Abstract
Bacterial symbionts of eukaryotes often give up generalist lifestyles to specialize to particular hosts. The eusocial honey bees and bumble bees harbor two such specialized gut symbionts, Snodgrassella alvi and Gilliamella apicola. Not only are these microorganisms specific to bees, but different strains of these bacteria tend to assort according to host species. By using in-vivo microbial transplant experiments, we show that the observed specificity is, at least in part, due to evolved physiological barriers that limit compatibility between a host and a potential gut colonizer. How and why such specialization occurs is largely unstudied for gut microbes, despite strong evidence that it is a general feature in many gut communities. Here, we discuss the potential factors that favor the evolution of host specialization, and the parallels that can be drawn with parasites and other symbiont systems. We also address the potential of the bee gut as a model for exploring gut community evolution.
Keywords: coevolution, gut symbionts, host specificity, insects, Snodgrassella
Bees serve critical ecological functions as plant pollinators. Some species, such as the Western honey bee (Apis mellifera), are indispensable for agriculture and have been prominent cultural icons in many human societies for thousands of years.1 Only recently have the microbiomes of these ubiquitous insects been described,2-4 and the bee gut has since emerged as an attractive model system for investigating gut community dynamics and host-microbe interactions. However, because of its novelty, the genomic and experimental data necessary for developing theoretical frameworks for this system have been lacking.5,6 In our paper,7 we sequenced the genomes of multiple strains of two common bee gut bacteria, Snodgrassella alvi and Gilliamella apicola, and showed that stringent host-symbiont compatibility is a characteristic property of this system.
The Specialized Gut Symbionts of Bees
Honey bees (Apis spp.) and bumble bees (Bombus spp) possess a distinctive gut microbiota dominated by about 8 bacterial phylotypes.2-4,8 Three groups, S. alvi, G. apicola, and Lactobacillus spp., form the majority of the gut community.9,10 Phylogenetic analyses indicate that they each comprise monophyletic clades of bee-associated bacteria, which is suggestive of an intimate symbiosis persisting over evolutionary time scales.4,11-14 The simplicity of the bee gut community, and its analogy to more complex mammalian models, offer a unique opportunity to study gut microbiomes from the perspective of microbial evolution and ecology in an experimentally tractable system.
Specialized microbial symbionts often exhibit host range restriction and co-diversification with host lineages.15-17 Indeed, various 16S rRNA surveys have consistently found patterns of correlation between bee gut symbiont strains and host species that cannot be explained by chance or geographic provenance alone.4,18 This is striking, as, unlike endosymbionts,19 gut associates possess greater avenues for dissemination, both through vertical transmission (e.g., from queen to daughter18), and through horizontal transmission (e.g., between workers20). Despite the capacity for transmission, different host species, including those living sympatrically, appear to harbor specific lineages of G. apicola and S. alvi that can be resolved through phylogenetic reconstruction.18
However, specificity of host association, defined here as the restriction of a microorganism to a particular host species or set of host species, does not imply specialization, which we define as the adaptation of a microorganism to a particular set of hosts and adaptation of the host to the microorganism. One can imagine scenarios in which extrinsic barriers such as geographic separation or niche segregation prevent hosts of different species from interacting in ways that allow for sharing of gut symbionts. This would result in apparent specificity, but not necessarily specialization. A phylogenetic correlation between host and symbiont is yet another aspect that can reflect long-term evolutionary associations, but may or may not result in specificity or specialization. For specialization, a host-microbe pair should display a direct preferential relationship in addition to any phylogenetic correlation. Perhaps the most straightforward method of testing this is through transplantation experiments, whereby cultured strains (or entire gut communities21) are introduced into gnotobiotic animals, and the microbial colonization load recorded as a proxy for host-microbe compatibility.
In our study, we inoculated S. alvi strains isolated from the honey bee A. mellifera and two bumble bees (Bombus bimaculatis and B. vagans) into lab-reared, germ-free adult workers of A. mellifera and B. impatiens.7 Consistent with the hypothesis of strain-level host-specialization, we observed higher levels of colonization in bees inoculated with their native S. alvi strains (Fig. 1). Although the Bombus-derived S. alvi strains were not isolated from B. impatiens, their hosts of origin were from the same subgenus (Pyrobombus), suggesting some flexibility within a general trend of decreasing compatibility with increasing host genetic distance. Cross-inoculation experiments with isolates from more distantly related Bombus will be needed to prove this.
Figure 1.
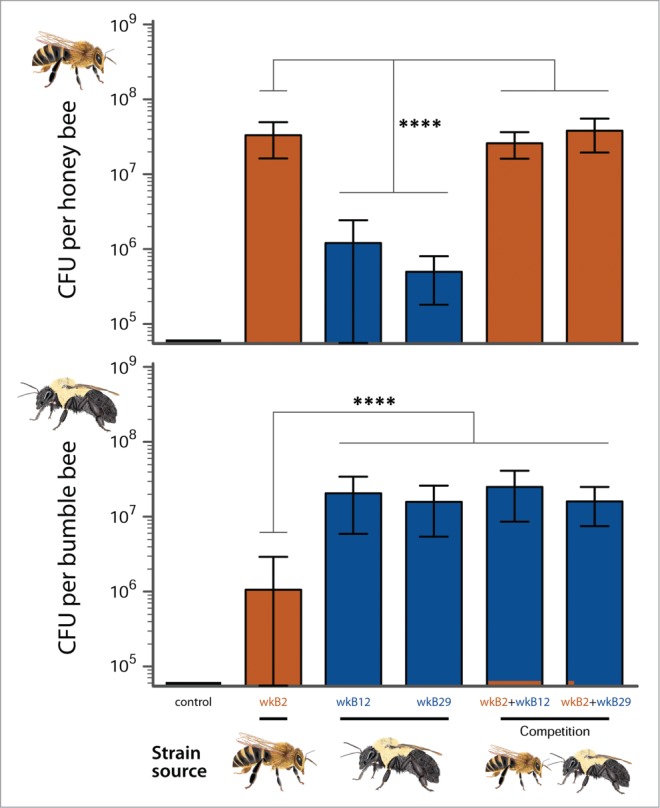
Host-specialized S. alvi strains, as demonstrated by transplantation and competition assays. In-vitro cultured strains were fed to sterile, newly-emerged adults, and total CFU counted from guts after 5 days. In competition assays, the inoculum consisted of a 1:10 ratio of native to non-native strain. Recovered proportions of each strain type are represented as bar colors: Orange, Apis-derived strain (wkB2); blue, Bombus-derived strains (wkB12, wkB29). ****P < 0.0001, bars denote 95% CI of means. Figure adapted from Kwong et al.7
We also conducted co-inoculations and found that the native S. alvi strains were able to become dominant in the gut despite an initial numerical disadvantage (Fig. 1). These kinds of competition assays are another simple way to indirectly test for host specialization. Because non-resident microorganisms may colonize opportunistically in the absence of the normal flora, the mere observation of colonization is insufficient to determine specialization. In a competition between a specialized community and an artificially introduced one, however, the one that has evolved to thrive in the gut of that particular host species will almost invariably win out.21
There are myriad reasons why we should care about host specialization. From a practical standpoint, specialized gut communities are indicative of intimate, evolved interactions between host and microbe, and hence are key to mediating symbiotic benefits that affect host biology. Gut microbial incompatibilities may lead to detrimental outcomes for host immunity and development.22,23 From an evolutionary perspective, specialized gut bacteria represent a unique but ubiquitous form of symbiosis that has thus far escaped close scientific scrutiny. The forces and mechanisms that shape symbioses in the gut remain largely unknown.
Evolution of Specialization
Host ecology, neutral genetic drift, and selective forces all likely contribute to host specialization in gut microorganisms. Disentangling these factors is challenging, but we suggest that the propensity for a gut bacterium to be specialized can be ascribed to 3 general characteristics: transmission mode, cost/benefit to host, and cost/benefit to the microbe. Systems in which vertical transmission dominates will enforce allopatry of microbial lineages in closely related hosts, enhancing divergence due to both drift and divergent selection reflecting distinct ecological niches of different host species. On the other hand, horizontal transmission between host species would lead to homogenization and fewer opportunities for specialization.
Beneficial microbes are expected to be preferentially retained by hosts due to selection, and thus will also be favored to become specialists. Microorganisms that harm their hosts, and thus threaten the persistence of their own microenvironment, would be unlikely to form the long-term associations needed for evolution of specialization, unless this is offset by a tremendous fitness advantage to the microbe. This would be the case for pathogens, for which the benefit of residing in a particular hostile host is greater than that of any other host or abiotic environment.
Ecological factors, chance, and selection are obviously not constant for a system, but shift through time. A host-microbe interaction that initially provides small benefits to the host or to the microbe may lead down the road to greater specialization, and would be aided by the establishment of a stable mode of transmission. Absent horizontal gene transfers, this would tend to be an irreversible process: genomic erosion, co-evolution with host immune function, and development of genetic incompatibilities (a Bateson–Dobzhansky–Muller model,24 but with incompatible loci between host and microbe genomes) would discourage promiscuity and host switching.
For the eusocial corbiculate bees, there appear to be at least 4 lineages of gut bacteria exhibiting host specificity: S. alvi, G. apicola, Lactobacillus spp, and Bifidobacterium spp.11,12,18 However, specialization to particular host lineages remains mostly untested by transplantation experiments, and 16S rRNA lacks sufficient resolution to reconstruct detailed phylogenetic histories of these bacteria at the strain level. New approaches leveraging the power of high-throughput genomics may help unravel the processes behind the evolution of specialization: shotgun metagenomics and metatranscriptomics enable functional profiling of whole communities,5,25 16S rRNA gene surveys allows broad assessment of community composition at the genus level,10 and an increasing number of sequenced strains and single cells7,26-31 permit analysis of diversity at the individual bacterium level.
These studies are beginning to reveal the intricate tapestry that is the history of the corbiculate bee gut microbiota, and point to a complex web of gene flow and recombination,9,30 as well as strong signals of specificity reflecting millions of years of host-microbe codivergence (Fig. 2).7,18 Within an individual host, deeply branching symbiont lineages also appear to coexist – cryptic species of gut bacteria that are all but invisible by 16S rRNA analysis.30 Such parallel lineages could reflect specialization of function to distinct ecological niches within the gut,5 a process likely common in gut microbes.32,33 The existence of reliable transmission routes, possible benefits to host, and an enriched habitat for gut symbionts may ultimately facilitate evolution of both sympatric diversification within hosts and specialization between hosts.
Figure 2.
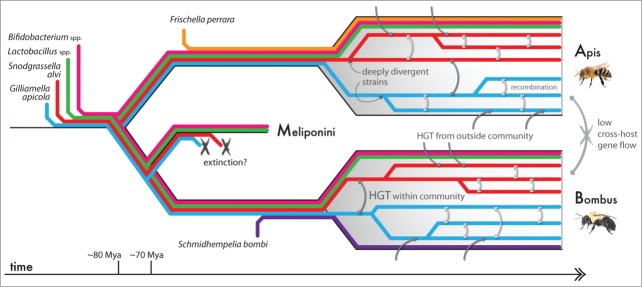
Evolution of the corbiculate bee gut microbiota. The eusocial origins of the coribiculate bees59 may have facilitated the development of a specialized gut microbiota by enlarging host reservoirs and providing a reliable transmission route. The symbiont genera Gilliamella, Snodgrassella, Lactobacillus, and Bifidobacterium may be ancestral to corbiculates (originating ca. 80 Mya60), and are presently all found in Apis and Bombus bees.4,12,14,18 Stingless bees (Meliponini) appear to have lost Gilliamella and Snodgrassella,18,61 but retain Lactobacillus and Bifidobacterium.14 Two bacteria related to Gilliamella (order: Orbales) were likely acquired sometime later: Frischella by the Apis lineage,31 and Schmidhempelia by Bombus.29 Within Apis and Bombus, Gilliamella and Snodgrassella strains have substantially diverged at many genomic loci, suggesting the existence of deeply-branching lineages co-existing within the same host.7,30 This may be due to niche differentiation in the gut.5 Recombination between some lineages still occurs, however, and likely explains the high 16S rRNA identity between strains.7,9,30 Horizontal gene transfer (HGT) between gut symbionts7 and from other/environmental sources62 may allow for dynamic gene repertoires in the bee gut microbiota. Nonetheless, it appears that gene flow between strains native to different bee hosts is generally limited.7 It is possible these evolutionary characteristics also extend to the other bee gut species, but, unlike Gilliamella and Snodgrassella, they have not yet been closely examined. Events other than that of known host splits (timings marked) are speculative and are for illustrative purposes only.
Mechanisms for Maintaining Specificity
Specialization, defined as adaptation through natural selection for the ability to use a host or to accept a symbiont, produces specific mechanisms that help establish and maintain the association. The molecular bases for symbioses are still poorly understood, particularly for gut microbes. Genome sequencing is now typically the first step in elucidating specificity determinants, and our genomic analysis of Snodgrassella and Gilliamella uncovered a large repertoire of cell-cell interaction genes which may perform such roles.7 These include RTX toxins, type VI secretion systems, type IV pili, capsular polysaccharides, and trimeric autotransporter adhesins.7 While these are the most promising candidates, the suite of host-specificity determinants undoubtedly extend beyond direct interaction genes and will require additional experimental evidence to be identified and validated.
Studies of bacterial symbionts (pathogens as well as mutualists) suggest that host specificity is mediated through at least 3 types of processes: host recognition and colonization, compatibility with host immune systems, and acquisition of nutrients specific to the host environment.34 In Vibrio fischeri, a bioluminescent symbiont of marine animals, specificity to the squid Euprymna scolopes critically depends on RscS, a sensor kinase that detects an as-yet unknown host factor and induces expression of exopolysaccharide that enables colonization.35 V. fisheri strains that colonize fish, in contrast, lack RscS.35 The mouse gut symbiont Lactobacillus reuteri also relies on biofilms for colonization, and the inability of L. reuteri strains from humans, pigs or chickens to establish in mice likely stems, in part, from the absence of particular genes for biofilm production.36
Human-specific bacterial pathogens can evade host defenses by utilizing proteases to break down antibodies or by binding down-regulators of complement-mediated immunity.34 Conversely, the host may develop specialized immune responses to encourage colonization of a beneficial microbiota, such as has been proposed for antimicrobial-peptide-mediated host specificity in Hydra.37 Nutritionally, each host presents a unique selective environment for a microbe. For example, cattle-specific Campylobacter strains tend to possess a vitamin B5 synthesis locus lacking in chicken-specific strains, presumably due to vitamin B5 scarcity in grasses compared to chicken feed.38 Microbes and their hosts may also have to compete for the same scarce resources. Opportunistic pathogens such as Neisseria and Haemophilus have evolved receptors to pick up iron in host-bound molecules of transferrin, leading to a co-evolutionary arms race and accelerated adaptive evolution at the responsible loci in both host and microbe.39
Like V. fischeri, host specificity for the nematode symbiont Xenorhabdus can be mediated by a single locus. Here, the genes nilABC are unique to strains infecting the nematode Steinernema carpocapsae but are absent in other Xenorhabdus; heterologous expression of nilABC in the other Xenorhabdus enable their colonization of S. carpocapsae.40 These findings beg the question as to whether single-locus dependent specificity, such as rscS and nilABC, are extreme outlier cases or, rather, represent a more general basis for host specialization. In Salmonella enterica, a widespread pathogen of mammals and birds, adaptation to hosts is thought to be multifactorial, with both gene gain and loss playing a part.41,42 However, it is unclear whether these events are the cause or consequence of specialization. A recent gene-swapping study of V. fischeri strains hosted by Australian or Hawaiian Euprymna squids suggested that multifactor-mediated host specificity is not incompatible with single loci of large effect: there may in fact be multiple genes in a genome capable of greatly altering host affinity.43
These studies demonstrate that horizontal gene transfer, whether by an experimenter or by natural processes (as proposed for rscS35 and nilABC40), can greatly alter a microbe's host range. In the plant pathogens Xanthomonas and Pseudomonas, type III secretion system effectors are likely important determinants of host specificity.44,45 The horizontal acquisition of the permissive effector genes can lead to effective colonization of the same host plant by distantly related pathogen strains, thus breaking apart the phylogenetic host-microbe correlations typically associated with co-evolved symbioses.44,45
Both horizontal gene transfer and genomic degradation probably play prominent roles in the evolution of specialization,42,46 but to what extent remains an unresolved question. There is also the host perspective to consider, as interplay between host immunity and the microbiota constitutes an ongoing dialog between partners that often have competing evolutionary interests.47 Behavioral mechanisms by the host (e.g. coprophagy, egg-smearing) may also evolve to facilitate symbiont maintenance. Delineating the diversity of mechanisms behind host specialization and the dominant forces influencing their evolution will be critical steps going forward, as will be the description of any general rules governing differences in these properties among mutualists, pathogens, and commensals, and between animal gut microbiotas and other types of host-microbe associations (Table 1).
Table 1.
Examples of host specificity in extracellular bacterial symbionts. The degree of specialization and the mechanisms involved remain areas of active investigation in these systems.
| Host ranges | Reported mechanisms of specificity | References | |
|---|---|---|---|
| Gut microbes | |||
| Campylobacter jejuni | mammals, birds | vitamin B5 biosynthesis | 38 |
| Lactobacillus reuteri | mammals, birds | biofilm production | 36 |
| Salmonella enterica | mammals, birds, reptiles | pathogenicity factors, loss of metabolic pathways | 42 |
| Snodgrassella alvi | honey bees and bumble bees | unknown | 7 |
| Other symbionts | |||
| Pseudomonas syringae, Xanthomonas spp. | plants | virulence factors, type 3 secretion effectors | 44,45 |
| Rhizobia | legume plants | Nod factors | 51,52 |
| Streptomyces philanthi | beewolf wasps | host response or behavior | 53 |
| Vibrio fischeri | squid, fish, environmental | biofilm production, bioluminescence | 35,43 |
| Xenorhabdus nematophila | nematodes | nilABC locus | 40 |
Methodologies to probe the genomic underpinnings of specialization are becoming ever more accessible due to advances in sequencing technologies. Genome-wide association,38 RNAseq,36 and TnSeq48 are now effective ways to quickly screen for candidate genes. Meanwhile, the toolbox for organismal genetic manipulation is also increasing rapidly.49,50 We anticipate that the development of new model systems, such as the bee gut community, will continue to accelerate in the years to come, and will provide much needed context toward understanding the diversity of gut microbial symbioses.
Conclusion and Perspective
Mounting evidence suggests that many gut microbes are host-specific,54-56 preferentially associating with a particular species over any other potential host or environment. Thus far, however, correlational data is in much greater abundance than elucidated causal mechanisms. Are these host-specific microbes really specialized to their hosts, or have circumstances simply produced the observed associations? In other words, given the chance, are these microbes able to colonize a range of other hosts? Specialization should be tested by transplantation and competition assays, and mechanisms need to be deduced from ‘–omics’ approaches and verified experimentally. Given the enormous plasticity of microbial genomes and propensity for horizontal gene transfer, greater scrutiny of strain-level variation at a genome-wide scale will also be essential to explain the evolution and diversification of gut microbes.
As a whole, gut microbes already comprise a highly derived group of organisms, distinct from their free-living predecessors. The forces driving ever-increasing specialization, down to the strain level, have yet to be clarified, but we predict that transmission mode and relative fitness benefits to the host and/or the microbe play a large part. Quantifying the contribution of fitness, over long time scales, to the development of specialization remains a challenge for the study of symbioses from an evolutionary perspective. Another open question is whether specialization destines microbes to an evolutionary dead-end due to the increased risk of extinction that result from highly restricted host ranges and the loss of functional capabilities from genome erosion. Intracellular symbionts can degenerate to the point where they are replaced,57 but for gut microbes, the prospect of gene flow may prevent this outcome.
The bee gut microbiota represents a system in which bacterial lineages have diversified within hosts and have evolved to specialize to distinct host species. These features parallel those apparent in the more complex microbiotas of mammals including humans, and the parallels reflect the fact that both are transmitted directly among individual hosts through social contact. The extent and nature of within-host and between-host diversification of such symbionts may have major implications for hosts.58 Thus, the bee gut community offers a simple model for investigating how coevolution of host-specialized gut symbionts affects host health and disease.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Kim Hammond for providing figures.
Funding
This work was supported by Yale University and the Canadian Natural Sciences and Engineering Research Council Postgraduate Scholarship (to WKK), and the US. National Science Foundation Dimensions of Biodiversity awards 1046153 and 1415604 (to NAM).
References
- 1.Crane E. The World History of Beekeeping and Honey Hunting. New York: Routledge; 1999 [Google Scholar]
- 2.Jeyaprakash A, Hoy MA, Allsopp MH. Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. J Invertebr Pathol 2003; 84(2):96-103; PMID:14615218; http://dx.doi.org/ 10.1016/j.jip.2003.08.007 [DOI] [PubMed] [Google Scholar]
- 3.Babendreier D, Joller D, Romeis JA, Bigler F, Widmer F. Bacterial community structures in honeybee intestines and their response to two insecticidal proteins. FEMS Microbiol Ecol 2007; 59:600-10; PMID:17381517; http://dx.doi.org/ 10.1111/j.1574-6941.2006.00249.x [DOI] [PubMed] [Google Scholar]
- 4.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol 2011; 20:619-28; PMID:21175905; http://dx.doi.org/ 10.1111/j.1365-294X.2010.04959.x [DOI] [PubMed] [Google Scholar]
- 5.Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci U S A 2012; 109:11002-7; PMID:22711827; http://dx.doi.org/ 10.1073/pnas.1202970109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinson VG, Moy J, Moran NA. Establishment of characteristic gut bacteria during development of the honey bee worker. Appl Environ Microbiol 2012; 78:2830-40; PMID:22307297; http://dx.doi.org/ 10.1128/AEM.07810-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwong WK, Engel P, Koch H, Moran NA. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc Natl Acad Sci U S A 2014; 111:11509-14; PMID:25053814; http://dx.doi.org/ 10.1073/pnas.1405838111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koch H, Schmid-Hempel P. Bacterial communities in central European bumblebees: low diversity and high specificity. Microb Ecol 2011; 62(1):121-33; PMID:21556885; http://dx.doi.org/ 10.1007/s00248-011-9854-3 [DOI] [PubMed] [Google Scholar]
- 9.Moran NA, Hansen AK, Powell JE, Sabree ZL. Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 2012; 7(4):e36393; PMID:22558460; http://dx.doi.org/ 10.1371/journal.pone.0036393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cariveau DP, Powell JE, Koch H, Winfree R, Moran NA. Variation in gut microbial communities and its association with pathogen infection in wild bumble bees (Bombus). ISME J 2014; 8(12):2369-79; PMID:24763369; http://dx.doi.org/ 10.1038/ismej.2014.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, et al.. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 2014; 80(20):6290-302; PMID:25085493; http://dx.doi.org/ 10.1128/AEM.02308-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McFrederick QS, Cannone JJ, Gutell RR, Kellner K, Plowes RM, Mueller UG. Specificity between lactobacilli and hymenopteran hosts is the exception rather than the rule. Appl Environ Microbiol 2013; 79:1803-12; PMID:23291551; http://dx.doi.org/ 10.1128/AEM.03681-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int J Syst Evol Microbiol 2013; 63:2008-18; PMID:23041637; http://dx.doi.org/ 10.1099/ijs.0.044875-0 [DOI] [PubMed] [Google Scholar]
- 14.Vásquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, Olofsson TC. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS One 2012; 7(3):e33188; PMID:22427985; http://dx.doi.org/ 10.1371/journal.pone.0033188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J 2010; 4(3):377-87; PMID:19924154; http://dx.doi.org/ 10.1038/ismej.2009.123 [DOI] [PubMed] [Google Scholar]
- 16.Moran NA, Tran P, Gerardo NM. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl Environ Microbiol 2008; 71(12):8802-10; PMID:16332876; http://dx.doi.org/ 10.1128/AEM.71.12.8802-8810.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kikuchi Y, Hosokawa T, Nikoh N, Meng XY, Kamagata Y, Fukatsu T. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol 2009; 7:2; PMID:19196451; http://dx.doi.org/ 10.1186/1741-7007-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch H, Abrol DP, Li J, Schmid-Hempel P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol Ecol 2013; 22:2028-44; PMID:23347062; http://dx.doi.org/ 10.1111/mec.12209 [DOI] [PubMed] [Google Scholar]
- 19.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 2011; 10:13-26; PMID:22064560; http://dx.doi.org/ 10.1038/nrmicro2670 [DOI] [PubMed] [Google Scholar]
- 20.Powell JE, Martinson VG, Urban-Mead K, Moran NA. Routes of acquisition of the gut microbiota of Apis mellifera. Appl Environ Microbiol 2014; pii:AEM.01861-14; PMID:25239900; http://dx.doi.org/ 10.1128/AEM.01861-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seedorf H, Griffin NW, Ridaura VK, Reyes A, Cheng J, Rey FE, Smith MI, Simon GM, Scheffrahn RH, Woebken D, et al.. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 2014; 159(2):253-66; PMID:25284151; http://dx.doi.org/ 10.1016/j.cell.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, et al.. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 2012; 149(7):1578-93; PMID:22726443; http://dx.doi.org/ 10.1016/j.cell.2012.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koch H, Schmid-Hempel P. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol Lett 2012; 15:1095-103; PMID:22765311; http://dx.doi.org/ 10.1111/j.1461-0248.2012.01831.x [DOI] [PubMed] [Google Scholar]
- 24.Orr HA. Dobzhansky, Bateson, and the genetics of speciation. Genetics 1996; 144(4):1331-5; PMID:8978022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee FJ, Rusch DB, Stewart FJ, Mattila HR, Newton IL. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ Microbiol 2015; 17(3):796-815; PMID:24905222 http://dx.doi.org/ 10.1111/1462-2920.12526 [DOI] [PubMed] [Google Scholar]
- 26.Bottacini F, Milani C, Turroni F, Sánchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, et al.. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 2012; 7(9):e44229; PMID:23028506; http://dx.doi.org/ 10.1371/journal.pone.0044229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson KE, Johansson A, Sheehan TH, Mott BM, Corby-Harris V, Johnstone L, Sprissler R, Fitz W. Draft genome sequences of two Bifidobacterium sp. from the honey bee (Apis mellifera). Gut Pathog 2013; 5(1):42; PMID:24350840; http://dx.doi.org/ 10.1186/1757-4749-5-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwong WK, Mancenido AL, Moran NA. Genome sequences of Lactobacillus sp. strains wkB8 and wkB10, members of the Firm-5 clade, from honey bee guts. Genome Announc 2014; 2(6) pii:e01176-14; PMID:25395644; http://dx.doi.org/ 10.1128/genomeA.01176-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinson VG, Magoc T, Koch H, Salzberg SL, Moran NA. Genomic features of a bumble bee symbiont reflect its host environment. Appl Environ Microbiol 2014; 80:3793-803; PMID:24747890; http://dx.doi.org/ 10.1128/AEM.00322-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engel P, Stepanauskas R, Moran NA. Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS Genet 2014; 10(9):e1004596; PMID:25210772; http://dx.doi.org/ 10.1371/journal.pgen.1004596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engel P, Vizcaino MI, Crawford JM. Gut symbionts from distinct hosts exhibit genotoxic activity via divergent colibactin biosynthesis pathways. Appl Environ Microbiol 2015; Epub ahead of print; PMID:25527542; http://dx.doi.org/ 10.1128/AEM.03283-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006; 124(4):837-48; PMID:16497592; http://dx.doi.org/ 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- 33.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, et al.. Genomic variation landscape of the human gut microbiome. Nature 2013; 493(7430):45-50; PMID:23222524; http://dx.doi.org/ 10.1038/nature11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan X, Yang Y, Zhang J-R. Molecular basis of host specificity in human pathogenic bacteria. Emerg Microbes Infect 2014; 3(3):e23; http://dx.doi.org/ 10.1038/emi.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature 2009; 458(7235):215-8; PMID:19182778; http://dx.doi.org/ 10.1038/nature07660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frese SA, Mackenzie DA, Peterson DA, Schmaltz R, Fangman T, Zhou Y, Zhang C, Benson AK, Cody LA, Mulholland F, et al.. Molecular characterization of host-specific biofilm formation in a vertebrate gut symbiont. PLoS Genet 2013; 9(12):e1004057; PMID:24385934; http://dx.doi.org/ 10.1371/journal.pgen.1004057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franzenburg S, Walter J, Künzel S, Wang J, Baines JF, Bosch TC, Fraune S. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc Natl Acad Sci U S A 2013; 110(39):E3730-8; PMID:24003149; http://dx.doi.org/ 10.1073/pnas.1304960110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheppard SK, Didelot X, Meric G, Torralbo A, Jolley KA, Kelly DJ, Bentley SD, Maiden MC, Parkhill J, Falush D. Genome-wide association study identifies vitamin B5 biosynthesis as a host specificity factor in Campylobacter. Proc Natl Acad Sci U S A 2013; 110(29):11923-7; PMID:23818615; http://dx.doi.org/ 10.1073/pnas.1305559110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barber MF, Elde NC. Escape from bacterial iron piracy through rapid evolution of transferrin. Science 2014; 346(6215):1362-6; PMID:25504720; http://dx.doi.org/ 10.1126/science.1259329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowles CE, Goodrich-Blair H. The Xenorhabdus nematophila nilABC genes confer the ability of Xenorhabdus spp to colonize Steinernema carpocapsae nematodes. J. Bacteriol 2008; 190:4121-8; PMID:18390667; http://dx.doi.org/ 10.1128/JB.00123-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foley SL, Johnson TJ, Ricke SC, Nayak R, Danzeisen J. Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol Mol Biol Rev 2013; 77(4):582-607; PMID:24296573; http://dx.doi.org/ 10.1128/MMBR.00015-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langridge GC, Fookes M, Connor TR, Feltwell T, Feasey N, Parsons BN, Seth-Smith HM, Barquist L, Stedman A, Humphrey T, et al.. Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc Natl Acad Sci U S A 2015; 112(3):863-8; PMID:25535353; http://dx.doi.org/ 10.1073/pnas.1416707112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chavez-Dozal AA, Gorman C, Lostroh CP, Nishiguchi MK. Gene-swapping mediates host specificity among symbiotic bacteria in a beneficial symbiosis. PLoS One 2014; 9(7):e101691; PMID:25014649; http://dx.doi.org/ 10.1371/journal.pone.0101691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hajri A, Brin C, Hunault G, Lardeux F, Lemaire C, Manceau C, Boureau T, Poussier S. A «repertoire for repertoire» hypothesis: repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PLoS One 2009; 4(8):e6632; PMID:19680562; http://dx.doi.org/ 10.1371/journal.pone.0006632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog 2011; 7(7):e1002132; PMID:21799664; http://dx.doi.org/ 10.1371/journal.ppat.1002132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ochman H, Moran NA. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 2001; 292(5519):1096-9; PMID:11352062; http://dx.doi.org/ 10.1126/science.1058543 [DOI] [PubMed] [Google Scholar]
- 47.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012; 336(6086):1268-73; PMID:22674334; http://dx.doi.org/ 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 2013; 11(7):435-42; PMID:23712350; http://dx.doi.org/ 10.1038/nrmicro3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perrimon N, Ni JQ, Perkins L. In vivo RNAi: today and tomorrow. Cold Spring Harb Perspect Biol 2010; 2(8):a003640; PMID:20534712; http://dx.doi.org/ 10.1101/cshperspect.a003640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014; 32(4):347-55; PMID:24584096; http://dx.doi.org/ 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perret X, Staehelin C, Broughton WJ. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev 2000; 64(1):180-201; PMID:10704479; http://dx.doi.org/ 10.1128/MMBR.64.1.180-201.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang D, Yang S, Tang F, Zhu H. Symbiosis specificity in the legume: rhizobial mutualism. Cell Microbiol 2012; 14(3):334-42; PMID:22168434; http://dx.doi.org/ 10.1111/j.1462-5822.2011.01736.x [DOI] [PubMed] [Google Scholar]
- 53.Kaltenpoth M, Roeser-Mueller K, Koehler S, Peterson A, Nechitaylo TY, Stubblefield JW, Herzner G, Seger J, Strohm E. Partner choice and fidelity stabilize coevolution in a Cretaceous-age defensive symbiosis. Proc Natl Acad Sci U S A. 2014; 111(17):6359-64; PMID:24733936; http://dx.doi.org/ 10.1073/pnas.1400457111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al.. Evolution of mammals and their gut microbes. Science 2008; 320(5883):1647-51; PMID:18497261; http://dx.doi.org/ 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, Hahn BH, Hugenholtz P. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 2010; 8(11):e1000546; PMID:21103409; http://dx.doi.org/ 10.1371/journal.pbio.1000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eren AM, Sogin ML, Morrison HG, Vineis JH, Fisher JC, Newton RJ, McLellan SL. A single genus in the gut microbiome reflects host preference and specificity. ISME J 2015; 9(1):90-100; PMID:24936765; http://dx.doi.org/ 10.1038/ismej.2014.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koga R, Moran NA. Swapping symbionts in spittlebugs: evolutionary replacement of a reduced genome symbiont. ISME J 2014; 8:1237-46; PMID:24401857; http://dx.doi.org/ 10.1038/ismej.2013.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engel P, Moran NA. The gut microbiota of insects – diversity in structure and function. FEMS Microbiol Rev 2013; 37:699-735; PMID:23692388; http://dx.doi.org/ 10.1111/1574-6976.12025 [DOI] [PubMed] [Google Scholar]
- 59.Cardinal S, Danforth BN. The antiquity and evolutionary history of social behavior in bees. PLoS One 2011; 6(6):e21086; PMID:21695157; http://dx.doi.org/ 10.1371/journal.pone.0021086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martins AC, Melo GA, Renner SS. The corbiculate bees arose from New World oil-collecting bees: implications for the origin of pollen baskets. Mol Phylogenet Evol 2014; 80:88-94; PMID:25034728; http://dx.doi.org/ 10.1016/j.ympev.2014.07.003 [DOI] [PubMed] [Google Scholar]
- 61.Leonhardt SD, Kaltenpoth M. Microbial communities of three sympatric Australian Stingless bee species. PLoS One 2014; 9(8):e105718; PMID:25148082; http://dx.doi.org/ 10.1371/journal.pone.0105718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian B, Fadhil NH, Powell JE, Kwong WK, Moran NA. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 2012; 3(6):e00377-12; PMID:23111871; http://dx.doi.org/ 10.1128/mBio.00377-12 [DOI] [PMC free article] [PubMed] [Google Scholar]


