Abstract
Nuclear pore complexes (NPCs) span the 2 membranes of the nuclear envelope (NE) and facilitate nucleocytoplasmic exchange of macromolecules. NPCs have a roughly tripartite structural organization with the so-called nuclear basket emanating from the NPC scaffold into the nucleoplasm. The nuclear basket is composed of the 3 nucleoporins Nup153, Nup50, and Tpr, but their specific role for the structural organization of this NPC substructure is, however, not well established. In this study, we have used thin-section transmission electron microscopy to determine the structural consequences of altering the expression of Nup153 in human cells. We show that the assembly and integrity of the nuclear basket is not affected by Nup153 depletion, whereas its integrity is perturbed in cells expressing high concentrations of the zinc-finger domain of Nup153. Moreover, even mild over-expression of Nup153 is coinciding with massive changes in nuclear organization and it is the excess of the zinc-finger domain of Nup153 that is sufficient to induce these rearrangements. Our data indicate a central function of Nup153 in the organization of the nucleus, not only at the periphery, but throughout the entire nuclear interior.
Keywords: electron microscopy, nuclear basket, nuclear pore complex, nucleoporin, Nup153, Nup50, Tpr
Abbreviations
- BIR
baculovirus IAP repeat
- DAPI
4',6-diamidino-2-phenylindole
- DMEM
Dulbecco's modified Eagle's medium
- EM
electron microscopy
- FBS
foetal bovine serum
- FG
phenylalanine-glycine
- GFP
green fluorescent protein
- IAP
inhibitor of apoptosis
- kDa
kilodalton
- MDa
megadalton
- MEM
minimal essential medium
- Min
minute
- NE
nuclear envelope
- NPC
nuclear pore complex
- Nup
nuclear pore protein, nucleoporin
- PBS
phosphate buffered saline
- PVDF
Polyvinylidene difluoride
- RT
room temperature
- siRNA
small interfering ribonucleic acid
- TBS
(Tris(hydroxymethyl)-aminomethan) buffered saline
- TEM
transmission electron microscopy
- Tpr
translocated promoter region
- XIAP
X-linked inhibitor of apoptosis
Introduction
Nuclear pore complexes (NPCs) control the molecular trafficking between the cytoplasm and the nucleus of interphase eukaryotic cells. NPCs are giant protein assemblies of about 100 MDa in vertebrates,1,2 arising from about 30 different nucleoporins (Nups), which occur in copy numbers of 8 or multiples thereof.2-4 NPCs reside in the double membrane of the nuclear envelope (NE) and they are characterized by a roughly tripartite architecture: the central framework (also known as spoke complex, spoke-ring complex, or scaffold-ring complex), which resides in the NE and which is flanked by the cytoplasmic filaments and the nuclear basket.2,5-10 The nuclear basket in turn is formed of 8 filaments that emanate from the nuclear ring moiety of the central framework and converge into a distal ring. The three major protein constituents of the nuclear basket are the nucleoporins Nup153, Nup50, and Tpr. Tpr is a ∼265 kDa protein that is characterized by a large coiled-coil domain spread over about 2-third of the protein and an acidic globular C terminus.11-13 Due to the extended coiled-coil region, Tpr is forming homodimers, which are thought to form the filaments of the nuclear basket. Immuno-electron microscopy (immuno-EM) studies in human cells and rat liver NEs in combination with domain-specific antibodies suggest that a part of the Tpr dimer folds back onto another part of itself, so that both the N-terminal and the C-terminal regions of Tpr are found at the distal end of the nuclear basket.14,15 Residues 436–606 are required and sufficient to localize Tpr to NPCs.11,12
Nup153 has a tripartite domain organization and topologically complex arrangement within the nuclear basket. The N-terminal domain of Nup153, which contains its NE- and NPC-targeting information (residues 2–144 and 39–339, respectively),16,17 anchors the protein to the nuclear ring moiety of the central framework.5,15,18-20 The N-terminal domain spans about 600 amino acids and appears to have little secondary structure.21,22 The human Nup153 harbors 4 zinc-fingers in its central region and this domain ties Nup153 to the distal ring of the nuclear basket.18 A C-terminal phenylalanine-glycine (FG)-repeat containing domain of Nup153 is characterized by a high flexibility and mobility and can locate all over the nuclear basket and reach out to the cytoplasmic periphery of the NPC's central pore.18,23-27 Nup153 binds Tpr via residues 228–439 and Nup50, the third component of the nuclear basket, via residues 337–611.11,28 Nup50 is a mobile nucleoporin that associates with the nuclear basket: it binds directly to Nup153, but exhibits no binding sites for Tpr.11,29-31 All 3 basket nucleoporins are evolutionary conserved with functional homologues in yeast, Drosophila, Xenopus and other species.32-37
While the biochemical interface between the 3 basket nucleoporins is well defined and their precise localization within the nuclear basket largely accepted, their respective importance for nuclear basket integrity is less well specified, especially the role of Nup153. EM analysis in human and yeast cells have shown that NPCs devoid of Tpr do not have nuclear baskets.38,39 An epifluorescence based study, on the other hand, suggested that overexpression of the C-terminal domain of Nup153 disrupts nuclear basket architecture by displacing Nup50 and Tpr from NPCs.40 It is unclear what exact role Nup153 plays for nuclear basket architecture, nor is it conclusively analyzed whether depletion of Nup153 by small interfering (si) RNAs displaced Tpr from NPCs or not.11,40–43 Depletion of Tpr by antibodies or siRNAs is not impairing Nup153 and Nup50 recruitment to NPCs.14,15
Here we have carried out a systematic ultrastructural analysis of human NPCs and we show that the absence of Nup153 does not interfere with nuclear basket assembly and integrity, while excess levels of Nup153 and its zinc-finger domain, but not its C-terminal domain, alter the basket architecture and lead to general changes in the organization of the nucleus.
Results
Nup153 is required for Nup50 but not for Tpr localization at nuclear pores
It has previously been reported that depletion of Nup153 by siRNAs displaced Tpr from NPCs,11,43 while other studies have shown that Tpr is present at NPCs even in the absence of Nup153.41,42,44 Therefore we first carried out immunofluorescence assays in HeLa cells that were depleted for Nup153 and analyzed the effect on Tpr and Nup50 recruitment to NPCs and vice versa. To do so, HeLa cells were transfected with siRNAs against Nup153, Nup50, and Tpr, respectively, and the reduction of the protein levels were determined by immunoblotting. As shown in Fig. 1A, each of the 3 basket nucleoporins was specifically and solely reduced by its respective siRNAs: depletion of Tpr did not affect the expression levels of Nup153 and Nup50, depletion of Nup153 did not affect Tpr and Nup50 expression levels, and depletion of Nup50 did not alter protein levels of Tpr and Nup153. We next analyzed whether or not the depletion of a given nuclear basket nucleoporin is influencing the localization of the other 2 at NPCs. We first examined the interplay between Nup153 and Nup50. As shown in Fig. 1B (top row), Nup153 and Nup50 resided at NPCs in HeLa cells treated with non-targeting siRNAs (scr siRNAs) and Nup50 was additionally found in the nucleoplasm. Nup153 depletion coincided with a displacement of Nup50 from NPCs, whereas the nucleoplasmic pool of Nup50 appeared unaffected (Fig. 1B, middle row). Depletion of Nup50 did not affected Nup153 localization at NPCs (Fig. 1B, bottom row). When examining the interplay between Nup153 and Tpr (Fig. 1C, top row), we revealed that depletion of Nup153 did not impaired Tpr localization at NPCs (Fig. 1C, middle row) and similarly Tpr depletion did not affected Nup153 location at NPCs (Fig. 1C, bottom row). Likewise, Nup50s localization at NPCs and in the nucleoplasm was not depending on Tpr (Fig. 1D, middle row) and Tpr localization was not influenced by a reduction of Nup50 (Fig. 1D, bottom row). Together, these data indicate that Nup153 is required for Nup50 recruitment to NPCs, whereas Nup153 and Tpr can be recruited independent of each other and of Nup50 to NPCs.
Figure 1.

Nup153 is required for Nup50 localization at nuclear pores. (A) Knockdown of Nup153 and Nup50 was verified 48 hours after transfection by Western blotting, while Tpr knockdown was verified 72 hours after transfection. HeLa T-Rex cellular extracts from cells treated with control, Nup153, Nup50 and Tpr siRNAs, respectively, were analyzed using antibodies directed against Nup153, Nup50, Tpr, and actin. Depletion of Nup153 resulted in the displacement of Nup50 from NPCs (B), whereas Tpr localization at NPCs remains unaffected (C). Knockdown of Nup50 did not compromised Nup153 (B) and Tpr (D) recruitment to NPCs. Similarly, Nup153 (C) and Nup50 (D) were localizing to NPCs in the absence of Tpr. Cells were analyzed by indirect fluorescence microscopy. Shown are confocal images. Scale bars, 10 μm.
Tpr is the major structural component of the nuclear basket
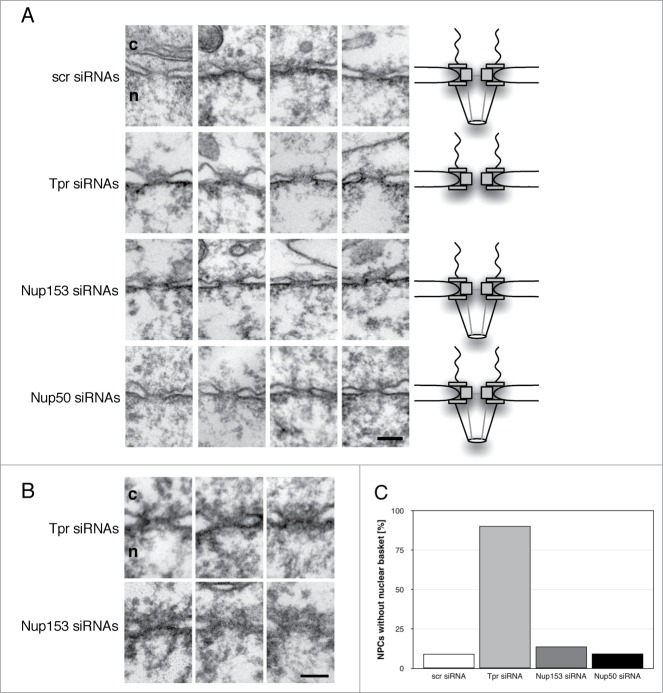
Next we examined the importance of each of the 3 basket nucleoporins for the structural integrity of the nuclear basket. To do so, we analyzed Epon-embedded HeLa T-Rex cells by thin-sectioning transmission electron microscopy (EM). HeLa T-Rex cells were employed because nuclear baskets are frequently visible in these cells (Fig. 2A), in contrast to our standard HeLa cell line (Fig. 2B), probably to a slightly compact chromatin. In HeLa T-Rex cells treated with non-targeting control siRNA (n = 58), the nuclei retained normal NPC morphology and filamentous, nuclear basket structures on the nuclear side of the NPC (Fig. 2A, scr siRNAs; the boundary of the nuclear baskets in the presented NPCs are highlighted in Fig. S1). In Tpr-siRNA treated cells these filamentous structures were reduced in ∼90% of the NPCs (Fig. 2A and C; n = 132), in agreement with previous studies in HeLa and yeast cells.38, 39 In contrast to Tpr depletion, depletion of Nup153 in the HeLa TRex cells led to a loss of the nuclear basket structure in only about 14% of the NPCs (Figs. 2A and C, Fig. S1; n = 87). Likewise, only about 6% of Nup50-depleted cells and the control cells had NPCs lacking the nuclear basket (Figs. 2A and C; n = 64 (Nup50 siRNA), n = 66 (scr siRNA)). Similar results for Tpr and Nup153 depletion, respectively, were obtained in our standard HeLa cell line (Fig. 2B), indicating that only Tpr is critical for nuclear basket assembly.
Figure 2.
The structural integrity of the nuclear basket depends exclusively on Tpr. (A) HeLa T-Rex cells were treated with control siRNAs as well as siRNAs against Nup153 and Nup50 for 48 hours and with siRNAs against Tpr for 72 hours. Cells were embedded in epoxy-based resin and examined by thin-section transmission electron microscopy (TEM). Shown are electron micrographs of selected, representative NPCs and a schematic model of the respective phenotype. (B) HeLa were treated with siRNAs against Nup153 for 48 hours and with siRNAs against Tpr for 72 hours and prepared and examined by thin-section TEM. (C) Quantification of the number of NPCs without a nuclear basket in HeLa T-Rex cells treated with the distinct siRNAs (n = 66 (scr siRNA), n = 132 (Tpr siRNA), n = 87 (Nup153 siRNA), n = 64 (Nup50 siRNA)). c, cytoplasm; n, nucleus. Scale bars, 100 nm.
The zinc-finger domain of Nup153 affects nuclear basket structure and nuclear organization
Based on epifluorescence analyses it has recently been speculated that overexpression of the C-terminal, FG-repeat domain of Nup153 interferes with nuclear basket assembly.40 We therefore decided to analyze the effect of the enhanced expression of Nup153 and its individual domains, respectively, on nuclear basket structure on the ultrastructural level. To do so, we first expressed Nup153 and 3 fragments fused to GFP (see Material and Methods) transiently in our HeLa T-Rex cells and determined their subcellular localization by direct immunofluorescence and confocal microscopy. As shown in Fig. 3A, full-length Nup153 (Nup153) and the N-terminal domain of Nup153 (Nup153-N) fused to GFP localized to NPCs and to the nucleoplasm, with some aggregation of the fusion proteins in the nucleoplasm, likely due to the over-expression. The zinc-finger domain of Nup153 (Nup153-Z) fused to GFP is lacking the NPC targeting cassette and the NLS of Nup153 and was consequently found dispersed in the cytoplasm and the nucleus of transfected cells. Similarly, the C-terminal domain of Nup153 is lacking Nup153s NPC targeting cassette and NLS and Nup153-C was found in the cytoplasm and the nucleus. Neither Nup153-Z nor Nup153-C were recruited to NPCs and, likely due to its larger size, less Nup153-C was found in the nucleus as compared to Nup153-Z. The transfection efficiency for all our constructs was verified by immunofluorescence microscopy and was higher than 90% for the Nup153 fragments and about 30–50% for the full-length protein (data not shown).
Figure 3.
Enhanced expression of Nup153-Z results in changes in nuclear basket structure. (A) HeLa cells were transiently transfected with GFP-Nup153 and the fragments GFP-Nup153-N, GFP-Nup153-Z, and GFP-Nup153-C, respectively, and analyzed by direct fluorescence microscopy 24 hours after transfection. Shown are confocal micrographs. Scale bars, 10 μm. (B) Electron microscopic images of nuclear pores in control HeLa T-Rex cells (MOCK) and in Nup153-Z and Nup153-C, respectively, expressing cells, 48 hours after transfection. Shown are representative examples of selected NPCs and a schematic model of the respective phenotype. (C) Quantification of the number of NPCs with nuclear baskets without a distal ring in HeLa T-Rex cells expressing Nup153-Z (n = 55) and Nup153-C (n = 87). c, cytoplasm; n, nucleus. Scale bar, 100 nm.
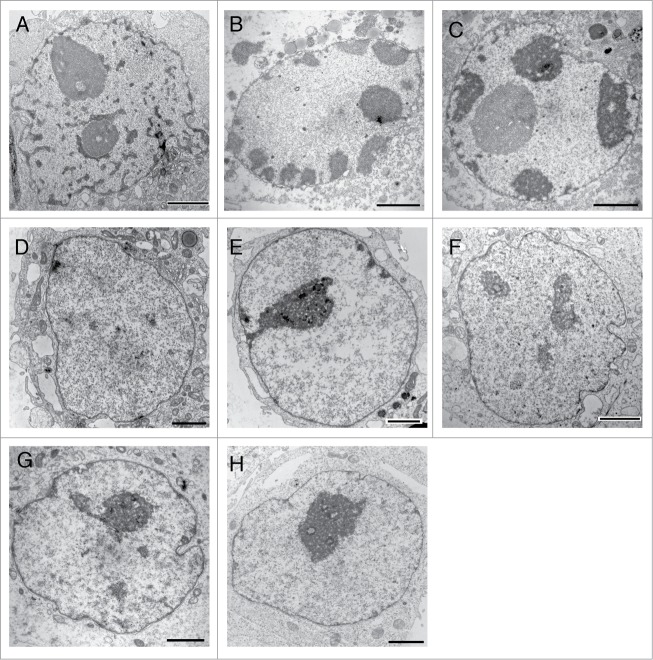
We next analyzed the integrity of the nuclear baskets in HeLa T-Rex cells expressing Nup153 fragments by thin-section EM. As shown in Fig. 3B, filamentous nuclear basket structures were observed in control cells (MOCK, transfected with the empty GFP plasmid), in cells over-expressing Nup153-Z as well as Nup153-C (the boundaries of the nuclear baskets in the depicted NPCs are highlighted in Fig. S2). Similarly, overexpression of full-length Nup153 and Nup153-N did not interfere with nuclear basket assembly (data not shown). By closer inspection of the nuclear baskets of Nup153-Z and Nup153-C expressing cells, however, it became evident that the baskets in Nup153-Z cells were not cone-shaped and appeared to lack the distal ring of the basket: only about 15% of the baskets had a distal ring, in contrast to ∼80% in cells expressing Nup153-C (Fig. 3C). The over-expression of the zinc-finger domain of Nup153 furthermore led to massive changes in the overall nuclear organization with heterochromatin formation at the NE and in the nucleoplasm (Figs. 4A–C) as well as a more frequent association of nucleoli with the NE (Fig. 4C). These effects are specific to the zinc-finger domain of Nup153: neither the expression of the zinc-finger domain of Nup358/RanBP2 (Fig. 4D) or of the zinc-finger of the inhibitor of apoptosis (IAP) protein XIAP (Fig. 4E) is affecting nuclear organization. Nup358 contains, as Nup153, a C2C2 zinc-finger motif, whereas the baculoviral IAP repeat (BIR) domain of XIAP contains a CCHC motif, which resembles the classical C2H2 zinc-finger motif.45,46 Nuclei of cells expressing high levels of Nup153-C (Figs. 4F and G) appeared indistinguishable from control nuclei (Fig. 4H). Together our data show that the zinc-finger but not the C-terminal domain of Nup153 is impacting NPC and nuclear architecture.
Figure 4.
Enhanced expression of Nup153-Z induces changes in nuclear organization. HeLa T-Rex cells were transiently transfected with GFP-Nup153-Z and GFP-Nup153-C, respectively, and prepared for thin-sectioning EM 48 hours after transfection. Overexpression of Nup513-Z led to formation of heterochromatin at the nuclear envelope (A, B, and C) and in the nucleoplasm ((A)and C) and to a peripheral localization of nucleoli (C). Overexpression of the zinc-finger domain of Nup358/RanBP2 (D) or XIAP (E) had no effect on nuclear organization. Nuclei from cells overexpressing Nup153-C ((F) and G) appeared indistinguishable from control nuclei (H). Scale bars, 2 μm.
Similar to the effect of Nup153-Z, enhanced levels of full-length Nup153 provoked nuclei with abnormal shape (Fig. 5B, S3B), heterochromatin formation (Figs. 5B and C, S3A and B), and enlarged, segregated nucleoli as well as the association of nucleoli with the NE (Fig. 5C), which was not seen in control cells (Fig. 5A). Additionally, Nup153 overexpression induced the formation of membranous structures in the cytoplasm (Fig. 5B, arrow and S3B) and inside the nucleus (Fig. S3A), in agreement with previously published data.16 Nup153-depleted cells (Fig. 5D), in contrast, had a nuclear architecture indistinguishable from control cells.
Figure 5.
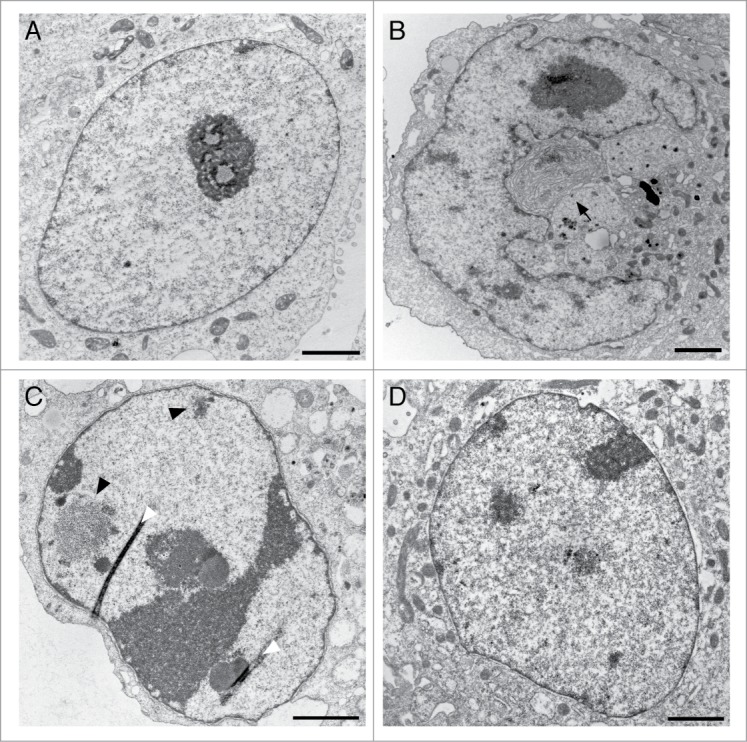
Enhanced expression of Nup153 alters nuclear organization. HeLa cells were transiently transfected with GFP-Nup153 and siRNAs against Nup153, respectively, and prepared for thin-sectioning EM 48 hours after transfection. Enhanced expression of Nup153 coincided with membranous structures in the cytoplasm (arrow in B), abnormally shaped nuclei (B), an increase in heterochromatin in the nucleoplasm (B, black arrowheads in C) and along the nuclear envelope (B) as well as to segregation of nucleoli and their association with the nuclear envelope (C). Nuclei from control cells and Nup153-depleted cells did not differ from each other. White arrowheads in (C) indicate section artifacts. Scale bars, 2 μm.
Transient transfection results in heterogeneous, often very high, expression levels of the exogenous protein in different cells. To study the impact of Nup153 over-expression on nuclear architecture in a more controlled manner, we next generated stable HeLa T-Rex cell lines expressing GFP-Nup153 upon tetracycline-induction. In doing so, we obtained a monoclonal cell line expressing GFP-Nup153 at levels similar to the endogenous protein (Fig. 6A). GFP-Nup153 in this cell line was expressed in all cells at equal level (Fig. 6B, top row) and the protein localized primarily to NPCs with a minor pool in the nucleoplasm (Fig. 6B, bottom row). On the ultrastructural level, we observed the same changes in nuclear organization as in HeLa T-Rex cells expressing GFP-Nup153 transiently with alterations in the nuclear shape (Figs. 6D-F), heterochromatin formation (Figs. 6D-F), and enlarged, segregated nucleoli as well as the association of nucleoli with the NE (Fig. 6D and E). In the absence of tetracycline and GFP-Nup153 expression, these cells had normal nuclear shape and organization (Fig. 6C). Together our data indicate that a moderate increase in Nup153 expression coincides with global changes in nuclear organization.
Figure 6.
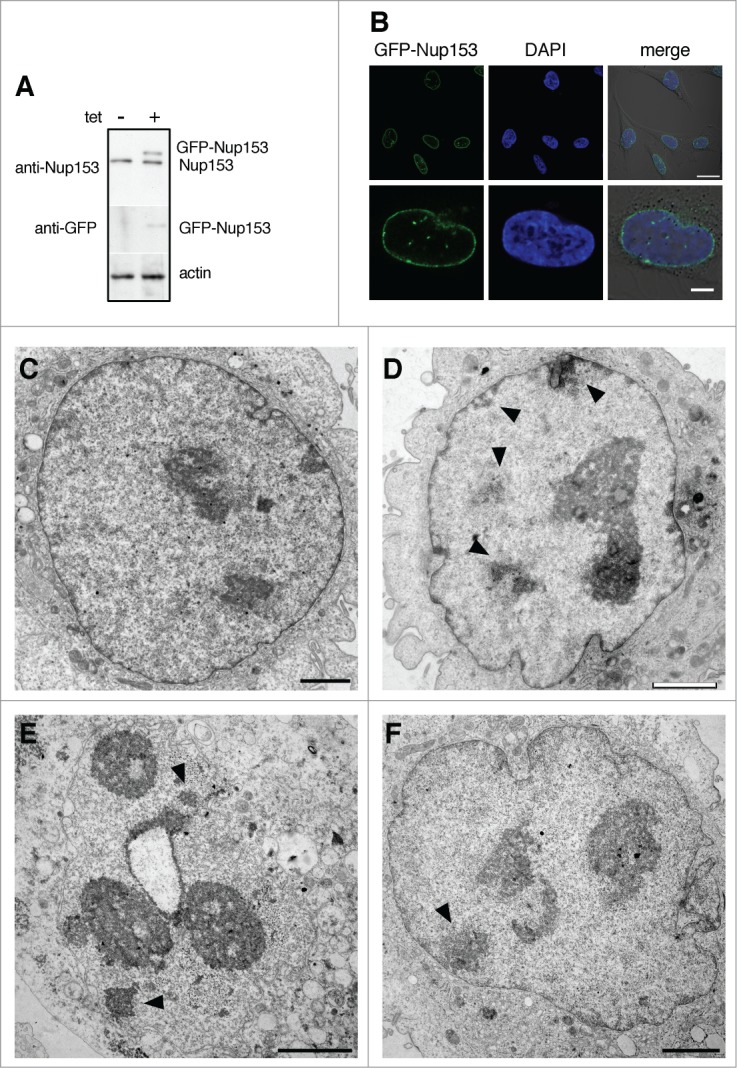
Induced expression of Nup153 alters nuclear organization. (A) Western blot analysis of tetracycline-inducible GFP-Nup153 expression in a monoclonal HeLa T-Rex cell line. Cells were treated with 0.1 μg/ml tetracycline for 24 hours and cell lysates were analyzed using antibodies directed against Nup153, GFP and actin, respectively. (B) Direct fluorescence analysis of tetracycline-inducible GFP-Nup153 in HeLa T-Rex cells by confocal microscopy. Scale bars, 20 μm (upper panel), 10 μm (lower panel). GFP-Nup153-inducible cells had normal nuclear organization and shape in the absence of tetracycline (C), whereas treatment with tetracycline and GFP-Nup153 expression led to altered nuclear shape (D-F), heterochromatin formation in the nucleoplasm and at the nuclear envelope (black arrowheads in D-F), as well as segregated and peripheral nucleoli (D, E, and F). Scale bars, 1 μm (C), 2 μm (D–F).
Discussion
Previous thin-sectioning EM studies have shown that Tpr and its homologues are critical components of the NPCs nuclear basket.38,39 Here we have carried out a systematic structural analysis using thin-sectioning EM to study the role of Nup153 for nuclear basket integrity and nuclear organization in human cells. The nuclear basket is a highly flexible structure and therefore difficult to study on high-resolution EM level, in particular in somatic cells. In Xenopus oocyte nuclei, nuclear baskets can be seen in cross-section along the NE15,18,19,23,47 or in samples prepared for scanning EM20,48 or freeze-drying.18,47 Tomographic studies of NPCs, however, usually lack information on the nuclear basket7–10,49 and only the distal ring of the basket had been somewhat resolved in NPCs from Xenopus oocyte nuclei.50 In somatic cells, electron microscopic visualization of the nuclear baskets is further hampered by its overlap with peripheral chromatin. Our study is therefore to the best of our knowledge the first systematic approach toward nuclear basket architecture in human cells on an ultrastructural level.
Nup153 and the nuclear basket
The NPCs nuclear basket is composed of 3 nucleoporins, i.e. Nup153, Nup50 and Tpr, and our data confirm that Tpr is the major structural component of the basket in human cells and required for its assembly (Fig. 2): only NPCs depleted of Tpr lack filamentous basket structures, while basket integrity remained unaffected by Nup153 and Nup50 depletion, respectively. Nuclear basket architecture also remained intact upon overexpression of the C-terminal domain of Nup153 (Fig. 3B), which refutes elsewhere epifluorescence-based interpretations40 and underscores the importance of ultrastructural approaches for qualitative structural statements. We provide further evidence that Nup153 is required for Nup50 localization at NPCs, but not for Tpr recruitment (Fig. 1C). This is in agreement with several previous studies40-42 and the notion that Nup153 is a rather mobile51,52 and largely unstructured protein.18, 21,22,53 It is furthermore supported by the fact that Nup153-depleted NPCs have a nuclear basket, while Tpr-depleted NPCs lack baskets (Fig. 2). Therefore, Nup153-depleted NPCs should be devoid of nuclear baskets, if Nup153 acts as the major anchor for Tpr at NPCs. The essential anchoring site for Tpr therefore remains to be identified, but a likely candidate is the Nup107–160 complex, which is the major constituent of the NPC scaffold.49 In this context, it has previously been shown that in the absence of Tpr, labeling of Nup107 at NPCs was significantly increased,11 suggesting that Tpr is binding and masking Nup107 to a certain extend. Similarly, it has been shown that the yeast Nup84 complex serves as an important anchoring partner for Mlp1 and Mlp2, the yeast homologues of Tpr.35,36,38
The zinc-finger domain of Nup153 and the nuclear basket
NPCs of cells that over-express the zinc-finger domain of Nup153 have baskets that appear to lack the distal ring: their basket filaments are visible, but they do not converge into a distal ring (Figs. 3 and S2). The zinc-fingers of Nup153 locate to the distal ring of the nuclear basket18 and it has previously been shown that the integrity of the nuclear basket54 as well as the interaction between Nup153 and Tpr depend on divalent cations.11 Since the zinc-finger domain of Nup153 alone is not targeted to NPCs but locates in the cytoplasm and the nucleus, our data suggest that excess levels of Nup153s zinc-fingers sequester divalent cations, so that the concentration of free divalent cations in the cell is reduced, which in turn somewhat affects the integrity of the nuclear basket's distal ring. Nup153s zinc-finger domain may also sequester something else or something in addition, which is important for the specific effect on nuclear architecture that coincides with the expression of this domain (Fig. 4). How the distal ring is formed exactly remains to be seen, but our data suggest that the zinc-fingers of Nup153 are a major constituent. As NPCs devoid of Nup153 have a basket with a distal ring (Figs. 2 and S1), Tpr is likely to be the second major constituent of the distal ring. In fact, both, the N-terminal and the C-terminal part of Tpr, have been detected in the proximity of the distal ring.15 To which extend the integrity of the distal ring is affected by Nup153 depletion is not assessable at the level of resolution that can be obtained by thin-section EM.
The zinc-finger domain of Nup153 and nuclear organization
Enhanced expression of Nup153s zinc-fingers not only affected the nuclear basket, but nuclear organization in more general, especially chromatin and nucleolar organization (Fig. 4). Similar changes in nuclear organization were induced by over-expression of full-length Nup153 (Figs. 5 and 6), whereas overexpression of the C-terminal domain of Nup153 or the zinc-finger domains of Nup358/RanBP2 and XIAP, respectively, had no obvious influence (Fig. 4). Therefore our data suggest that the zinc-finger domain of Nup153 is responsible for these alterations in nuclear organization. The zinc-fingers of Nup153 are known to bind the small GTPase Ran, which appears to be important to maintain a high local concentration of Ran in the vicinity of the NPC.53,55,56 On the other hand, it had been shown that the zinc-fingers of Nup153 recognize specific DNA sequences in vitro and it has been suggested that this might play a role in genome organization.57 Our data presented here support this idea and they further underscore the importance of the nuclear basket in chromatin organization, as also the absence of Tpr provoked severe changes in chromatin organization, in human and in yeast cells.38,58 Nup153 is known to bind chromatin in Drosophila and human cells,43,59 whether or not this occurs via its zinc-finger region remains to be seen. A further characterization of Nup153s zinc-finger domain and the identification of novel interacting partners will be therefore of high interest for future investigations.
As a summary, we have shown here that Tpr acts as structural backbone of the NPCs nuclear basket, in contrast to Nup153 and Nup50. The zinc-finger domain of Nup153 is important for the structural integrity of the distal ring of the nuclear basket and also implicated in the organization of the nuclear architecture. Future studies will be required to expand our current understanding of the Nup153-chromatin interface.
Materials and Methods
Cell culture and transfections
HeLa T-Rex cells (Life Technologies Invitrogen, Gent, Belgium) were grown in Minimal Essential Medium (MEM) and HeLa cells in Dulbecco's modified Eagle's medium (DMEM), both supplemented with 10% fetal bovine serum (FBS) plus penicillin and streptomycin. Cells were transfected with plasmids using Turbofect transfection reagent (Thermo Scientific Fermentas, St. Leon-Rot, Germany) and with siRNAs using Lipofectamine RNAiMAX (Life Technologies Invitrogen, Gent, Belgium) following the instructions of the manufacturer.
Plasmids
For all constructs human NUP153 was amplified by PCR. N-terminally tagged GFP-Nup153 (residues 1–1475) was produced as described previously.18 N-terminally tagged GFP-Nup153-N (residues 1–650) and GFP-Nup153-Z (residues 657–879) were cloned into HindIII/BamHI cut pEGFP-C3 (Clontech, Palo Alto, CA) and GFP-Nup153-C (residues 880–1475) was inserted into HindIII/XmaI cut pEGFP-C3. Tetracycline-inducible GFP-Nup153 was produced in pcDNA4/TO (Life Technologies Invitrogen, Gent, Belgium). To do so, full-length NUP153 was inserted into the HindIII/NotI sites of pcDNA4/TO. The EGFP encoding sequence was amplified from pEGFP-C3 and inserted at the HindIII site at the 5′ end of NUP153. In-frame insertion was confirmed by sequencing.
Nup358/RanBP2 fragments (residues1351–1810) and XIAP (residues 450–485) were amplified by PCR and inserted into XhoI/BamHI cut pEGFP-C3.
Antibodies
The following antibodies were used: mouse monoclonal anti-Nup153, clone SA1 (IF 1:800 (Fig.1), WB 1:200 (Fig. 1), WB 1:50 (Fig. 6); hybridoma supernatant), mouse monoclonal anti-Tpr (IF 1:6000, WB 1:2000; Abnova, H00007175-M01), rabbit polyclonal anti-Nup50 (IF 1:1000, WB 1:250; Bethyl, A301–782A), rabbit polyclonal anti-α-tubulin (WB 1:2000; Abcam, ab18251), rat monoclonal anti-GFP, clone 3H9 (WB 1:1000; Chromotek), rabbit polyclonal anti-actin (1:1000; Sigma). Secondary anti-mouse IgG-Alexa 488 and anti-rabbit IgG-Alexa 568 antibodies were from Molecular Probes (Paisley, UK) and used at 1:1000. Secondary alkaline phosphatase coupled antibodies for Western blot were from Sigma/Aldrich and used at 1:20000.
Generation of stable cell lines
To generate inducible HeLa T-Rex cell lines, HeLa T-Rex cells were transfected with pcDNA4/TO-GFP-NUP153 and positive clones were selected by treatment with 5 μg/ml blasticidin and 200 μg/ml zeocin. Individual clones were isolated, expanded and cultured in MEM (Life Technologies Gibco, Gent, Belgium) containing 10% FBS, blasticidin, zeocin, penicillin and streptomycin. In order to induce GFP-Nup153 expression, cells were treated with 0.1 μg/ml tetracycline for 24 hours.
Electron Microscopy
HeLa T-Rex cells and HeLa cells were treated with scrambled siRNAs, siRNAs against Nup153 and Nup50 for 48 hours and with siRNAs against Tpr for 72 hours, respectively. Alternatively, HeLa T-Rex cells were transfected with GFP-Nup153 variants for 48 hours. Cells were harvested using a cell scraper, pelleted and washed once in PBS. Cells were fixed in Karnofski solution (3% paraformaldehyde, 0.5% glutaraldehyde in 10 mM PBS, pH 7.4) for 1 hour, washed once in PBS and post-fixed first in 1% reduced osmium tetroxide (containing 1.5% potassium ferricyanide) for 40 minutes and subsequently in 1% osmium tetroxide for another 40 minutes. After washing in water, fixed samples were dehydrated, embedded in Epon resin, and processed for EM as described.18 EM micrographs were recorded on a Phillips CM-100 transmission electron microscope equipped with a CCD camera at an acceleration voltage of 80 kV. Images were recorded using the systems software and processed using Adobe Photoshop.
Immunofluorescence
HeLa cells were grown on coverslips and transfected with smart-pool siRNAs against Nup50 (L-00012369–01, Dharmacon), Nup153 (L-005283–00), Tpr (L-010548–00) and non-targeting siRNAs (D-001810–10), respectively, as described for electron microscopy. Cells were fixed in 4% paraformaldehyde for 15 min at RT and permeabilized for 5 min at RT in PBS containing 0,3% Triton X-100. After blocking 60 min in PBS containing 5% BSA, cells were incubated with the respective primary antibodies overnight at 4°C. After washing in PBS containing 5% BSA, cells were incubated with appropriate secondary antibodies for 45 min at RT and mounted with Mowiol 4–88 (Sigma) containing 1μg/ml DAPI. Images were acquired with a Zeiss LSM 710 confocal microscope. Images were recorded using the microscope system software and processed using Image J and Adobe Photoshop (Adobe Systems, Mountain View, CA)
Immunoblotting
Whole-cell lysates were obtained from siRNA-treated HeLa cells lysed in lysis buffer (50 mM Tris-HCl, pH 7.8, 150 mM NaCl, 1% Nonidet-P40 and protease inhibitor cocktail tablets (Roche, Basel Switzerland). Twenty μg of protein were loaded and separated by sodium dodecyl sulfate-polyacrylamide (7%) gel electrophoresis (SDS-PAGE). The proteins were transferred onto a PVDF membrane (Immobilon-P, Millipore) and the membranes were blocked with TBS containing 0.1% Tween 20 and 5% non-fat dry milk for 1 hour. The membranes were then incubated for 1 hour in blocking solution containing a primary antibody followed by washing 3x in TBS containing 0.1% Tween 20 and 5% non-fat dry. The membranes were next incubated with alkaline phosphatase labeled secondary antibodies for 1 hour, washed 3x in TBS and developed using Lightning CDP Star Chemiluminescence reagent (Applied Biosystem) and an X-ray film. Films were scanned and processed using Adobe Photoshop software.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Drs. Denis Lafontaine and Ueli Aebi for insightful discussions.
Funding
This work was supported by grants from the Fonds de la Recherche Scientifique-FNRS Belgium (grants T.0237.13, 1.5019.12, and F.6006.10), the Fonds Brachet and the Fonds Van Buuren.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website: http://www.tandfonline.com/kncl
References
- 1. Reichelt R, Holzenburg A, Buhle EL, Jr., Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol 1990; 110:883-94; PMID:2324201; http://dx.doi.org/ 10.1083/jcb.110.4.883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ori A, Banterle N, Iskar M, Andres-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, et al. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol 2013; 9:648; PMID:23511206; http://dx.doi.org/ 10.1038/msb.2013.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 2002; 158:915-27; PMID:12196509; http://dx.doi.org/ 10.1083/jcb.200206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 2000; 148:635-51; PMID:10684247; http://dx.doi.org/ 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fahrenkrog B, Aebi U. The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol 2003; 4:757-66; PMID:14570049; http://dx.doi.org/ 10.1038/nrm1230 [DOI] [PubMed] [Google Scholar]
- 6. Lim RY, Aebi U, Fahrenkrog B. Towards reconciling structure and function in the nuclear pore complex. Histochem Cell Biol 2008; 129:105-16. PMID:18228033; http://dx.doi.org/ 10.1007/s00418-007-0371-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beck M, Forster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 2004; 306:1387-90; PMID:15514115; http://dx.doi.org/ 10.1126/science.1104808 [DOI] [PubMed] [Google Scholar]
- 8. Beck M, Lucic V, Forster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature 2007; 449:611-5; PMID:17851530; http://dx.doi.org/ 10.1038/nature06170 [DOI] [PubMed] [Google Scholar]
- 9. Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J Mol Biol 2009; 395:578-86; PMID:19913035; http://dx.doi.org/ 10.1016/j.jmb.2009.11.010 [DOI] [PubMed] [Google Scholar]
- 10. Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure 2012; 20:998-1006; PMID:22632834; http://dx.doi.org/ 10.1016/j.str.2012.03.025 [DOI] [PubMed] [Google Scholar]
- 11. Hase ME, Cordes VC. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 2003; 14:1923-40; PMID:12802065; http://dx.doi.org/ 10.1091/mbc.E02-09-0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hase ME, Kuznetsov NV, Cordes VC. Amino acid substitutions of coiled-coil protein Tpr abrogate anchorage to the nuclear pore complex but not parallel, in-register homodimerization. Mol Biol Cell 2001; 12:2433-52; PMID:11514627; http://dx.doi.org/ 10.1091/mbc.12.8.2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Snow CJ, Paschal BM. Roles of the nucleoporin tpr in cancer and aging. Adv Exp Med Biol 2014; 773:309-22; PMID:24563354; http://dx.doi.org/ 10.1007/978-1-4899-8032-8_14 [DOI] [PubMed] [Google Scholar]
- 14. Frosst P, Guan T, Subauste C, Hahn K, Gerace L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biol 2002; 156:617-30; PMID:11839768; http://dx.doi.org/ 10.1083/jcb.200106046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krull S, Thyberg J, Bjorkroth B, Rackwitz HR, Cordes VC. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell 2004; 15:4261-77; PMID:15229283; http://dx.doi.org/ 10.1091/mbc.E04-03-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of nuclear pore complex protein Nup153. J Cell Biol 1996; 134:1141-56; PMID:8794857; http://dx.doi.org/ 10.1083/jcb.134.5.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Enarson P, Enarson M, Bastos R, Burke B. Amino-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma 1998; 107:228-36; PMID:9745047; http://dx.doi.org/ 10.1007/s004120050301 [DOI] [PubMed] [Google Scholar]
- 18. Fahrenkrog B, Maco B, Fager AM, Koser J, Sauder U, Ullman KS, Aebi U. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J Struct Biol 2002; 140:254-67; PMID:12490173; http://dx.doi.org/ 10.1016/S1047-8477(02)00524-5 [DOI] [PubMed] [Google Scholar]
- 19. Pante N, Thomas F, Aebi U, Burke B, Bastos R. Recombinant Nup153 incorporates in vivo into Xenopus oocyte nuclear pore complexes. J Struct Biol 2000; 129:306-12; PMID:10806081; http://dx.doi.org/ 10.1006/jsbi.2000.4232 [DOI] [PubMed] [Google Scholar]
- 20. Walther TC, Fornerod M, Pickersgill H, Goldberg M, Allen TD, Mattaj IW. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. Embo J 2001; 20:5703-14; PMID:11598013; http://dx.doi.org/ 10.1093/emboj/20.20.5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lim RYH, Aebi U, Stoffler D. From the trap to the basket: getting to the bottom of the nuclear pore complex. Chromosoma 2006; 115:15-26; PMID:16402261; http://dx.doi.org/ 10.1007/s00412-005-0037-1 [DOI] [PubMed] [Google Scholar]
- 22. Schwartz TU. Modularity within the architecture of the nuclear pore complex. Curr Opin Struct Biol 2005; 15:221-6. PMID:15837182; http://dx.doi.org/ 10.1016/j.sbi.2005.03.003 [DOI] [PubMed] [Google Scholar]
- 23. Paulillo SM, Phillips EM, Koser J, Sauder U, Ullman KS, Powers MA, Fahrenkrog B. Nucleoporin domain topology is linked to the transport status of the nuclear pore complex. J Mol Biol 2005; 351:784-98; PMID:16045929; http://dx.doi.org/ 10.1016/j.jmb.2005.06.034 [DOI] [PubMed] [Google Scholar]
- 24. Paulillo SM, Powers MA, Ullman KS, Fahrenkrog B. Changes in nucleoporin domain topology in response to chemical effectors. J Mol Biol 2006; 363:39-50; PMID:16962132; http://dx.doi.org/ 10.1016/j.jmb.2006.08.021 [DOI] [PubMed] [Google Scholar]
- 25. Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. Embo J 1999; 18:1982-95; PMID:10202161; http://dx.doi.org/ 10.1093/emboj/18.7.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ando Y, Tomaru Y, Morinaga A, Burroughs AM, Kawaji H, Kubosaki A, Kimura R, Tagata M, Ino Y, Hirano H, et al. Nuclear pore complex protein mediated nuclear localization of dicer protein in human cells. PLoS One 2011; 6:e23385; PMID:21858095; http://dx.doi.org/ 10.1371/journal.pone.0023385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soop T, Ivarsson B, Bjorkroth B, Fomproix N, Masich S, Cordes VC, Daneholt B. Nup153 affects entry of messenger and ribosomal ribonucleoproteins into the nuclear basket during export. Mol Biol Cell 2005; 16:5610-20. PMID:16195343; http://dx.doi.org/ 10.1091/mbc.E05-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Makise M, Mackay DR, Elgort S, Shankaran SS, Adam SA, Ullman KS. The Nup153-Nup50 protein interface and its role in nuclear import. J Biol Chem 2012; 287:38515-22; PMID:23007389; http://dx.doi.org/ 10.1074/jbc.M112.378893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smitherman M, Lee K, Swanger J, Kapur R, Clurman BE. Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol Cell Biol 2000; 20:5631-42; PMID:10891500; http://dx.doi.org/ 10.1128/MCB.20.15.5631-5642.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arnaoutov A, Dasso M. Ran-GTP regulates kinetochore attachment in somatic cells. Cell Cycle 2005; 4:1161-5; PMID:16082212; http://dx.doi.org/ 10.4161/cc.4.9.1979 [DOI] [PubMed] [Google Scholar]
- 31. Ogawa Y, Miyamoto Y, Asally M, Oka M, Yasuda Y, Yoneda Y. Two isoforms of Npap60 (Nup50) differentially regulate nuclear protein import. Mol Biol Cell 2010; 21:630-8; PMID:20016008; http://dx.doi.org/ 10.1091/mbc.E09-05-0374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol 2005; 171:955-65; PMID:16365162; http://dx.doi.org/ 10.1083/jcb.200509061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol 2001; 153:1465-78; PMID:11425876; http://dx.doi.org/ 10.1083/jcb.153.7.1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol Biol Cell 2009; 20:2146-59; PMID:19225157; http://dx.doi.org/ 10.1091/mbc.E08-08-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol 1999; 144:839-55; PMID:10085285; http://dx.doi.org/ 10.1083/jcb.144.5.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kosova B, Pante N, Rollenhagen C, Podtelejnikov A, Mann M, Aebi U, Hurt E. Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with nic96p. J Biol Chem 2000; 275:343-50; PMID:10617624; http://dx.doi.org/ 10.1074/jbc.275.1.343 [DOI] [PubMed] [Google Scholar]
- 37. Bogerd AM, Hoffman JA, Amberg DC, Fink GR, Davis LI. nup1 mutants exhibit pleiotropic defects in nuclear pore complex function. J Cell Biol 1994; 127:319-32; PMID:7929578; http://dx.doi.org/ 10.1083/jcb.127.2.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell 2013; 24:3920-38; PMID:24152732; http://dx.doi.org/ 10.1091/mbc.E13-07-0412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Funasaka T, Tsuka E, Wong RW. Regulation of autophagy by nucleoporin Tpr. Sci Rep 2012; 2:878; PMID:23170199; http://dx.doi.org/ 10.1038/srep00878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mackay DR, Makise M, Ullman KS. Defects in nuclear pore assembly lead to activation of an Aurora B-mediated abscission checkpoint. J Cell Biol 2010; 191:923-31; PMID:21098116; http://dx.doi.org/ 10.1083/jcb.201007124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lussi YC, Shumaker DK, Shimi T, Fahrenkrog B. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus 2010; 1:71-84; PMID:21327106; http://dx.doi.org/ 10.4161/nucl.1.1.10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Umlauf D, Bonnet J, Waharte F, Fournier M, Stierle M, Fischer B, Brino L, Devys D, Tora L. The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci 2013; 126:2656-67; PMID:23591820; http://dx.doi.org/ 10.1242/jcs.118000 [DOI] [PubMed] [Google Scholar]
- 43. Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 2006; 21:811-23; PMID:16543150; http://dx.doi.org/ 10.1016/j.molcel.2006.02.007 [DOI] [PubMed] [Google Scholar]
- 44. Mackay DR, Elgort SW, Ullman KS. The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol Biol Cell 2009; 20:1652-60; PMID:19158386; http://dx.doi.org/ 10.1091/mbc.E08-08-0883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci 2007; 32:63-70; PMID:17210253; http://dx.doi.org/ 10.1016/j.tibs.2006.12.007 [DOI] [PubMed] [Google Scholar]
- 46. Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol 2001; 11:39-46; PMID:11179890; http://dx.doi.org/ 10.1016/S0959-440X(00)00167-6 [DOI] [PubMed] [Google Scholar]
- 47. Schwarz-Herion K, Maco B, Sauder U, Fahrenkrog B. Domain Topology of the p62 Complex Within the 3-D Architecture of the Nuclear Pore Complex. J Mol Biol 2007; 370:796-806; PMID:17544442; http://dx.doi.org/ 10.1016/j.jmb.2007.05.030 [DOI] [PubMed] [Google Scholar]
- 48. Allen TD, Rutherford SA, Murray S, Drummond SP, Goldberg MW, Kiseleva E. Scanning electron microscopy of nuclear structure. Methods Cell Biol 2008; 88:389-409; PMID:18617044; http://dx.doi.org/ 10.1016/S0091-679X(08)00420-2 [DOI] [PubMed] [Google Scholar]
- 49. Bui KH, von Appen A, Diguilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andres-Pons A, Glavy JS, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell 2013; 155:1233-43; PMID:24315095; http://dx.doi.org/ 10.1016/j.cell.2013.10.055 [DOI] [PubMed] [Google Scholar]
- 50. Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J Mol Biol 2003; 328:119-30; PMID:12684002; http://dx.doi.org/ 10.1016/S0022-2836(03)00266-3 [DOI] [PubMed] [Google Scholar]
- 51. Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol 2001; 154:71-84; PMID:11448991; http://dx.doi.org/ 10.1083/jcb.200101089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell 2004; 15:1991-2002; PMID:14718558; http://dx.doi.org/ 10.1091/mbc.E03-10-0743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Partridge JR, Schwartz TU. Crystallographic and biochemical analysis of the Ran-binding zinc finger domain. J Mol Biol 2009; 391:375-89; PMID:19505478; http://dx.doi.org/ 10.1016/j.jmb.2009.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jarnik M, Aebi U. Toward a more complete 3-D structure of the nuclear pore complex. J Struct Biol 1991; 107:291-308; PMID:1725493; http://dx.doi.org/ 10.1016/1047-8477(91)90054-Z [DOI] [PubMed] [Google Scholar]
- 55. Higa MM, Alam SL, Sundquist WI, Ullman KS. Molecular characterization of the ran-binding zinc finger domain of NUP153. J Biol Chem 2007; 282; 17090-100; PMID:17426026; http://dx.doi.org/ 10.1074/jbc.M702715200 [DOI] [PubMed] [Google Scholar]
- 56. Schrader N, Koerner C, Koessmeier K, Bangert JA, Wittinghofer A, Stoll R, Vetter IR. The crystal structure of the Ran-Nup153ZnF2 complex: a general Ran docking site at the nuclear pore complex. Structure 2008; 16:1116-25; PMID:18611384; http://dx.doi.org/ 10.1016/j.str.2008.03.014 [DOI] [PubMed] [Google Scholar]
- 57. Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell 1993; 72:29-38; PMID:8422679; http://dx.doi.org/ 10.1016/0092-8674(93)90047-T [DOI] [PubMed] [Google Scholar]
- 58. Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. Embo J 2010; 29:1659-73; PMID:20407419; http://dx.doi.org/ 10.1038/emboj.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet 2010; 6:e1000846; PMID:20174442; http://dx.doi.org/ 10.1371/journal.pgen.1000846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.