology and a series of systematic analyses, we suggest that lncRNAs also play critical roles in the regulation of noncanonical Hedgehog/GLI 2 signal transduction pathways in cancer cells, which further broadens the scope of known lncRNA functions and aids in the discovery and design of more effective and evidence-based therapeutic targets for the treatment of human cancers and other diseases.
Keywords: lncrna, bcar4, breast cancer, metastasis, signal transduction, Hedgehog/GLI, locked nucleic acids (LNA)
Classification of Distinct LncRNAs Based on Their Genomic Context
It is quite surprising that the human genome produces such a vast number of long non coding RNAs (lncRNAs), the study of which has benefitted greatly from many powerful technologies and approaches to characterize the essence of these non-protein coding transcripts, which include the following: high-throughput RNA-sequencing (RNA-seq) technologies, large-scale whole genome sequencing, tiling arrays (ChIP-chip, transcriptome mapping), serial analysis of gene expression (SAGE), as well as profiling of specific histone marks (such as H3K4-H3K36 domains) in human cells.1,2 In addition, considerable efforts have been made to combine the analysis of large-scale sequencing data and experimental validation approaches in order to annotate new RNA species. Benefiting from the progress made by the RefSeq, Ensembl, and GENCODE Consortium within the framework of the ENCODE Project, many lncRNAs have been comprehensively standardized and annotated, resulting in an integrated and curated lncRNA database that represents an invaluable resource for future studies of lncRNAs.3,4
LncRNAs can be classified into distinct groups based on their different features such as genomic location, molecular function/effects, mechanisms/modes of action.5 Currently, one of the most broadly used and relatively convenient ways of classification relies on the corresponding genomic context, i.e., the position in the chromosome where the lncRNA is transcribed. Additionally, it is becoming clear that the residing genomic localization also helps predict the functional roles of a category of lncRNAs. With this perspective, lncRNAs can be characterized as: 1) lincRNAs (large intergenic noncoding RNAs), including well studied and cancer associated Xist,6,7 H19,8 HOTAIR,9,10 NEAT2/MALAT1,11-13 lincRNA-RoR,14 lincRNA-P21.15 Many lincRNAs are initially discovered by analyzing intergenic (non-protein-coding) chromatin-state maps that mark actively transcribed regions that are initiated by RNA Pol II (K4-K36 domain: H3K4me3 at the promoter and H3K36me3 along the whole transcripts), which is also the typical histone modification pattern for actively transcribed protein-coding genes. LincRNAs can be transcribed from thousands of genomic loci and it is estimated that the total number of human lincRNAs is around 3,300 with high evolutionary conservation.16,17 Furthermore, it was revealed that nearly 20% of lincRNAs associate with the chromatin-modifying complex PRC2 and affect gene expression programs.18 Another study found that nearly 30% of lincRNAs expressed in mice embryonic stem cells are associated with at least one of 11 chromatin modifying factors,19 suggesting potentially similar functional mechanisms for many lincRNAs.16,18 Two) Natural antisense transcripts. They are transcribed with the opposite orientation to the sense DNA strand of their mRNA/lncRNA counterparts at the same or separate genomic loci and form perfect/imperfect pairs.20,21 They are prevalent in eukaryotes, including humans, mice, yeast, Drosophila, and Aragidopsis.22 Functionally, they have been proposed to control multiple layers of gene regulation including transcription, mRNA splicing, and translation,22 particularly regulating neighboring conjugated sense gene expression.21,22 For example, Kcnq1ot1 antisense silences its flanking genes via deposition of inactive chromatin-specific histone modifications.23 Currently, the highest estimated number of human NATs is around 6,000.24 Three) Pseudogenes. By definition, pseudogenes are dysfunctional counterparts of genes that have lost protein-coding potential due to accumulation of mutations during genome evolution.25 They are generally identified by computational analysis of genomic sequences using complex algorithms and are characterized by homology to a known gene and nonfunctionality.26 It is estimated that the human genome contains more than 18,000 pseudogenes.27 More and more evidence suggests that pseudogenes may have physiological significance by their direct interactions with DNA or transcripts of the parental protein-coding genes.25 Recently, it was revealed that the mRNA levels of the tumor suppressor PTEN and oncogenic KRAS are regulated by their pseudogenes PTENP1 and KRASP1, attributing a novel biological role to expressed pseudogenes in cancer progression.28 Four) Long intronic ncRNAs. They are transcribed from intronic regions of protein-coding genes and it has been revealed that about 81% and 70% of all spliced human and mouse protein-coding genes, respectively, have transcriptionally active introns.29 LncRNAs transcribed from introns are generally produced through the post-splicing process and are indicative of gene transcription events, which affects many other genes and regulates their expression.30 For example, it was reported that a number of intronic RNA sequences are capable of binding to the core component EZH2, and in another case, overexpression of the intronic RNA for the gene SMYD3 was sufficient to reduce endogenous SMYD3 mRNA and protein levels in human cancer cells.31 Five) Other uncharacterized and divergent transcripts. It was reported that the human genome also produces many diverse and heterogeneous RNA species from transcription start sites and even the regulatory enhancer regions. Usually, these classes of lncRNAs have extremely low abundance in cells and their biological function remain largely elusive. Although lncRNAs can be classified into different groups based on the above criteria, it is still difficult to know the exact total number of distinct human lncRNAs. The combination of several well-established high-confidence lncRNA databases estimated that the total number of lncRNAs (lincRNAs+NATs+intronic lncRNAs+ pseudogenes+others) is at around 111,000 transcripts (integrated statistics from LNCipedia, Feb. 2015).
Functional Significance of LncRNAs and Their Underlying Molecular Mechanisms
It is now clear that the human genome encodes numerous lncRNAs and are now recognized as another crucial layer of the functional outputs of the mammalian genome with bona fide, widespread biological functions across diverse biological processes;32,33 however, compared to protein-coding genes, there is still little knowledge regarding the biological roles of lncRNAs due to technical limitations and the intrinsic properties of lncRNAs, such as short half-life and extremely low levels. Several well-investigated cases have reported on the implicated roles of lncRNAs in X-chromosome inactivation,6,7 imprinting,34,35 control of pluripotency in lineage differentiation,14,19 as well as some diseases such as cancer.32,36,37 1). X-chromosome inactivation (XCI) is an early developmental process by which one X chromosome is transcriptionally silenced in female mammals. It is now well known that the lncRNA Xist acts as a major effector during the XCI process. The inactive X chromosome is coated with Xist, which is essential for the initiation and maintenance of XCI.6 Another lncRNA Tsix is a gene antisense to Xist located at the X-inactivation center and has a role in regulating the early steps of X-inactivation but not the silencing step.38,39 Recently, one study has suggested that the RNA Xist silences X-chromosome transcription by directly interacting with SHARP, recruiting SMRT, activating HDAC3, and deacetylating histones to exclude Pol II across the X chromosome.7 2) Genomic imprinting affects 1% of genes in mammals and results in a monoallelic, parental-specific expression pattern, which is achieved by putting epigenetic marks, such as DNA methylation, at specific gene loci in gametes.40 The majority of imprinted clusters contain a lncRNA, which is crucial for maintaining imprinted gene signatures. For example, the lncRNAs Kcnq1/Kcnq1ot1 and Airn are involved in the imprinting of related genomic loci by occupying the chromatin and recruiting the chromatin remodeling complex to achieve the imprinting effects.41-43 3) Previous studies have identified the governing transcription factors required for maintaining pluripotency, namely, Oct4, Nanog, Sox2, Klf4, and c-Myc.44 By performing a shRNA-mediated loss-of-function screening, one recent study showed that 26 lincRNAs are required for the maintenance of pluripotency of mES cells. The authors found that knockdown of dozens of lincRNAs resulted in a departure from the pluripotent state and upregulation of differentiation programs.19
Mechanistically, a handful of studies have implicated lncRNAs in recruiting/directing the chromatin modifying complexes at specific genomic loci to modify chromatin structures and further regulate the gene expression program. Indeed, RNA has been speculated to be an integral component of chromatin in addition to proteins and DNA since long ago. The discovery of lncRNAs and further understanding of their biology greatly helps us to appreciate how RNA species are involved in pathways of chromatin modification. Benefitting from the development and standardization of new techniques such as lncRNA-pulldown, RNA immunoprecipitation (RIP), Crosslinking and RNA Immunoprecipitation (CLIP), and Chromatin Isolation by RNA Purification (ChIRP),36,45,46 the list of chromatin modification complexes associated with lncRNAs is growing steadily.47 For example, HOTAIR associates with 2 different chromatin modification complexes PRC2 and LSD1/CoREST/REST and functions as a scaffold/guider to assemble these factors to genomic DNA to repress gene expression in the HOXD locus.9 HOTTIP binds the adaptor protein WDR5 directly and targets WDR5/MLL complexes across HOXA to maintain active chromatin.48 The lateral mesoderm-specific lncRNA Fendrr is essential for proper heart and body wall development in the mouse. Fendrr associates with the PRC2 and TrxG/MLL complexes to regulate H3K27 trimethylation and H3K4 trimethylation at the promoter regions of several transcription factors.49 Beyond the aforementioned major theme regarding lncRNAs in chromatin states regulation, which has been extensively studied, lncRNAs can also directly regulate transcription by interference with RNA Pol II50,51 by acting as decoys for transcription factors52 or by affecting the localization of transcription factors to achieve fine-tuned gene expression programs.53 Intriguingly, at the transcriptional level, a set of lncRNAs can function same with defined chromatin enhancers to promote the expression of neighboring protein-coding genes.54 In addition, it has been reported that the lncRNAs TUG1 and NEAT2 are involved in gene activation or repression through organization of distinct nuclear substructures, such as Polycomb bodies and interchromatin granules in response to growth signals, with lncRNAs as the key functional players that bind either methylated or unmethylated Pc2.13 LncRNAs also regulate mRNA processing including splicing55 and editing,56 as well as post-transcriptional events such as controlling the initiation of translation and mRNA stability via direct base pairing.57 Recently, there is evidence supporting mutual regulation between lncRNAs and miRNAs. For example, the lncRNAs PTENP1 and KRASP1 have been found to serve as “sponges” to bind miRNA, thereby sequestering miRNAs away from their mRNA targets.28 The lncRNA H19 has been found to be a developmental reservoir of miR-675 that suppresses growth and Igf1r.58,59
Although lncRNAs function through a variety of interesting mechanisms, it is obvious that the current emphasis is still on how lncRNAs regulate transcription or post-transcriptional processes.32 Most assuredly, lncRNAs should be involved in a wide array of tasks in cells given their biochemical versatility, various cellular localizations, as well as large amounts of uncharacterized candidates. For example, a substantial proportion of lncRNAs resides within, or is dynamically shuttled, to the cytoplasm where they may regulate protein localization, modification, and even intrinsic enzymatic activity. A recent study found that lnc-DC bound directly to STAT3 in the cytoplasm, which promoted STAT3 phosphorylation by preventing STAT3 binding with SHP1 as well as subsequent dephosphorylation by SHP1 in the regulation of dendritic cell differentiation.60
The Implication of LncRNAs in Cancer
Compared to mRNA levels in cells, most types of lncRNA are present at relatively low levels. However, many of the lncRNAs show tissue specific expression patterns. Further lncRNA profiling in multiple cancer cell lines and clinical tissues has made it increasingly clear that many lncRNAs are expressed in a disease-, or developmental stage-specific manner, suggesting that they have specific biological significance, human disease relevance, and diagnostic applicability.32,61. Specifically, it has been reported that the expression levels of dozens of lncRNAs are correlated with cancer development and progression (Table 1); furthermore, gain-/loss-of-function analyses in various models indicate the importance of these lncRNAs in many cancer types.32,62 For example, the lncRNAs PCGEM1 and PRNCR1 are highly expressed in prostate cancers and regulate androgen dependent or independent cancer cell growth, revealing their potential as therapeutic targets by targeting lncRNA-dependent regulatory networks in human prostate cancers.36 HOTAIR is a highly expressed lncRNA in metastatic breast, liver, colorectal, gastrointestinal, and pancreatic cancer cells/tissues and its expression level in primary tumors is a powerful predictor of eventual metastasis and death.10,63 Other lncRNAs have been found to be tumor suppressors by operating as a transcriptional repressor. For example, Linc-p21 is induced by p53 and mediates p53-dependent gene repression in mice cells15 and acts in a reciprocal way with HIF-1α to modulate the Warburg effects in human cells.64 In addition, lncRNAs have been shown to have prognostic and diagnostic value in a number of cancers.65 In spite of these advances, the contributions of most lncRNAs to the hallmarks of cancer biology continues to be poorly characterized.66 In addition, mechanism studies of lncRNAs are still heavily focused on the interaction between lncRNAs and epigenetic modifiers or other chromatin associated factors. However, more exciting and even surprising findings should be anticipated after the development and utilization of new open-ended techniques and systematic assays in the fields of lncRNA and cancer research. This will also help us understand the molecular details about the interaction between lncRNAs and their protein partners, which are required for the design of effective therapeutic targets for the treatment of human cancers. In the next section, we will introduce how we have combined many open-ended and systematic methods to investigate the in vitro and in vivo functions of the lncRNA BCAR4 in regulating a non-canonical Hedgehog pathway. We will further discuss the potential of targeting the lncRNA BCAR4 for the treatment of human cancers.
Table 1.
The involvement of lncRNAs in various cancers
| lncRNA | Function/mechanisms | Cancer type | Refs |
|---|---|---|---|
| 7SK | Targeting/associated with HEXIM | Gastric | 91 |
| ANRIL | Epigenetic regulation of CDKN2A/B | Prostate | 92 |
| BANCR | N/A | Non-small lung cancer | 93 |
| BCAR4 | Required for GLI2-dependent transcription | Breast/Prostate | 83 |
| BCYRN1 | N/A | Breast/Esophagus/Ovarian | 94 |
| CCAT1 | N/A | Gastric/colorectal | 95,96 |
| H19 | Gene regulation/miRNA sponges | Breast/liver/prostate | 8,97–99 |
| HOTAIR | Epigenetic regulation/chromatin targeting | Breast/colorectal | 10,100 |
| LncRNA-LALR1 | Activating Wnt/β-catenin | Liver cancer | 101 |
| MALAT1 | Sequester/gene expression | Lung/colorectal | 102,103 |
| MEG3 | N/A | Cervical | 104 |
| MIR31HC | Regulation of HSP90 | Hepatocellular/colorectal | 105 |
| PCAT1 | Regulating BRCA2 and homologous recombination | Prostate | 106 |
| PVT1 | N/A | Breast/ovarian | 107 |
| PCGEM1 | Regulating AR receptor | Prostate | 36,108 |
| PRNCR1 | Regulating AR receptor | Prostate | 36 |
| TUG1 | Epigenetic regulation | Non-small lung cancer | 109 |
| UCA1 | Gene regulation | Breast | 110 |
| XIST | Imprinting | Hematologic cancer | 111 |
Potential Involvement of LncRNAs in Hedgehog Pathways in Cancer
The evolutionarily conserved canonical Hedgehog (Hh) system plays an important role in organogenesis and the pattering phase of normal early development from Drosophila to humans but has also been linked to tumorigenesis.67 In the canonical Hh pathway, 3 secreted ligands have been identified, namely Sonic Hedgehog (SHH), Indian Hedgehog (IHH), and Desert Hedgehog (DHH). These ligands bind to the negative regulatory receptor Patched (PTCH), a 12-transmembrane protein receptor, which inhibits the activation of Smoothened (SMO), a 7-transmembrane effector, by preventing its surface translocation into the cilium in the absence of Hh ligands. The binding between the ligands and PTCH receptors results in abolishment of the repression effect of PTCH on SMO and the activation of SMO orchestrates a signaling cascade, eventually leading to the final activation of GLI zinc finger transcriptional factors containing GLI1/2/368. In addition, protein kinase A, glycogen synthase kinase 3β, and casein kinase 1α also form a complex with other signaling components to regulate the activation of GLI transcription factors.69
In adults, the canonical Hh pathway is either largely inactive or essentially undetectable in most cells but is implicated and reactivated in the development of cancers including brain, lung, breast, prostate, and skin cancers.70 This reactivation is due either to ligand-dependent (autocrine or paracrine mechanisms to Hh ligands from tumor cells or stromal cells) or ligand-independent (a variety of mutations in the downstream components) mechanisms.67 GLI gene amplification was first reported in malignant glioma.71 Subsequently, inactivating mutations in PTCH or activating mutations in SMO were identified in nearly 90% of sporadic basal cell carcinoma.72,73 In addition, genetic mice models further shed light on the functional importance of the Hh pathway in tumorigenesis; for example, heterozygous loss of Ptch1 results in an increased tendency to develop basaloid tumors while transgenic expression of GLI1 in the epidermis results in skin tumors in mice.74,75 Aside from the aberrant reactivation of the canonical Hh pathway in cancers, the effector of GLI also directly interacts with other distinct cancer-associated signaling pathways, acting independently of the HH-PTCH1-SMO-GLI paradigm. For example, in esophageal adenocarcinomas, tumor necrosis factor-α (TNF-α) induced activation of the mammalian target of rapamycin (mTOR)-S6 kinase (S6K) via direct phosphorylation, results in GLI1 activation in a Hh ligand independent manner.76 Several studies have also demonstrated that TGF-β-SMAD3, RAF-MEK-MAPK, and PI3K-AKT cascades also lead to stabilization or increased expression of GLI in distinct cancer types, inducing the expression of Hh ligand-independent and GLI-dependent genes.77-79 This kind of crosstalk constitutes noncanonical Hh/GLI pathway activation.67 Genomic analysis has identified hundreds GLI target genes, many of which show tissue-/cell-specific patterns. However, a portion of them are common targets in distinct cell lines. These include genes involved in cell proliferation and survival (CCND1/2, MYCN, IGFBP6, BCL2) and genes involved in angiogenesis and metastasis (VEGF, TGF-β, SNAIL, MUC5AC).68,70 In breast cancer, the potential roles of Hh signaling have not been well defined. It has been reported that the PTCH2 mutation exists in both the primary tumor and brain metastasis of a patient with aggressive basal-like breast cancer (BLBC).80 There is also evidence that SMO and Hh are ectopically expressed in BLCB or invasive ductal carcinoma (IDC).81,82 Also, there have been reports of increased GLI activity in breast cancers; however, this activity occurs in the absence of Hh ligands, which raises the possibility that there might be other regulators, such as lncRNAs, that might contribute to GLI activation, especially since lncRNAs are known to directly bind transcription factors.
The LncRNA BCAR4 Regulates Non-canonical Hh/GLI2 Pathway in Breast Cancer Cells
Recent work from our lab demonstrated how a lncRNA, BCAR4, functions to orchestrate a noncanonical Hh cascade to activate GLI2-dependent gene transcription and to promote cancer cell metastasis83 (Fig. 1). Initially, we sought to identify human breast cancer relevant lncRNAs by analyzing clinical breast cancer tissue samples with a lncRNA array. BCAR4 ranked first in the candidate list and showed the most dramatic upregulation in breast cancer tissues. RNAScope analysis further supported the conclusion that higher BCAR4 expression is correlated with advanced lymph node metastasis stage and shorter survival time for breast cancer patients. Additionally, Oncomine data-mining showed that elevated BCAR4 expression is not only correlated to invasive breast cancer but also to ERBB2/ER/PR negative breast cancer status. The assortment of unbiased and compelling evidence strongly supports BCAR4 as a driving force in the process of breast cancer progression and metastasis.
Figure 1.
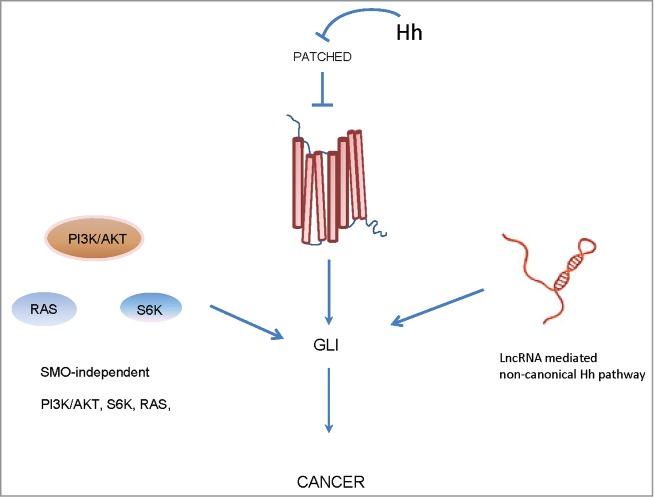
LncRNA mediated non-canonical Hh pathway.
Since most lncRNAs act in-trans to functional in cells, understanding the interacting protein partners of a lncRNA will by necessity reveal pertinent information to help determine the roles of a specific lncRNA. Interestingly, the putative protein complex, which associates with BCAR4, contains 2 RNA-binding proteins (SNIP1 and PNUTS), one kinase (CIT), and one transcription factor (GLI2). Furthermore, we performed a series of robust and thorough tests including EMSA, to study in vitro RNA-protein binding, as well as an RNA pull-down-Dot Blot assay to characterize the protein binding domains of the lncRNA. Our data support the notion that the primary RNA sequence (50–100 nt) may play a critical role in determining the specific RNA-protein interaction. This also suggests the potential explanation for the low sequence conservation of the lncRNA across different species as only a 50–100 nt fragment in the whole lncRNA gene appears to be essential for its interaction with proteins. Indeed, as it has been suggested that the secondary structure of lncRNAs mediates their functional interactions with protein factors,84 we speculated that this short fragment might possess highly structured regions critical for its function.
GLI2 is one of the 3 effectors (GLI1/2/3) that are downstream of the Hh pathway and its post-translational modifications, such as phosphorylation, have been reported to be critical for its activity. Our data show that CIT kinase can directly phosphorylate GLI2 at S149, which induces its nuclear translocation, binding to promoters of downstream genes, and activating transcription. The importance of GLI2 (S149) phosphorylation is supported by its correlation with invasive breast cancer status and its widespread existence in other tested cancer types (including lung, liver, colorectal, and ovarian cancers) as revealed by immunohistochemistry in a large number of clinical tissue samples. In addition, treatment cells with several cancer metastasis-related chemokines or growth factors including CCL21, CXCL21, IGF-1, or TGF-β induced GLI2 phosphorylation at S149 to differing degrees. To examine the genomic occupancy of BCAR4, we performed a Chromatin Isolation by RNA Purification (ChIRP) assay and found that after nuclear translocation and activation of phosphor-GLI2 (S149), BCAR4 is recruited to the promoters of GLI2-dependent downstream genes. Consistently, knockdown of BCAR4 dramatically suppresses CCL21-induced expression of GLI downstream target genes. Recent findings suggest that the Hh/GLI pathway is critical for cancer metastasis.68,70 Our loss-of-function studies in cells showed that knockdown of BCAR4 significantly inhibits the metastatic ability of multiple breast cancer cell lines; consistently, overexpression of full-length BCAR4, but not the deletion mutants which abolished SNIP1 and PNUTS binding, respectively, dramatically increased cell invasion and GLI2 target gene expression in a non-metastatic breast cancer cell line. These data strongly suggest the importance of BCAR4 in the phospho-GLI2-mediated transcriptional activation of a subset of genes, which contribute to breast cancer cell migration and invasion.
Mechanistically, we found that BCAR4 acts as a “double-unlock-switch” which is required for GLI2-dependent gene activation. Our research, and that of others, has found that the DUF domain of SNIP1 binds to the catalytic domains of p300 to inhibit its HAT activity;83,85 however, CCL21-induced binding of BCAR4 to the DUF domain of SNIP1 releases its interaction with the catalytic domain of p300, leading to the activation of p300. Subsequently, the activated p300 enhances the acetylation of H3K18 on the promoters of GLI2 target transcription units, which further releases the inhibitory roles of PNUTS on PP1 phosphatase enzymatic activity; this consequently modulates the phosphorylation of Pol II Ser5 at GLI2 target gene promoter regions and activates transcription. Therefore, on a molecular level, these findings demonstrate how a lncRNA, BCAR4, through its direct interactions with RNA binding proteins SNIP1 and PNUTS, acts as a key to bridge signal-induced epigenetic regulation of general transcription machinery during the activation of GLI2 target genes in breast cancer cells.
While targeted therapies against selected proteins in breast cancer are promising, they are limited due to the development of resistance. Our goal was to uncover clues for a more efficacious breast cancer treatment from a lncRNA point-of-view. There has been a longstanding interest in targeting noncoding RNAs by using a Locked Nucleic Acids (LNA)-based antisense oligonucleotides strategy, as there have been several successful applications that have targeted miRNAs in cancer.86 To evaluate the therapeutic potential of BCAR4, we used in vitro synthesized LNAs to knockdown endogenous BCAR4 expression. Significantly, 2 individual LNA treatments substantially reduced lung metastases, providing the first demonstration of the pharmacologic value of lncRNAs in human cancers.
Concluding Remarks and Future Perspectives
In summary, our findings are the first to show how a lncRNA directs cooperative epigenetic regulation downstream of specific chemokine signals, thereby contributing to breast cancer metastasis.83 Cell signaling, mediated mostly by membrane receptors, intracellular kinases, and nuclear transcriptional factors, is part of a complex system of communication that governs and coordinates basic cellular activities. The active involvement of lncRNA in this process makes it more likely that lncRNAs are innate components of the classical protein-mediated signal transduction pathways in cells. Intriguingly, we observed that BCAR4 is also highly expressed in other organ malignancies including lung squamous cell carcinoma, skin malignant melanoma, kidney clear cell carcinoma, colon adenocarcinoma, and rectum adenocarcinoma. In addition, knockdown of BCAR4 in HCT116, H1299, HepG2, and Hey8 cells significantly impaired the migration and invasion of these cells;83 this suggests that the lncRNA BCAR4 may also contribute to the metastatic potential of these cancers by regulating GLI2-dependent gene activation. Indeed, the TCGA database showed that elevated levels of the BCAR4 transcript are correlated with prostate cancer metastasis. It is necessary to investigate, in cancer cell lines from other cancer types, whether or not BCAR4 directly binds SNIP1 and PNUTS. In addition, it is important to examine the potential extracellular signals which induce GLI2 (S149) phosphorylation to comprehend the general human cancer relevance of this BCAR4-dependent GLI2 transcriptional program. Here we posit that a large proportion of cancer susceptibility may be the result of dysregulated lncRNAs. Examination of these lncRNA genes and their functional mechanisms will broaden our understanding of their biology, human cancer relevance, as well as their contributions to the hallmarks of cancers.66 Most importantly, these studies will accelerate their integration into clinical applications for diagnosis, prevention, and therapeutic treatment of human cancers. We are still in the early stages of the expanding field of lncRNA research and there is no doubt that development of new technologies and methods for characterizing the functions of lncRNAs will continue to accelerate the pace of exciting discoveries. For example, a new approach has been developed that allows for the direct evaluation of RNA structure in living cells and the assessment of dynamic changes in RNA structure in different cell states.87 In addition, RNA aptamers have been developed that bind fluorophores resembling the fluorophore in GFP, named Spinach, and is markedly resistant to photobleaching, which can be used to examine the localization of lncRNAs in cells.88
The development of acquired resistance to the targeting of specific cancer signals is clearly a complex phenomenon involving multiple pathways and has frequently been a challenge in cancer therapeutics; the Hh pathway is no exception to this phenomenon. Currently, the major antagonists against the Hh pathway target SMO, such as vismodegib, sonidegib, BMS-833923, PF04449913, and LY2940680, which have been studied in clinical trials for many cancers types.67 For example, the strategy of blocking SMO with vismodegib alone has been approved clinically for the treatment of advanced BCC,89 and responses have been observed for BCC treatment with either sonidegib or BMS-833923 alone.90 However, acquired resistance has been observed in both the preclinical and clinical settings due to the activation of noncanonical Hh pathway, amplification of downstream Hh target genes, or resistance mutations of SMO.67 One of the biggest challenges in treating drug-resistant tumors is the underlying complexity of the networking pathways; a detailed understanding of the mechanisms leading to GLI activation (canonical, noncanonical, and crosstalk) would allow for the development of the appropriate targeted therapies and improved outcomes. While consideration of lncRNAs in this pathway does add additional complexity, it provides the rational basis for the targeting of either lncRNA alone or as part of a combination therapy with established inhibitors to overcome drug resistance.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014; 15, 7-21; PMID:24296535; http://dx.doi.org/ 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- 2.Clark MB, Mercer TR, Bussotti G, Leonardi T, Haynes KR, Crawford J, Brunck ME, Cao KA, Thomas GP, Chen WY, et al.. Quantitative gene profiling of long noncoding RNAs with targeted RNA sequencing. Nat Meth 2015; 12, 339-42; PMID:25751143; http://dx.doi.org/ 10.1038/nmeth.3321 [DOI] [PubMed] [Google Scholar]
- 3.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al.. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 2012; 22, 1775-89; PMID:22955988; http://dx.doi.org/ 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie C, Yuan J, Li H, Li M, Zhao G, Bu D, Zhu W, Wu W, Chen R, Zhao Y. NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic Acids Res 2014; 42, D98-D103; PMID:24285305; http://dx.doi.org/ 10.1093/nar/gkt1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. Rna Biol 2013; 10, 924-33; http://dx.doi.org/ 10.4161/rna.24604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 2013; 504, 465-9; PMID:24162848; http://dx.doi.org/ 10.1038/nature12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al.. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015; 521:232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monnier P, Martinet C, Pontis J, Stancheva I, Ait-Si-Ali S, Dandolo L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD1. Proc Natl Acad Sci 2013; 110, 20693-8; http://dx.doi.org/ 10.1073/pnas.1310201110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai M-C, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329, 689-93; PMID:20616235; http://dx.doi.org/ 10.1126/science.1192002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al.. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464, 1071-6; PMID:20393566; http://dx.doi.org/ 10.1038/nature08975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al.. MALAT-1, a novel noncoding RNA, and thymosin [β]4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003;22, 8031-41; PMID:12970751; http://dx.doi.org/ 10.1038/sj.onc.1206928 [DOI] [PubMed] [Google Scholar]
- 12.Zhang B, Arun G, Mao YS, Lazar Z, Hung G, Bhattacharjee G, Xiao X, Booth CJ, Wu J, Zhang C, et al.. The lncRNA malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2012; 2, 111-23; PMID:22840402; http://dx.doi.org/ 10.1016/j.celrep.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011; 147, 773-88; PMID:22078878; http://dx.doi.org/ 10.1016/j.cell.2011.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al.. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 2010; 42, 1113-7; PMID:21057500; http://dx.doi.org/ 10.1038/ng.710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al.. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010; 142, 409-19; PMID:20673990; http://dx.doi.org/ 10.1016/j.cell.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al.. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009; 458, 223-7; PMID:19182780; http://dx.doi.org/ 10.1038/nature07672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al.. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotech 2010; 28, 503-10; http://dx.doi.org/ 10.1038/nbt.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al.. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009; 106, 11667-72; http://dx.doi.org/ 10.1073/pnas.0904715106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al.. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477, 295-300; PMID:21874018; http://dx.doi.org/ 10.1038/nature10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol 2012; 30, 453-9; http://dx.doi.org/ 10.1038/nbt.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Chung PJ, Liu J, Jang IC, Kean MJ, Xu J, Chua NH. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res 2014; 24, 444-53; PMID:24402519; http://dx.doi.org/ 10.1101/gr.165555.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapidot M, Pilpel Y. Genome‐wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep 2006; 7(12):1216-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 2008; 32, 232-46; PMID:18951091; http://dx.doi.org/ 10.1016/j.molcel.2008.08.022 [DOI] [PubMed] [Google Scholar]
- 24.Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense–antisense pairs. Nucleic Acids Res 2004; 32, 4812-20; PMID:15356298; http://dx.doi.org/ 10.1093/nar/gkh818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poliseno L. Pseudogenes: Newly Discovered Players in Human Cancer. Sci Signal 2012; 5:re5; PMID:22990117 [DOI] [PubMed] [Google Scholar]
- 26.Brosch M, Saunders GI, Frankish A, Collins MO, Yu L, Wright J, Verstraten R, Adams DJ, Harrow J, Choudhary JS, et al.. Shotgun proteomics aids discovery of novel protein-coding genes, alternative splicing, and “resurrected” pseudogenes in the mouse genome. Genome Res 2011; 21, 756-67; PMID:21460061; http://dx.doi.org/ 10.1101/gr.114272.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Gerstein M. Large-scale analysis of pseudogenes in the human genome. Curr Opin Genet Dev 2004; 14, 328-35; PMID:15261647; http://dx.doi.org/ 10.1016/j.gde.2004.06.003 [DOI] [PubMed] [Google Scholar]
- 28.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010; 465, 1033-8; PMID:20577206; http://dx.doi.org/ 10.1038/nature09144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Louro R, El-Jundi T, Nakaya HI, Reis EM, Verjovski-Almeida S. Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci. Genomics 2008; 92, 18-25; PMID:18495418; http://dx.doi.org/ 10.1016/j.ygeno.2008.03.013 [DOI] [PubMed] [Google Scholar]
- 30.Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics 2009; 93, 291-8; PMID:19071207; http://dx.doi.org/ 10.1016/j.ygeno.2008.11.009 [DOI] [PubMed] [Google Scholar]
- 31.Guil S, Soler M, Portela A, Carrère J, Fonalleras E, Gómez A, Villanueva A, Esteller M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol 2012; 19, 664-70; PMID:22659877; http://dx.doi.org/ 10.1038/nsmb.2315 [DOI] [PubMed] [Google Scholar]
- 32.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Bio 2013; 14, 699-712; http://dx.doi.org/ 10.1038/nrm3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012; 81, 145-66; PMID:22663078; http://dx.doi.org/ 10.1146/annurev-biochem-051410-092902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santoro F, Mayer D, Klement RM, Warczok KE, Stukalov A, Barlow DP, Pauler FM. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development 2013; 140, 1184-95; PMID:23444351; http://dx.doi.org/ 10.1242/dev.088849 [DOI] [PubMed] [Google Scholar]
- 35.Lee Jeannie T, Bartolomei Marisa S. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 2013; 152, 1308-23; PMID:23498939; http://dx.doi.org/ 10.1016/j.cell.2013.02.016 [DOI] [PubMed] [Google Scholar]
- 36.Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al.. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 2013; 500, 598-602; PMID:23945587; http://dx.doi.org/ 10.1038/nature12451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer 2013; 108, 2419-25; PMID:23660942; http://dx.doi.org/ 10.1038/bjc.2013.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 1999; 21, 400-4; PMID:10192391; http://dx.doi.org/ 10.1038/7734 [DOI] [PubMed] [Google Scholar]
- 39.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev 2009; 23, 1831-42; PMID:19684108; http://dx.doi.org/ 10.1101/gad.1811209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet 2014; 15, 517-30; PMID:24958438; http://dx.doi.org/ 10.1038/nrg3766 [DOI] [PubMed] [Google Scholar]
- 41.Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat Genet 2000; 25, 19-21; PMID:10802648; http://dx.doi.org/ 10.1038/75546 [DOI] [PubMed] [Google Scholar]
- 42.Barlow DP, Bartolomei M. S. Genomic Imprinting in Mammals. Cold Spring Harb Perspect Biol 2014; 6:1-20; PMID:24492710; http://dx.doi.org/ 10.1101/cshperspect.a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mancini-DiNardo D, Steele SJS, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 2006; 20, 1268-82; PMID:16702402; http://dx.doi.org/ 10.1101/gad.1416906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126, 663-76; PMID:16904174; http://dx.doi.org/ 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 45.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al.. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007; 129, 1311-23; PMID:17604720; http://dx.doi.org/ 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu C, Qu K, Zhong Franklin L, Artandi Steven E, Chang Howard Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 2011; 44, 667-78; PMID:21963238; http://dx.doi.org/ 10.1016/j.molcel.2011.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakagawa S, Kageyama Y. Nuclear incRNAs as epigenetic regulators—beyond skepticism. Bioch Biophys Acta 2014; 1839, 215-22; http://dx.doi.org/ 10.1016/j.bbagrm.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 48.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al.. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011; 472, 120-4; PMID:21423168; http://dx.doi.org/ 10.1038/nature09819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M, et al.. The tissue-specific lncRNA fendrr is an essential regulator of heart and body wall development in the mouse. dev Cell 24, 206-14; PMID:23369715; http://dx.doi.org/ 10.1016/j.devcel.2012.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human alu RNA Is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 2008; 29, 499-509; PMID:18313387; http://dx.doi.org/ 10.1016/j.molcel.2007.12.013 [DOI] [PubMed] [Google Scholar]
- 51.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007; 445, 666-70; PMID:17237763; http://dx.doi.org/ 10.1038/nature05519 [DOI] [PubMed] [Google Scholar]
- 52.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al.. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011; 43, 621-9; PMID:21642992; http://dx.doi.org/ 10.1038/ng.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005; 309, 1570-3; PMID:16141075; http://dx.doi.org/ 10.1126/science.1115901 [DOI] [PubMed] [Google Scholar]
- 54.Ørom UA, et al.. Long non-coding RNAs with enhancer-like function in human. Cell 2010; 143, 46-58; http://dx.doi.org/ 10.1016/j.cell.2010.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al.. The Nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010; 39, 925-38; PMID:20797886; http://dx.doi.org/ 10.1016/j.molcel.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci 2010; 35, 377-83; PMID:20382028; http://dx.doi.org/ 10.1016/j.tibs.2010.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong CG, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3 ′ UTRs via alu elements. Nature 2011; 470, 284-8; PMID:21307942; http://dx.doi.org/ 10.1038/nature09701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007; 13, 313-6; PMID:17237358; http://dx.doi.org/ 10.1261/rna.351707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and igf1r. Nat Cell Biol 2012; 14, 659-65; PMID:22684254; http://dx.doi.org/ 10.1038/ncb2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014; 344, 310-3; PMID:24744378; http://dx.doi.org/ 10.1126/science.1251456 [DOI] [PubMed] [Google Scholar]
- 61.Gutschner T, Diederichs S. The hallmarks of cancer A long non-coding RNA point of view. Rna Biol 2012; 9, 703-19; PMID:22664915; http://dx.doi.org/ 10.4161/rna.20481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov 2011; 1, 391-407; PMID:22096659; http://dx.doi.org/ 10.1158/2159-8290.CD-11-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics 2013; 193, 651-69; PMID:23463798; http://dx.doi.org/ 10.1534/genetics.112.146704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang F, Zhang H, Mei Y, Wu M. reciprocal regulation of HIF-1α and LincRNA-p21 modulates the warburg effect. Molecular Cell 2014; 53, 88-100; PMID:24316222; http://dx.doi.org/ 10.1016/j.molcel.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 65.Yang G, Lu X, Yuan L. LncRNA: A link between RNA and cancer. Biochim Biophys Acta 2014; 1839, 1097-109; http://dx.doi.org/ 10.1016/j.bbagrm.2014.08.012 [DOI] [PubMed] [Google Scholar]
- 66.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell 2011; 144, 646-74; PMID:21376230; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 67.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the hedgehog pathway in cancer. Nat Med 2013; 19, 1410-22; PMID:24202394; http://dx.doi.org/ 10.1038/nm.3389 [DOI] [PubMed] [Google Scholar]
- 68.Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev 2008; 22, 2454-72; PMID:18794343; http://dx.doi.org/ 10.1101/gad.1693608 [DOI] [PubMed] [Google Scholar]
- 69.Merchant M, Vajdos FF, Ultsch M, Maun HR, Wendt U, Cannon J, Desmarais W, Lazarus RA, de Vos AM, de Sauvage FJ. Suppressor of fused regulates gli activity through a dual binding mechanism. Mol Cell Biol 2004; 24, 8627-41; PMID:15367681; http://dx.doi.org/ 10.1128/MCB.24.19.8627-8641.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scales SJ, de Sauvage FJ. Mechanisms of hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci 2009; 30, 303-12; PMID:19443052; http://dx.doi.org/ 10.1016/j.tips.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 71.Kinzler KW, Bigner SH, Bigner DD, Trent JM, Law ML, O'Brien SJ, Wong AJ, Vogelstein B. Identification of an amplified, highly expressed gene in a human glioma. Science 1987; 236, 70-3; PMID:3563490; http://dx.doi.org/ 10.1126/science.3563490 [DOI] [PubMed] [Google Scholar]
- 72.Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer 2008; 8, 743-54; PMID:18813320; http://dx.doi.org/ 10.1038/nrc2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lam CW, Xie J, To KF, Ng HK, Lee KC, Yuen NW, Lim PL, Chan LY, Tong SF, McCormick F. A frequent activated smoothened mutation in sporadic basal cell carcinomas. Oncogene 1999; 18, 833-6; PMID:9989836; http://dx.doi.org/ 10.1038/sj.onc.1202360 [DOI] [PubMed] [Google Scholar]
- 74.Kimura H, Stephen D, Joyner A, Curran T. Gli1 is important for medulloblastoma formation in Ptc1+//- mice. Oncogene 2005; 24, 4026-36; PMID:15806168; http://dx.doi.org/ 10.1038/sj.onc.1208567 [DOI] [PubMed] [Google Scholar]
- 75.Nilsson M, Undèn AB, Krause D, Malmqwist U, Raza K, Zaphiropoulos PG, Toftgård R. Induction of basal cell carcinomas and trichoepitheliomas in mice overexpressing GLI-1. Proc Natl Acad Sci U S A 2000; 97, 3438-43; http://dx.doi.org/ 10.1073/pnas.97.7.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, Li CW, Hsu JL, Miller SA, Wang X, et al.. The Crosstalk of mTOR/S6K1 and hedgehog pathways. Cancer Cell 2012; 21, 374-87; PMID:22439934; http://dx.doi.org/ 10.1016/j.ccr.2011.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dennler S, André J, Alexaki I, Li A, Magnaldo T, ten Dijke P, Wang XJ, Verrecchia F, Mauviel A. Induction of sonic hedgehog mediators by transforming growth factor-β: smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res 2007; 67, 6981-6; PMID:17638910; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-0491 [DOI] [PubMed] [Google Scholar]
- 78.Ji Z, Mei FC, Xie J, Cheng X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem 2007; 282, 14048-55; PMID:17353198; http://dx.doi.org/ 10.1074/jbc.M611089200 [DOI] [PubMed] [Google Scholar]
- 79.Riobó NA, Lu K, Ai X, Haines GM, Emerson CP. Phosphoinositide 3-kinase and akt are essential for sonic hedgehog signaling. Proc Natl Acad Sci U S A 2006; 103, 4505-10; http://dx.doi.org/ 10.1073/pnas.0504337103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al.. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 2010; 464, 999-1005; PMID:20393555; http://dx.doi.org/ 10.1038/nature08989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O'Toole SA, Machalek DA, Shearer RF, Millar EK, Nair R, Schofield P, McLeod D, Cooper CL, McNeil CM, McFarland A, et al.. Hedgehog overexpression is associated with stromal interactions and predicts for poor outcome in breast cancer. Cancer Res 2011; 71, 4002-14; PMID:21632555; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-3738 [DOI] [PubMed] [Google Scholar]
- 82.Moraes RC, Zhang X, Harrington N, Fung JY, Wu MF, Hilsenbeck SG, Allred DC, Lewis MT. Constitutive activation of smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development 2007; 134, 1231-42; PMID:17287253; http://dx.doi.org/ 10.1242/dev.02797 [DOI] [PubMed] [Google Scholar]
- 83.Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH, et al.. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159, 1110-25; PMID:25416949; http://dx.doi.org/ 10.1016/j.cell.2014.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ulitsky I, Bartel David P. lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26-46; PMID:23827673; http://dx.doi.org/ 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim RH, Wang D, Tsang M, Martin J, Huff C, de Caestecker MP, Parks WT, Meng X, Lechleider RJ, Wang T, et al.. A novel smad nuclear interacting protein, SNIP1, suppresses p300-dependent TGF-β signal transduction. Genes Dev 2000; 14, 1605-16; PMID:10887155 [PMC free article] [PubMed] [Google Scholar]
- 86.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res 2006; 34, 2294-304; PMID:16690972; http://dx.doi.org/ 10.1093/nar/gkl183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Spitale RC, Crisalli P, Flynn RA, Torre EA, Kool ET, Chang HY. RNA SHAPE analysis in living cells. Nat Chem Biol 2013; 9, 18-20; PMID:23178934; http://dx.doi.org/ 10.1038/nchembio.1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science 2011; 333, 642-6; PMID:21798953; http://dx.doi.org/ 10.1126/science.1207339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, Solomon JA, Yoo S, Arron ST, Friedlander PA, et al.. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med 2012; 366, 2171-9; PMID:22670903; http://dx.doi.org/ 10.1056/NEJMoa1113713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jimeno A, Weiss GJ, Miller WH Jr, Gettinger S, Eigl BJ, Chang AL, Dunbar J, Devens S, Faia K, Skliris G, et al.. Phase I study of the hedgehog pathway inhibitor IPI-926 in adult patients with solid umors. Clin Cancer Res 2013; 19, 2766-74; PMID:23575478; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med 2013; 11, 225; PMID:24063685; http://dx.doi.org/ 10.1186/1479-5876-11-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bochenek G, Häsler R, El Mokhtari NE, König IR, Loos BG, Jepsen S, Rosenstiel P, Schreiber S, Schaefer AS. The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Hum Mol Genet 2013; 22, 4516-27; PMID:23813974; http://dx.doi.org/ 10.1093/hmg/ddt299 [DOI] [PubMed] [Google Scholar]
- 93.Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, et al.. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer 2014; 13, 68; PMID:24655544; http://dx.doi.org/ 10.1186/1476-4598-13-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen W, Böcker W, Brosius J, Tiedge H. expression of neural BC200 RNA in human tumours. J Pathol 1997; 183, 345-51; http://dx.doi.org/ 10.1002/(SICI)1096-9896(199711)183:3%3c345::AID-PATH930%3e3.0.CO;2-8 [DOI] [PubMed] [Google Scholar]
- 95.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol 2013; 139, 437-45; PMID:23143645; http://dx.doi.org/ 10.1007/s00432-012-1324-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xiang J-F, Yin QF, Chen T, Zhang Y, Zhang XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al.. Human colorectal cancer-specific CCAT1-L incRNA regulates long-range chromatin interactions at the MYC locus. Cell Res 2014; 24, 513-31; PMID:24662484; http://dx.doi.org/ 10.1038/cr.2014.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Berteaux N, Aptel N, Cathala G, Genton C, Coll J, Daccache A, Spruyt N, Hondermarck H, Dugimont T, Curgy JJ, et al.. A Novel H19 antisense RNA overexpressed in breast cancer contributes to paternal IGF2 expression. Mol Cell Biol 2008; 28, 6731-45; PMID:18794369; http://dx.doi.org/ 10.1128/MCB.02103-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, et al.. The imprinted H19 LncRNA antagonizes Let-7 microRNAs. Mol Cell 52, 101-12; PMID:24055342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, et al.. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. Febs J 2014; 281, 3766-75; PMID:24988946; http://dx.doi.org/ 10.1111/febs.12902 [DOI] [PubMed] [Google Scholar]
- 100.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al.. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011; 71, 6320-6; PMID:21862635; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-1021 [DOI] [PubMed] [Google Scholar]
- 101.Xu D, Yang F, Yuan JH, Zhang L, Bi HS, Zhou CC, Liu F, Wang F, Sun SH. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/β-Catenin signaling. Hepatology 2013; 58, 739-51; PMID:23483581; http://dx.doi.org/ 10.1002/hep.26361 [DOI] [PubMed] [Google Scholar]
- 102.Gutschner T, Hämmerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al.. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013; 73, 1180-9; PMID:23243023; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, et al.. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer 2014; 111, 736-48; PMID:25025966; http://dx.doi.org/ 10.1038/bjc.2014.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qin R, Chen Z, Ding Y, Hao J, Hu J, Guo F. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma 2013; 60, 486-92; PMID:23790166; http://dx.doi.org/ 10.4149/neo_2013_063 [DOI] [PubMed] [Google Scholar]
- 105.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone Deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell 2013; 49, 1083-96; PMID:23395002; http://dx.doi.org/ 10.1016/j.molcel.2013.01.010 [DOI] [PubMed] [Google Scholar]
- 106.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S, Sahu A, Malik R, Wilder-Romans K, Navone N, et al.. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res 2014; 74, 1651-60; PMID:24473064; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guan Y, Kuo WL, Stilwell JL, Takano H, Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al.. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res 2007; 13, 5745-55; PMID:17908964; http://dx.doi.org/ 10.1158/1078-0432.CCR-06-2882 [DOI] [PubMed] [Google Scholar]
- 108.Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, Livezey JR, Connell T, Sesterhenn IA, Yoshino K, et al.. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci U S A 2000; 97, 12216-21; PMID:11050243; http://dx.doi.org/ 10.1073/pnas.97.22.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, et al.. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis 2014; 5, e1243; http://dx.doi.org/ 10.1038/cddis.2014.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, Mo YY. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis 2014; 5, e1008; http://dx.doi.org/ 10.1038/cddis.2013.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152, 727-42; PMID:23415223; http://dx.doi.org/ 10.1016/j.cell.2013.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]


