Abstract
Chronic hepatitis B virus (HBV) infection is a major risk factor for developing hepatocellular carcinoma (HCC), and HBV X protein (HBx) acts as cofactor in hepatocarcinogenesis. In liver tumors from animals modeling HBx- and HBV-mediated hepatocarcinogenesis, downregulation of chromatin regulating proteins SUZ12 and ZNF198 induces expression of several genes, including epithelial cell adhesion molecule (EpCAM). EpCAM upregulation occurs in HBV-mediated HCCs and hepatic cancer stem cells, by a mechanism not understood. Herein we demonstrate HBx induces EpCAM expression via active DNA demethylation. In hepatocytes, EpCAM is silenced by PRC2 and ZNF198/LSD1/Co-REST/HDAC1 chromatin modifying complexes. Cells with stable knockdown of SUZ12, an essential PRC2 subunit, upon HBx expression demethylate a CpG dinucleotide located adjacent to NF-κB/RelA half-site. This NF-κB/RelA site is in a CpG island downstream from EpCAM transcriptional start site (TSS). Chromatin immunoprecipitation (ChIP) assays demonstrate HBx-dependent RelA occupancy of NF-κB half-site, while RelA knockdown suppresses CpG demethylation and EpCAM expression. TNF-α activates RelA, propagating demethylation to nearby CpG sites, shown by sodium bisulfite sequencing. RelA-dependent demethylation occurring upon HBx expression requires methyltrasferase EZH2, TET2 a key player in cytosine demethylation, and inactive DNMT3L, shown by knockdown assays and sodium bisulfite sequencing. Co-immunoprecipitations and sequential ChIP assays demonstrate that RelA in the presence of HBx forms a complex with EZH2, TET2 and DNMT3L, although the role of DNMT3L remains to be understood. Interestingly, the human EpCAM gene also has a CpG island downstream from its TSS, and a NF-κB binding site flanked by CpGs. HepG2 cells derived from human HCC exhibit demethylation of these NF-κB-flanking CpG sites, and HBV replication propagates demethylation to nearby CpG sites. DLK1, another PRC2 target gene, also upregulated in HBV-mediated HCCs, is demethylated in liver tumors at CpG dinucleotides flanking the NF-kB binding sequence, supporting that this active DNA demethylation mechanism functions during oncogenic transformation.
Keywords: HBx, EpCAM, EZH2 /PRC2, TET2, DNMT3L, RelA/NF-κB
INTRODUCTION
Chronic infection by the hepatitis B virus (HBV) is linked to hepatocellular carcinoma (HCC).1 The multifunctional HBx protein encoded by HBV2, 3 is essential for the viral life cycle4, and implicated in HCC pathogenesis acting as a cofactor in hepatocarcinogenesis5, 6 by a mechanism not well-understood. Regarding this mechanism, our earlier studies7-9 employed a genome-wide shRNA library screen and identified down-regulation of SUZ12 and ZNF198 proteins has role in HBx-mediated oncogenic transformation4.
SUZ12 is a component of the Polycomb Repressive Complex 2 (PRC2), comprised of essential subunits EZH2, SUZ12 and EED, that mediates the repressive trimethylation of H3 on lysine27 (ref. 10). LSD1/Co-REST/HDAC1 complex, stabilized by ZNF198 (ref. 11), erases activating histone modifications, namely methylations of H3 on lysine4 and acetylations. Moreover, the repressive chromatin modifying activity of these two complexes is coupled by binding the long noncoding RNA (lncRNA) HOTAIR.12 In animal models of HBx- and HBV-mediated hepatocarcinogenesis down-regulation of ZNF198 and/or SUZ12 in liver tumors is associated with elevated expression of a subset of SUZ12/PRC2 target genes including the epithelial cell adhesion molecule EpCAM.13 EpCAM expression characterizes both bi-potential liver progenitors14, 15 and hepatic cancer stem cells16. Here, we investigate the mechanism of EpCAM gene reactivation, using an immortalized mouse liver cell line expressing HBx via the Tet-off expression system17, in combination with stable knockdown cell lines of SUZ12 and ZNF198 (ref. 9).
EpCAM is a 40kDa transmembrane protein highly expressed in most epithelial cancers.18, 19 The elevated expression of EpCAM in rapidly proliferating tumor cells, together with its role in the activation of Wnt signaling20, 21, and the enhanced expression of c-myc and cyclins22, 23, support a role for EpCAM in tumor progression and cancer initiating/stem cells24. However, despite the frequent and elevated expression of EpCAM in tumors and cancer initiating cells the molecular mechanism mediating EpCAM transcription is not understood. In Hep3B cells, Wnt/β-catenin signaling was shown to up-regulate EpCAM-luciferase reporter expression.16 In human ovarian cancer cell lines EpCAM expression was correlated with changes in DNA methylation and histone modifications25, and in human embryonic stem cells (hESCs) the EpCAM gene was identified as a PRC2 target26. Our studies also identified EpCAM expression to be epigenetically silenced by the PRC2 and LSD1/Co-REST/HDAC1 chromatin modifying complexes in untransformed hepatocytes.13
Another epigenetic mechanism directly involved in regulating transcription is DNA methylation of cytosine residues at CpG dinucleotides, generating 5-methylcytosine.27 This epigenetic modification is mediated by de novo DNA methyltransferases DNMT3A and DNMT3B, and propagated to daughter cells during DNA replication by the maintenance enzyme DNMT1.28 The DNMT3 family also includes the catalytically inactive DNMT3L. In ESCs, DNMT3L activates DNA methylation at gene bodies and suppresses DNA methylation at promoters of bivalent genes.29 DNA demethylation also has a crucial role in activation of silenced genes. The Ten-Eleven Translocation (TET) enzymes, TET1, TET2 and TET3 catalyze oxidation of 5mC to hydroxylated 5hmC form, leading to cytosine demethylation.30 Recent studies have identified direct interaction of TETs with specific transcription factors as a mechanism for targeting demethylation of specific cytosine residues, referred to as active DNA demethylation.31-33
Herein, we demonstrate that EpCAM transcription is dependent on cellular context. In SUZ12 knockdown cells expressing HBx, a CpG site located in proximity to an A/T-centric NF-κB half-site34, downstream from the transcriptional start site (TSS) of EpCAM, becomes demethylated. This demethylation is necessary for EpCAM expression. We show EpCAM expression by HBx requires the transcription factor RelA, the methyltransferase EZH2, the TET2 enzyme catalyzing the conversion of 5-methylcytosine to 5-hydroxymethylcytosine, and the catalytically inactive DNMT3L. Although the role of DNMT3L in this complex is not understood, we interpret these results to mean that RelA binds specifically to the NF-κB half-site, 5’GGGAAT3’ (ref. 35, 36), and via interaction with EZH2, recruits TET2 and DNMT3L to the EpCAM sequence. Thus, our results identify a novel mechanism of active DNA demethylation, directed by RelA binding to its half-site.
RESULTS
PRC2 and LSD1/Co-REST/HDAC1 complexes coordinately regulate activity of the EpCAM promoter
Our earlier studies observed a correlation between loss of function of PRC2 and LSD1/Co-REST/HDAC1 chromatin modifying complexes and EpCAM expression in liver tumors of animals modeling HBx- and HBV-mediated hepatocarcinogenesis.13 To understand the molecular mechanism deregulating EpCAM expression in HBx- and HBV-mediated HCCs, we employed the tetracycline regulated HBx-expressing 4pX-1GIPZ cell line17, derived from the immortalized mouse hepatocyte AML12 cell line37, and knockdown cell lines for SUZ12 and ZNF198, namely 4pX-1-SUZ12kd and 4pX-1-ZNF198kd, described earlier4 (Fig. S1A). Indeed, EpCAM expression exhibited HBx-dependent induction and enhanced expression in SUZ12 and ZNF198 knockdown cell lines (Fig. S1B).
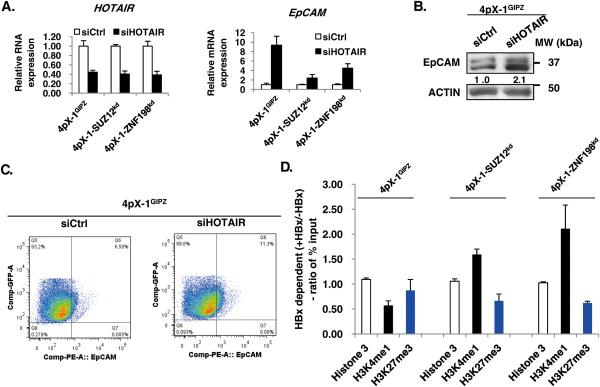
To directly link regulation of EpCAM expression to PRC2 and LSD1/Co-REST/HDAC1 complexes, first, we examined the effect of knockdown of lncRNA HOTAIR known to tether together these complexes.12 HOTAIR knockdown in 4pX-1GIPZ cells increased EpCAM mRNA by nearly 10-fold (Fig. 1A), while in 4pX-1-SUZ12kd and 4pX-1-ZNF198kd cells the increase was only 2-fold and 4-fold, respectively (Fig. 1A). EpCAM immunoblots and flow cytometry with EpCAM antibody of HBx-expressing 4pX-1GIPZ cells further confirm EpCAM induction by HOTAIR knockdown (Fig. 1B and C). To further demonstrate coordinated effects of PRC2 and LSD1/Co-REST/HDAC1 complexes on the EpCAM promoter, we performed ChIP assays with antibodies for H3K27me3 and H3K4me1 using 4pX-1-SUZ12kd and 4pX-1-ZNF198kd cell lines. HBx expression had no effect on the association of H3 with the EpCAM promoter. However, the histone modifications of the EpCAM promoter in the SUZ12 and ZNF198 knockdown cell lines exhibited an HBx-dependent increase in the activating modification H3K4me1, and a reduction of the repressive modification H3K27me3, in comparison to 4pX-1GIPZ cells (Fig. 1D and Fig. S2). Together, these results support the PRC2 and LSD1/Co-REST/HDAC1 complexes coordinately regulate histone modifications of the EpCAM promoter.
Figure 1. The PRC2 and ZNF198/LSD1/Co-REST/HDAC1 complexes coupled by the noncoding RNA HOTAIR repress EpCAM Expression.
A. (Left panel) PCR quantification of HOTAIR following transfection of control siRNA (siCtrl) or HOTAIR siRNA (siHOTAIR) in 4pX-1GIPZ, 4pX-SUZ12kd and 4pX-1-ZNF198kd cell lines. (Right panel) PCR quantification of EpCAM following transfection of siCtrl or siHOTAIR. B. Immunoblot of EpCAM following transfection of siCtrl or siHOTAIR in 4pX-1GIPZ cells. C. Quantification of EpCAM-positive cells (upper right quadrant) by flow cytometry following transfection of siCtrl or siHOTAIR in 4pX-1GIPZ cells. D. ChIP assays with indicated antibodies in 4pX-1GIPZ, 4pX-SUZ12kd and 4pX-1-ZNF198kd cells. Data represent ratio of % input62, quantified by ChIP assays performed with (+) or without (−) HBx in indicated cell lines (Fig. S2 A-D).
HBx induces DNA demethylation of EpCAM CpG sites in a cell context-dependent manner
Since histone modifications correlate with DNA methylation of CpG sites in gene regulatory sequences38-40, and the PRC2 complex interacts with DNMTs41, we examined whether DNA methylation regulates EpCAM expression. Indeed, treatment with 5-Aza-2-deoxycytidine, an inhibitor of DNMTs enhanced EpCAM expression (Fig. S3A). Furthermore, quantification of the levels of 5-hydroxymethylcytosine (5hmC) of the EpCAM sequence by hydroxymethylated DNA immunoprecipitation assays (hMeDIP) demonstrated an increase in cytosine hydroxymethylation by HBx in 4pX-1GIPZ and 4pX-1-SUZ12kd cells (Fig. S3B), suggesting that active DNA demethylation regulates EpCAM expression.
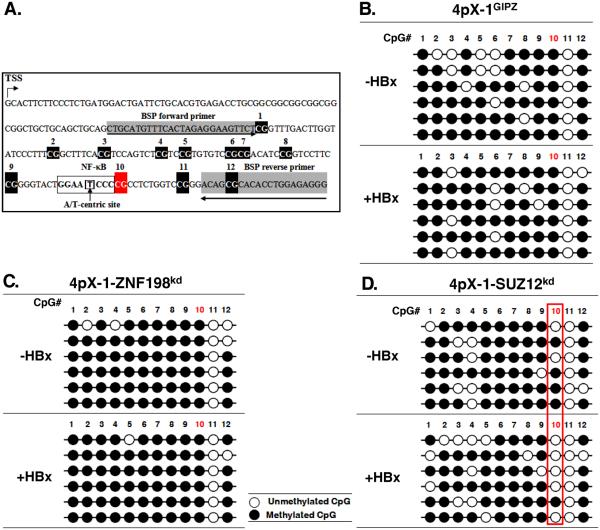
Next, we employed sodium bisulfite sequencing to investigate effects of SUZ12 and ZNF198 knockdown on the DNA methylation pattern of the EpCAM gene. Employing the Methprimer program, we identified a CpG island comprised of 12 CpG dinucleotides downstream from the transcriptional start site (TSS) of EpCAM (Fig. 2A). Sodium bisulphite treatment of DNA from 4pX-1GIPZ cells (Fig. 2B) and 4pX-1-ZNF198kd cells (Fig. 2C) showed the DNA methylation pattern of the 12 CpG dinucleotides did not exhibit significant change due to HBx expression. Interestingly, in SUZ12 knockdown cells (Fig. 2D) an HBx-dependent demethylation was observed at CpG dinucleotide #10, in nearly 90% of sequenced clones. CpG site #10 is adjacent to an A/T-centric RelA half-site.34 Earlier X-ray crystallographic studies of the RelA homodimer demonstrated its binding to the A/T centric half-site.34
Figure 2. HBx induces EpCAM CpG demethylation in SUZ12 knockdown cells.
A. Diagram of CpG dinucleotides in mouse EpCAM gene. The transcriptional start site (TSS), numbered CpG dinucleotides, an A/T centric NF-κB site, and the position of bisulfite sequencing primers (BSP) are indicated. B.-D. Bisulfite sequencing results of EpCAM clones, using DNA from indicated cell lines. Open and closed circles denote absence and presence, respectively, of cytosine modifications including methylation and hydroxymethylation.
RelA together with EZH2 regulate EpCAM expression in SUZ12 knockdown cells
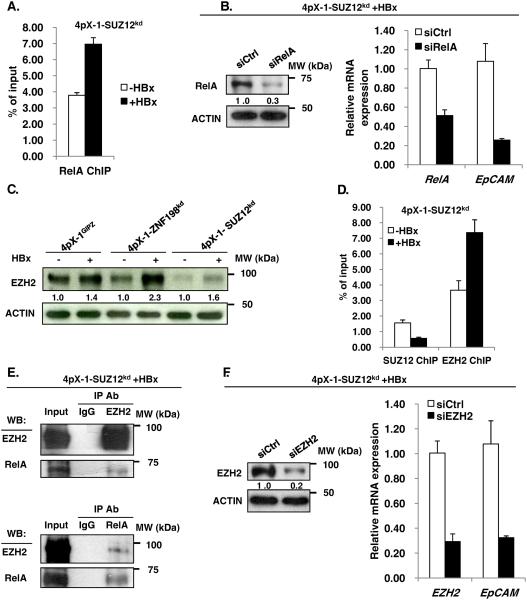
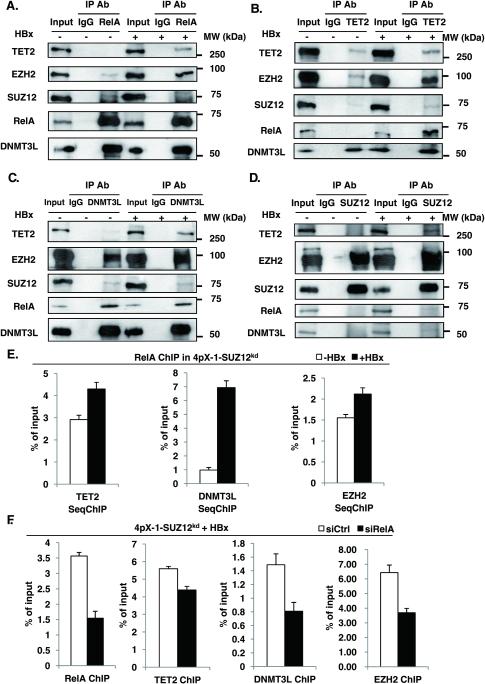
To determine whether DNA demethylation of CpG site #10, located in proximity to an A/T-centric RelA half-site34, is functional in EpCAM expression, first, we examined by ChIP assays whether RelA binds to the EpCAM sequence spanning the 12 CpG sites. In 4pX-1-SUZ12kd cells, HBx increased by nearly 50% the occupancy of the EpCAM CpG island by RelA (Fig. 3A). Next, we knocked-down RelA by siRNA transfection in 4pX-1-SUZ12kd cells and monitored EpCAM expression by PCR. We observed a 5-fold reduction in EpCAM mRNA levels by RelA knockdown, supporting the functional significance of RelA in EpCAM transcription (Fig. 3B). By contrast, RelA knockdown in 4pX-1-ZNF198kd cells resulted only in 1.5-fold decrease in EpCAM mRNA levels (Fig. S4).
Figure 3. RelA and EZH2 regulate EpCAM expression in SUZ12 knockdown cells.
A. ChIP assays with RelA antibody, employing 4pX-1-SUZ12kd cells grown +/− HBx expression by tetracycline removal for 18h, and EpCAM primers spanning CpG sites shown in Fig. 2A. B. PCR quantification of EpCAM mRNA following transfection of control siRNA (siCtrl) or RelA siRNA (siRelA). Immunoblot of RelA after transfection of siRelA vs. siCtrl. C. Immunoblots of EZH2 in indicated cell lines +/− HBx expression by tetracycline removal for 18h. D. ChIP assays with SUZ12 or EZH2 antibody, employing 4pX-1-SUZ12kd cells grown +/− HBx expression by tetracycline removal for 18h, and EpCAM primers spanning CpG sites shown in Fig. 2A. E. Co-immunoprecipitations of EZH2 and RelA, employing lysates from 4pX-1-SUZ12kd cells grown with (+) HBx for 18h. F. PCR quantification of EpCAM mRNA following EZH2 knockdown by siRNA transfection (siEZH2) in 4pX-1-SUZ12kd cells, with (+) HBx. Immunoblot of EZH2 following transfection of siEZH2 vs siCtrl.
Recent studies have shown direct interaction of EZH2 with RelA/RelB heterodimer in promoting expression of NF-κB targets in breast cancer.42 Therefore, we investigated whether EZH2 has a similar role in EpCAM expression. We determined EZH2 protein levels in the three cell lines26 as a function of HBx expression. Interestingly, HBx increased protein level of EZH2 in all three cell lines (Fig. 3C). Next, we examined by ChIP assays association of EZH2 with the EpCAM sequence containing the RelA half-site. In 4pX-1-SUZ12kd cells, EZH2 exhibited enhanced occupancy at the EpCAM sequence (Fig. 3D); by contrast absence of SUZ12 occupancy at that site (Fig. 3D) suggests that EZH2 interacts alone, not in complex with the other subunits of PRC2. Reciprocal co-immunoprecipitations of RelA and EZH2 demonstrate that these proteins interact with each other (Fig. 3E), in agreement with similar observations by others.42 To determine whether association of RelA and EZH2 with the EpCAM sequence has functional significance, we knocked-down EZH2 and quantified by PCR EpCAM mRNA levels. In 4pX-1-SUZ12kd cells, transfection of EZH2 siRNA reduced expression of EpCAM (Fig. 3F), thereby demonstrating that the interaction between RelA and EZH2 (Fig. 3E) is functionally important for EpCAM expression.
TET2 and DNMT3L are required for DNA demethylation of EpCAM
The HBx-dependent demethylation of CpG dinucleotide #10, observed selectively in SUZ12 knockdown cells, suggested an active DNA demethylation mechanism. To investigate this mechanism, we determined by PCR mRNA levels of enzymes involved in DNA demethylation. The dioxygenases TET1-3 directly convert 5mC to 5hmC.43, 44 On the other hand, the catalytically inactive DNMT3L indirectly maintains low levels of DNA methylation at gene promoters/regulatory sequences by displacing DNMT3A and DNMT3B from binding the PRC2 complex.29 TET1 (ref. 45) and DNMT3L29 were reported to interact with EZH2. However whether TET2 and TET3 enzymes interact with EZH2 is unknown.
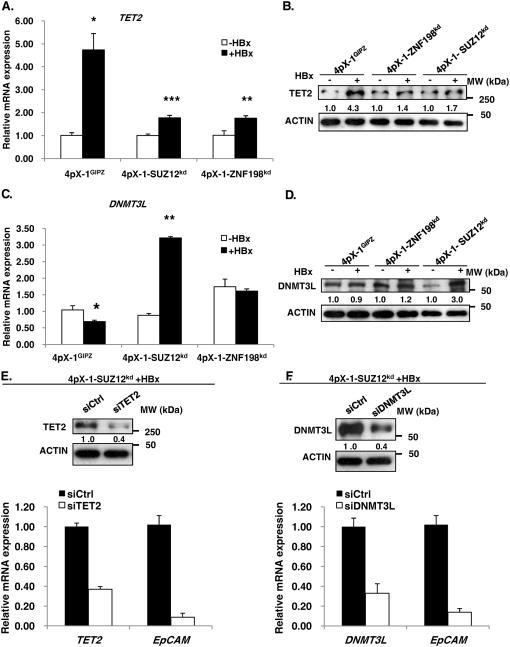
Employing RT-PCR we quantified mRNA levels of TET1-3 as a function of HBx expression (Fig. 4A and Fig. S5). TET1 did not exhibit HBx-dependent induction in the three cell lines (Fig. S5A). TET2 and TET3 mRNAs exhibited statistically significant HBx-dependent induction in both 4pX-1GIPZ and 4pX-1-SUZ12kd cells, whereas in 4pX-1-ZNF198kd cells only TET2 was induced (Fig. 4A and Fig. S5B). Interestingly DNMT3L was induced by HBx only in 4pX-1-SUZ12kd cells (Fig. 4C). Immunoblots of TET2 and DNMT3L confirmed this HBx-dependent induction (Fig. 4B and D). Since TET2 lacks the CXXC motif required for DNA binding, present in TET1 and TET3 (ref. 30), we reasoned TET2 may interact with the RelA/EZH2 complex (Fig. 3E). Therefore, we knocked-down TET2 in 4pX-1-SUZ12kd cells by siRNA transfection and quantified expression levels of EpCAM. Knockdown of TET2 in the presence of HBx significantly reduced EpCAM expression in 4pX-1-SUZ12kd cells (Fig. 4E). By contrast, siRNA knockdown of TET3 had no effect on EpCAM mRNA levels (Fig. S5C). Also, knockdown of DNMT3L by siRNA transfection reduced EpCAM expression in 4pX-1-SUZ12kd cells (Fig. 4F).
Figure 4. TET2 and DNMT3L are required for EpCAM expression.
A. PCR quantification of TET2 in indicated cell lines +/− HBx expression by tetracycline removal for 18h. B. Immunoblot of TET2 in indicated cell lines +/− HBx. C. PCR quantification of DNMT3L in indicated cell lines +/− HBx. D. Immunoblot of DNMT3L in indicated cell lines +/− HBx. E. PCR quantification of EpCAM mRNA following transfection of TET2 siRNA (siTET2) vs. control siRNA (siCtrl), in 4pX-1-SUZ12kd cells expreesing HBx. TET2 knockdown by siTET2 transfection (50pM) quantified by PCR and confirmed by TET2 immunoblots. F. PCR quantification of EpCAM mRNA following transfection of DNMT3L siRNA (siDNMT3L) vs. siCtrl, in 4pX-1-SUZ12kd cells expressing HBx. DNMT3L knockdown by siDNMT3L transfection (80pM) quantified by PCR and confirmed by DNMT3L immunoblots.
TET2 and DNMT3L are in complex with EZH2 and RelA in HBx expressing cells
Next, we examined whether RelA, EZH2, DNMT3L and TET2 are in complex (Fig. 5). Indeed, RelA co-immunoprecipitated DNMT3L and EZH2, while expression of HBx enhanced the amount of co-immunoprecipitated EZH2 and TET2 (Fig. 5A). Conversely, TET2 co-immunoprecipitated EZH2, and in the presence of HBx we detected increased levels of TET2, RelA and DNMT3L (Fig. 5B). In turn, DNMT3L co-immunoprecipitated EZH2 and RelA, while HBx expression increased the amount of co-immunoprecipitated EZH2 and TET2 (Fig. 5C). Importantly, this complex comprised of RelA, EZH2, TET2 and DNMT3L does not contain SUZ12, since immunoprecipitation of SUZ12 does not co-immunoprecipitate these proteins (Fig. 5D). To demonstrate the functional assembly of the RelA-directed EZH2/TET2/DNMT3L complex, we employed two approaches (Fig. 5E and F). First, we performed ChIP assays with the RelA antibody and sequential ChIP assays (SeqChIP) with antibody for TET2, DNMT3L and EZH2. HBx expression in 4pX-1-SUZ12kd cells increased the association of TET2, DNMT3L and EZH2 with RelA-bound to the EpCAM CpG island (Fig. 5E). Conversely, siRNA knockdown of RelA reduced the amount of TET2, DNMT3L and EZH2 that immunoprecipitated in ChIP assays with the EpCAM sequence (Fig. 5F). Taken together, these results demonstrate the requirement for RelA in the formation of the EZH2/TET2/DNMT3L complex, and the association of these proteins with the EpCAM chromatin.
Figure 5. TET2 and DNMT3L are in complex with EZH2 and RelA.
A.-D. Immunoprecipitations (IP) with IgG and indicated antibodies using WCE from 4pX-1-SUZ12kd cells +/− HBx expression for 18h. IPs were immunoblotted with TET2, EZH2, SUZ12, RelA and DNMT3L antibodies. E. ChIP assays with RelA antibody, employing 4pX-1-SUZ12kd cells grown +/− HBx expression by tetracycline removal for 18h; ChIPed DNA was used in sequential seqChIP assays employing indicated antibodies and EpCAM primers spanning CpG sites shown in Fig. 2A. F. ChIP assays performed with indicated antibody, employing 4pX-1-SUZ12kd cells transfected with siRNAs for RelA (siRelA) or control (siCtrl), in the presence of HBx expression. EpCAM primers spanning CpG sites shown in Fig.2A were used.
RelA and EZH2 are necessary for DNA demethylation of CpG site #10 of EpCAM
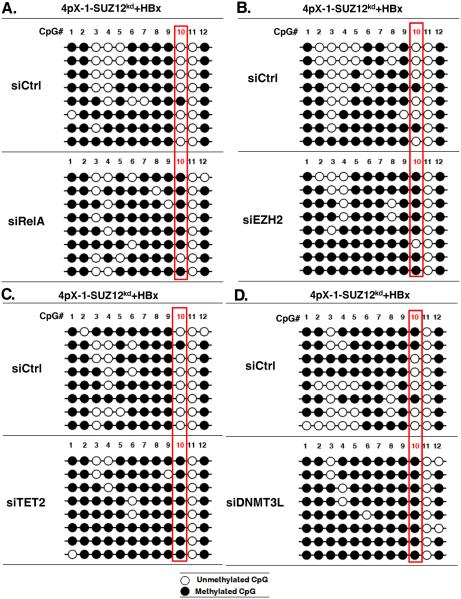
To directly demonstrate the role of the complex comprised of RelA/EZH2/TET2/DNMT3L in DNA demethylation of EpCAM CpG site #10, we knocked-down by siRNA transfection each of these proteins in HBx-expressing 4pX-1-SUZ12kd cells. Sodium bisulfite sequencing of DNA isolated from HBx-expressing 4pX-1-SUZ12kd cells following knockdown of RelA, EZH2, TET2 or DNMT3L demonstrated that CpG site #10 was now mostly methylated (Fig. 6).
Figure 6. RelA, EZH2, TET2 and DNMT3L are required for HBx-induced EpCAM gene demethylation.
A.-D. Bisulfite sequencing results of EpCAM clones, using DNA from 4pX-1-SUZ12kd cells expressing HBx and transfected with siRNAs for RelA (siRelA), EZH2 (siEZH2), TET2 (siTET2), DNMT3L (siDNMT3L) and control (siCtrl).
TNF-α treatment and HBV replication propagate DNA demethylation to additional CpG sites in proximity to the RelA/NF-κB site
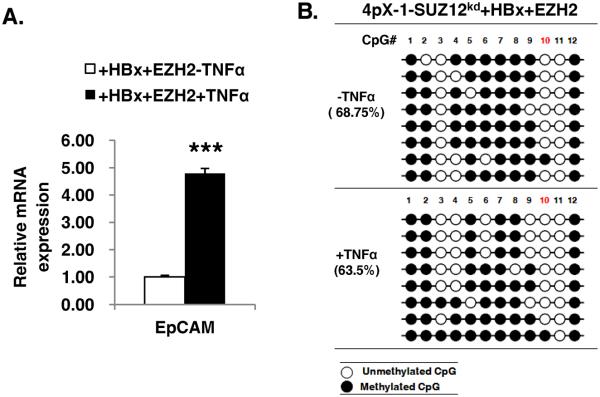
To confirm the role of RelA in demethylation of the EpCAM sequence, we activated RelA by treatment with TNF-α (0.1 ng/ml), in HBx expressing cells transfected with EZH2. Although HBx activates NF-κB signaling46, we reasoned treatment with TNF-α will further increase or prolong RelA activation. Indeed, under these conditions, EpCAM expression was further elevated (Fig. 7A). Interestingly, sodium bisulfite sequencing detected DNA demethylation of additional CpG sites, sites #3, 4, 6 and 9 (Fig. 7B), supporting that the level/duration of RelA activation influences demethylation of additional CpG sites.
Figure 7. TNF-α increases EpCAM expression and propagates demethylation of nearby CpG sites.
A. PCR quantification of EpCAM mRNA following transfection of EZH2 expression vector in 4pX-1-SUZ12kd cells expressing HBx and treated with TNF-α (0.1ng/ml) for 18h. B. Bisulfite sequencing results of EpCAM clones, using DNA from 4pX-1-SUZ12kd cells expressing HBx, transfected with EZH2 expression vector, and treated with TNF-α (0.1ng/ml) for 18h.
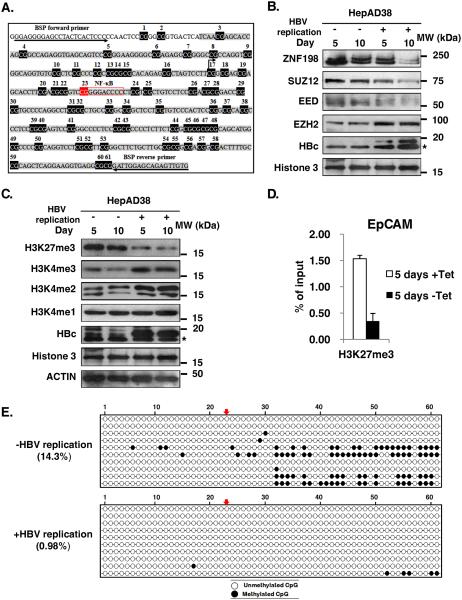
To link these findings to the regulation of the human EpCAM promoter, we determined by sodium bisulfite sequencing the methylation pattern of a 450bp sequence downstream from the TSS of the human EpCAM gene, containing 61 CpG dinucleotides (Fig. 8A). Interestingly CpG site #23 is adjacent to an NF-κB site (Fig. 8A). We employed a human liver cancer cell line derived from HepG2 cells, named HepAD38 cell line.47 HepAD38 cells support HBV replication via the stable integration of the HBV genome under control of the tetracycline-regulated (Tet-off) expression system.47 Under conditions of HBV replication by tetracycline removal, monitored by the increased protein levels of the viral HBc antigen (Fig. 8B), EpCAM expression increases13, SUZ12 levels decreased13, while EZH2 levels increased (Fig. 8B). In turn, the downregulation of SUZ12 and ZNF198 proteins observed in HBV replicating cells resulted in reduction of the repressive H3K27me3 modification, whereas H3K4 modifications associated with transcriptional activation were increased (Fig. 8C). ChIP assays show reduction of the repressive H3K27me3 associated with the EpCAM promoter as a function of HBV replication (Fig. 8D). In agreement with the tumor origin of this cell line, sodium bisulfite sequencing showed that CpG site #23 was unmethylated. Interestingly, upon induction of HBV replication for two days, additional CpG sites became demethylated (Fig. 8E), as with EZH2 overexpression and TNF-α treatment in 4pX-1-SUZ12kd cells, suggesting that active DNA demethylation is involved in the expression of the human EpCAM gene, in HBV replicating cells.
Figure 8. Increased EpCAM demethylation in HBV replicating HepAD38 cells.
A. Diagram of CpG dinucleotides in human EpCAM gene. The transcriptional start site (TSS), numbered CpG dinucleotides, a NF-κB site, and the position of bisulfite sequencing primers (BSP) are indicated. B. Immunoblots of nuclear extracts isolated from HepAD38 cells grown with (+) or without (−) HBV replication by tetracycline removal for 5 and 10 days, employing antibodies for ZNF198, SUZ12, EED, EZH2, HBc, and Histone3 serving as loading control. * indicates non-specific band. C. Immunoblots of HepAD38 lysates from cells grown with (+) or without (−) Tet for 5 and 10 days, employing the indicated antibodies. D. ChIP assays with H3K27me3 antibody employing primers for the human EpCAM CpG island. E. Bisulfite sequencing results of EpCAM clones, using DNA from HepAD38 cells gown in the absence (−) of HBV replication or with (+) HBV replication for 2 days, by tetracycline removal. % methylation is indicated. Arrows indicate CpG dinucleotide #23 adjacent to NF-κB site.
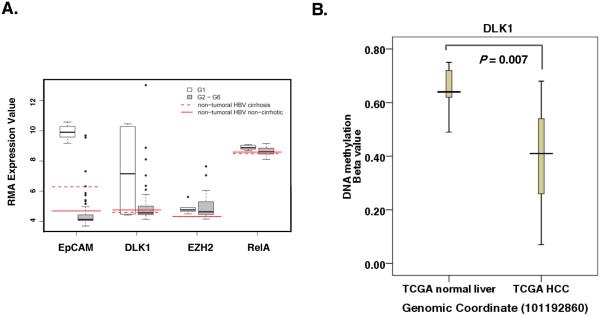
In further support of this mechanism, we re-analyzed the HCC transcriptome data of Boyault et al48 which classified HCCs in six subgroups (G1-G6). Of interest is the G1 group of tumors associated with low copy number of HBV and overexpression of genes expressed in fetal liver. Indeed, the G1 group exhibits robust expression of hepatic progenitor genes EpCAM and DLK1 (ref. 49) (Fig. 9A and Supplementary Table S1), both genes are targets of the PRC2 and LSD1/Co-REST/HDAC1 complexes13. The G1 group also exhibits a small but statistically significant induction of EZH2 and RelA (Fig. 9A and Supplementary Table S1), key players in our mechanism of active DNA demethylation (Fig. 6).
Figure 9. NF-κB site-driven DNA demethylation in human HCCs.
A. Boxplots showing Robust Multiarray Average (RMA) expression values for genes of interest in HCC Group 1 (n = 6) and the pooled value for HCC Groups 2 – 6 (n = 53). The RMA value for control microarrays hybridized with pooled RNA samples from non-tumoral, HBV-infected liver either without cirrhosis (solid red lines) or with cirrhosis (dashed red lines). RMA values for EpCAM, DLK1, EZH2 and RelA are shown. Statistical significance of the expression difference is shown in Table S1. B. DNA methylation beta value derived from the human HCC methylome available through The Cancer Genome Atlas (TCGA). The difference in beta values between normal vs. HCC is statistically significant (p=0.007).
To evaluate the methylation status of the EpCAM CpG island under study (Fig. 8A) in human HCCs, we examined the methylation data (Illumina Human Methylation 450 array) from The Cancer Genome Atlas (TCGA), by evaluating the beta value (β) defined as the ratio of intensities between methylated vs. non-methylated DNA. However, beta values for the EpCAM CpG island under study are not represented in TCGA data. Since DLK1 is also overexpressed in the G1 group of HCCs, and is the target of PRC2 and LSD1/Co-REST/HDAC1 complexes13, we examined the DNA methylation status of CpG sites located within NF-κB binding sites. Employing TCGA human HCC methylome, we identified in the DLK1 promoter a CpG dinucleotide within an NF-κB binding site. Importantly, beta values indicate that indeed this CpG site is demethylated in human HCCs in comparison to peritumoral tissue (Fig. 9B). We interpret these data to mean that the DNA demethylation mechanism we identified in our model cell lines is also functional in human HCCs.
DISCUSSION
In this study we provide evidence for a novel mechanism of active DNA demethylation induced by HBx that allows re-expression of the EpCAM gene observed in HBV-mediated HCCs.13 We demonstrate that this mechanism is directed by the transcription factor RelA. RelA binds to an A/T centric RelA half-site35, 36 located downstream from the TSS of the EpCAM gene (mouse and human) and directs DNA demethylation by TET2. Our results identify a novel bridging function for EZH2 in the formation of the DNA demethylation complex comprised of TET2 and the catalytically inactive DNMT3L. Although in this study we have not determined the mechanism by which DNMT3L functions in this complex, studies by others have shown that DNMT3L maintains the demethylated state in bivalent promoters in ESCs.29
Specifically, while DNMT3L enhances DNA methylation at gene bodies of active genes, it prevents DNA methylation at the promoters of H3K27me3-positive genes, like EpCAM. Our results show that DNMT3L interacts directly with EZH2 and TET2. We speculate that following DNA demethylation, DNMT3L, recruited to the EpCAM promoter via the RelA complex, counteracts the activity of DNMT3A and DNMT3B, thereby maintaining hypomethylation. Importantly, we demonstrate this DNA demethylation complex is induced by HBx in the context of SUZ12 downregulation, i.e., EZH2 acts independently of the other core subunits of the PRC2 complex (Figs. 3D and 5D). The cellular context of the 4pX-1-SUZ12kd cell line resembles down-regulation of SUZ12 observed in liver tumors of animals modeling HBx- and HBV-mediated hepatocarcinogenesis, as well as the host cell during HBV replication13 (Fig. 8B), all of which exhibit elevated expression of EpCAM13. Our recent studies (Zhang et al, pending) have determined the mechanism by which HBx promotes SUZ12 downregulation, involving proteasomal degradation induced by Polo-like-kinase1 (Plk1)-mediated phosphorylation of SUZ12. In turn, Plk1 is overexpressed in many human cancers including HBV-induced HCC, and is activated by HBx.7
How HBx induces formation of the RelA-directed DNA demethylation complex is not understood. RelA undergoes posttranslational modifications including methylation.50 Whether EZH2 methylates RelA or other members of the complex remains to be determined. Based on recent findings that RelA activity is regulated by methylation50, and by analogy to the ability of EZH2 to methylate the transcription factor STAT3 (ref. 51), it is reasonable to propose that EZH2 is a likely methyltransferase for RelA. HBx could alter the methyltransferase specificity of EZH2 by inducing its phosphorylation, as it has been shown to occur for the EZH2-mediated methylation of STAT3 (ref. 51). It is well-documented that HBx induces activation of cellular signal transduction pathways2, 3 as well as activation of the mitotic Plk1 (ref. 7). Interestingly, EZH2 contains several Plk1 consensus phosphorylation sites. We speculate HBx-driven post-translational modifications of EZH2 alter its methyltransferase specificity towards methylation of RelA or the other components of the DNA demethylation complex.
The involvement of RelA in this active DNA demethylation process is quite interesting, because RelA/NF-κB is constitutively activated in many human cancers including liver cancer.52 HBx45 as well as inflammatory stimuli originating from the microenviroment52 activate NF-κB. Our results demonstrating propagation of DNA demethylation to nearby CpG sites in the EpCAM gene upon treatment of HBx-expressing cells with TNF-α, identify another mechanism by which chronic inflammation could be involved in cancer pathogenesis. We speculate RelA/NF-κB, activated by HBx and TNF-α secreted by cytotoxic T lymphocytes53, integrates effects of chronic inflammation and HBx action in mediating HCC pathogenesis in chronically infected HBV patients. Furthermore, recent studies have shown that an inverse relationship exists between enhanced RelA expression and decreased HNF4 expression in human liver tumors54, thereby linking RelA expression to less differentiated HCCs exhibiting elevated expression of EpCAM16. Although NF-κB regulates various classes of genes involved in tumor development, including proliferation, survival and metastasis52, the repertoire of genes regulated via this active DNA demethylation mechanism directed by RelA and EZH2 requires further studies. Our results have identified the hepatic progenitor DLK1, another PRC2 target gene, to be demethylated in human HCCs at a CpG site located within NF-κB cis-acting element (Fig. 9B). Other genes that could undergo a similar DNA demethylation mechanism include the PRC2 target gene BAMBI13, or genes expressed in early hepatic progenitors or during hepatic lineage determination such as SOX9 (ref. 55) and SALL4 (ref. 56). Interestingly, these genes have CpG sites located within NF-κB cis-acting elements. Further studies are needed to determine the mechanism by which this active DNA demethylation, associated with the NF-κB cis-acting elements, regulates transcription.
Regarding the involvement of EZH2 in this DNA demethylation mechanism, our results show that EZH2 acts independently of the other subunits of the PRC2 complex (Fig. 5D). Whether the methyltransferase activity of EZH2 is required for its involvement in the RelA-driven DNA demethylation is also unknown. Since EZH2 is overexpressed in many human cancers including liver cancer57 our studies identify a novel role for EZH2, independent of its histone3 methyltransferase function, in oncogenesis.
MATERIALS AND METHODS
Cell lines The 4pX-1 cell line is a tetracycline regulated HBx-expressing cell line, derived from immortalized mouse hepatocyte AML12 cells.37 4pX-1GIPZ is derived from the 4pX-1 cell line17 containing a stable insertion of GIPZ vector and thus serving as vector control for the stable knockdown (kd) cell lines 4pX-1-SUZ12kd and 4pX-1-ZNF198kd. All 4pX-1-derived cell lines express HBx by tetracycline removal for 18h and grown as described.9, 17 HBx expression is confirmed by RT-PCR. Treatment of indicated cell lines with 10μM 5-aza-2’-Deoxycytidine (#11166, Cayman Chemical) was for 48h. The HepAD38 cell line that supports HBV replication by removal of tetracycline was grown as described.47
Sodium bisulfite sequencing employed the following reagents and protocols described by the manufacturers: PureLink Genomic DNA Mini Kit (K1820-01, Invitrogen) for genomic DNA isolation; EZ DNA Methylation-Direct Kit (D5020, Zymo) for bisulfate treatment of DNA. Mouse EpCAM bisulfate sequencing primers were designed by Methprimer.58
Chromatin Immunoprecipitation (ChIP) assays, Immunoprecipitations and immunoblot analyses performed using standard protocols.13 The following reagents and antibodies were used: Lysis Buffer (#9803, Cell Signaling); Normal Rabbit IgG (#2729, Cell Signaling); Protease inhibitor cocktail (P8340, SIGMA); ChIP assay kit (17-295, Millipore). Protein A/G PLUS-agarose immunoprecipitation reagent (sc-2003, SANTA CRUZ); Dynabeads Protein G (10003D, Invitrogen); EED antibody (#61203, Active Motif); EZH2 antibody (#5246, Cell Signaling); ZNF198 antibody (PA1-41457, Thermo fisher); Tri-Methyl-Histone H3 (Lys4) antibody (#9727S, Cell Signaling); Di-Methyl-Histone H3 (Lys4) antibody (#9725S, Cell Signaling); Mono-Methyl-Histone H3 (Lys4) antibody (#9723S, Cell Signaling); Anti-Histone H3 (tri methyl K27) antibody (ab6002, Abcam); anti-EpCAM antibody (ab71916, Abcam); Histone H3 antibody (61277, Active Motif) Cell ; RelA antibody (sc-109, SANTA CRUZ); SUZ12 antibody (ab12073, Abcam); RNase Cocktail Enzyme Mix (AM2286, Ambion); TET2 antibody (MABE462, Millipore); DNMT3L antibody (ab3493, Abcam); 5-hmC antibody (MAb-31HMC, Diagenode).
Reverse Transcription and Quantitative Real-Time PCR: RNA was isolated employing PureLink RNA Mini Kit (12183018A, Invitrogen). cDNA was synthesized from 2.0 μg total RNA isolated using iScript™ cDNA Synthesis Kit (170-8891, Bio-Rad). Quantitative real-time PCR reactions were performed in triplicates and normalized to GAPDH employing FastStart Essential DNA Green Master (06924204001, Roche), SYBR green (Roche), and Roche LightCycler 96. The 2−ΔΔCt method was used for analysis.59 Primer sequences are listed in Table S2, supplementary information.
Statistical analyses: Statistical significance was assessed by Student’s t-test. Data were expressed as mean ± standard deviation (SD) and *P < 0.05, **P < 0.01 and ***P < 0.001 were considered significant.
Re-analysis of microarray data. CEL files and metadata for the microarray study of Boyault, et al48 were downloaded from ArrayExpress (accession number E-TABM-36). CEL files were processed with R and Bioconductor60 using the Robust Multi-array Average (RMA) function in the affy package (v 1.42.3)61. Probe set annotation for the Affymetrix Human Genome microarray U133A was obtained from the hgu133a.db (v 2.14.0) package.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant DK044533 to OA. Shared Resources (flow cytometry and DNA sequencing) are supported by NIH grant P30CA023168 to Purdue Cancer Research Center. The authors thank Dr. R. Hullinger for critical review of this manuscript.
Footnotes
CONFLICT OF INTEREST: The authors have no competing financial interest in relation to the work described in this manuscript.
References
- 1.Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 2.Andrisani OM, Barnabas S. The transcriptional function of the hepatitis B virus X protein and its role in hepatocarcinogenesis (Review) International journal of oncology. 1999;15:373–379. doi: 10.3892/ijo.15.2.373. [DOI] [PubMed] [Google Scholar]
- 3.Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. Journal of virology. 2004;78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. Journal of virology. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Madden CR, Finegold MJ, Slagle BL. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. Journal of virology. 2001;75:3851–3858. doi: 10.1128/JVI.75.8.3851-3858.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terradillos O, Billet O, Renard CA, Levy R, Molina T, Briand P, et al. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 7.Studach L, Wang WH, Weber G, Tang J, Hullinger RL, Malbrue R, et al. Polo-like kinase 1 activated by the hepatitis B virus X protein attenuates both the DNA damage checkpoint and DNA repair resulting in partial polyploidy. The Journal of biological chemistry. 2010;285:30282–30293. doi: 10.1074/jbc.M109.093963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Studach LL, Rakotomalala L, Wang WH, Hullinger RL, Cairo S, Buendia MA, et al. Polo-like kinase 1 inhibition suppresses hepatitis B virus X protein-induced transformation in an in vitro model of liver cancer progression. Hepatology. 2009;50:414–423. doi: 10.1002/hep.22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang WH, Studach LL, Andrisani OM. Proteins ZNF198 and SUZ12 are down-regulated in hepatitis B virus (HBV) X protein-mediated hepatocyte transformation and in HBV replication. Hepatology. 2011;53:1137–1147. doi: 10.1002/hep.24163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gocke CB, Yu H. ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PloS one. 2008;3:e3255. doi: 10.1371/journal.pone.0003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Studach LL, Menne S, Cairo S, Buendia MA, Hullinger RL, Lefrancois L, et al. Subset of Suz12/PRC2 target genes is activated during hepatitis B virus replication and liver carcinogenesis associated with HBV X protein. Hepatology. 2012;56:1240–1251. doi: 10.1002/hep.25781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Boer CJ, van Krieken JH, Janssen-van Rhijn CM, Litvinov SV. Expression of Ep-CAM in normal, regenerating, metaplastic, and neoplastic liver. The Journal of pathology. 1999;188:201–206. doi: 10.1002/(SICI)1096-9896(199906)188:2<201::AID-PATH339>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Schmelzer E, Zhang L, Bruce A, Wauthier E, Ludlow J, Yao HL, et al. Human hepatic stem cells from fetal and postnatal donors. The Journal of experimental medicine. 2007;204:1973–1987. doi: 10.1084/jem.20061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tarn C, Bilodeau ML, Hullinger RL, Andrisani OM. Differential immediate early gene expression in conditional hepatitis B virus pX-transforming versus nontransforming hepatocyte cell lines. The Journal of biological chemistry. 1999;274:2327–2336. doi: 10.1074/jbc.274.4.2327. [DOI] [PubMed] [Google Scholar]
- 18.Schnell U, Cirulli V, Giepmans BN. EpCAM: structure and function in health and disease. Biochimica et biophysica acta. 2013;1828:1989–2001. doi: 10.1016/j.bbamem.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Went P, Vasei M, Bubendorf L, Terracciano L, Tornillo L, Riede U, et al. Frequent high-level expression of the immunotherapeutic target Ep-CAM in colon, stomach, prostate and lung cancers. British journal of cancer. 2006;94:128–135. doi: 10.1038/sj.bjc.6602924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hachmeister M, Bobowski KD, Hogl S, Dislich B, Fukumori A, Eggert C, et al. Regulated intramembrane proteolysis and degradation of murine epithelial cell adhesion molecule mEpCAM. PloS one. 2013;8:e71836. doi: 10.1371/journal.pone.0071836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nature cell biology. 2009;11:162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 22.Chaves-Perez A, Mack B, Maetzel D, Kremling H, Eggert C, Harreus U, et al. EpCAM regulates cell cycle progression via control of cyclin D1 expression. Oncogene. 2013;32:641–650. doi: 10.1038/onc.2012.75. [DOI] [PubMed] [Google Scholar]
- 23.Munz M, Kieu C, Mack B, Schmitt B, Zeidler R, Gires O. The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene. 2004;23:5748–5758. doi: 10.1038/sj.onc.1207610. [DOI] [PubMed] [Google Scholar]
- 24.Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer research. 2009;69:5627–5629. doi: 10.1158/0008-5472.CAN-09-0654. [DOI] [PubMed] [Google Scholar]
- 25.van der Gun BT, de Groote ML, Kazemier HG, Arendzen AJ, Terpstra P, Ruiters MH, et al. Transcription factors and molecular epigenetic marks underlying EpCAM overexpression in ovarian cancer. British journal of cancer. 2011;105:312–319. doi: 10.1038/bjc.2011.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & development. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HJ, Hore TA, Reik W. Reprogramming the Methylome: Erasing Memory and Creating Diversity. Cell stem cell. 2014;14:710–719. doi: 10.1016/j.stem.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO reports. 2011;12:647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neri F, Krepelova A, Incarnato D, Maldotti M, Parlato C, Galvagni F, et al. Dnmt3L antagonizes DNA methylation at bivalent promoters and favors DNA methylation at gene bodies in ESCs. Cell. 2013;155:121–134. doi: 10.1016/j.cell.2013.08.056. [DOI] [PubMed] [Google Scholar]
- 30.Delatte B, Deplus R, Fuks F. Playing TETris with DNA modifications. The EMBO journal. 2014;33:1198–1211. doi: 10.15252/embj.201488290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de la Rica L, Rodriguez-Ubreva J, Garcia M, Islam AB, Urquiza JM, Hernando H, et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome biology. 2013;14:R99. doi: 10.1186/gb-2013-14-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujiki K, Shinoda A, Kano F, Sato R, Shirahige K, Murata M. PPARgamma-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nature communications. 2013;4:2262. doi: 10.1038/ncomms3262. [DOI] [PubMed] [Google Scholar]
- 33.Ko M, An J, Bandukwala HS, Chavez L, Aijo T, Pastor WA, et al. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang VY, Huang W, Asagiri M, Spann N, Hoffmann A, Glass C, et al. The transcriptional specificity of NF-kappaB dimers is coded within the kappaB DNA response elements. Cell reports. 2012;2:824–839. doi: 10.1016/j.celrep.2012.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen YQ, Ghosh S, Ghosh G. A novel DNA recognition mode by the NF-kappa B p65 homodimer. Nature structural biology. 1998;5:67–73. doi: 10.1038/nsb0198-67. [DOI] [PubMed] [Google Scholar]
- 36.Chen YQ, Sengchanthalangsy LL, Hackett A, Ghosh G. NF-kappaB p65 (RelA) homodimer uses distinct mechanisms to recognize DNA targets. Structure. 2000;8:419–428. doi: 10.1016/s0969-2126(00)00123-4. [DOI] [PubMed] [Google Scholar]
- 37.Wu JC, Merlino G, Fausto N. Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:674–678. doi: 10.1073/pnas.91.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meissner A. Guiding DNA methylation. Cell stem cell. 2011;9:388–390. doi: 10.1016/j.stem.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 39.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature genetics. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 41.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 42.Lee ST, Li Z, Wu Z, Aau M, Guan P, Karuturi RK, et al. Context-specific regulation of NF-kappaB target gene expression by EZH2 in breast cancers. Molecular cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 43.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cartron PF, Nadaradjane A, Lepape F, Lalier L, Gardie B, Vallette FM. Identification of TET1 Partners That Control Its DNA-Demethylating Function. Genes & cancer. 2013;4:235–241. doi: 10.1177/1947601913489020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su F, Schneider RJ. Hepatitis B virus HBx protein activates transcription factor NF-kappaB by acting on multiple cytoplasmic inhibitors of rel-related proteins. Journal of virology. 1996;70:4558–4566. doi: 10.1128/jvi.70.7.4558-4566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ladner SK, Otto MJ, Barker CS, Zaifert K, Wang GH, Guo JT, et al. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrobial agents and chemotherapy. 1997;41:1715–1720. doi: 10.1128/aac.41.8.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka M, Okabe M, Suzuki K, Kamiya Y, Tsukahara Y, Saito S, et al. Mouse hepatoblasts at distinct developmental stages are characterized by expression of EpCAM and DLK1: drastic change of EpCAM expression during liver development. Mechanisms of development. 2009;126:665–676. doi: 10.1016/j.mod.2009.06.939. [DOI] [PubMed] [Google Scholar]
- 50.Lu T, Yang M, Huang DB, Wei H, Ozer GH, Ghosh G, et al. Role of lysine methylation of NF-kappaB in differential gene regulation. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13510–13515. doi: 10.1073/pnas.1311770110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nature reviews Drug discovery. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakamoto Y, Guidotti LG, Pasquetto V, Schreiber RD, Chisari FV. Differential target cell sensitivity to CTL-activated death pathways in hepatitis B virus transgenic mice. J Immunol. 1997;158:5692–5697. [PubMed] [Google Scholar]
- 54.Ning BF, Ding J, Liu J, Yin C, Xu WP, Cong WM, et al. Hepatocyte nuclear factor 4alpha-nuclear factor-kappaB feedback circuit modulates liver cancer progression. Hepatology. 2014 doi: 10.1002/hep.27177. [DOI] [PubMed] [Google Scholar]
- 55.Sun L, Mathews LA, Cabarcas SM, Zhang X, Yang A, Zhang Y, et al. Epigenetic regulation of SOX9 by the NF-kappaB signaling pathway in pancreatic cancer stem cells. Stem Cells. 2013;31:1454–1466. doi: 10.1002/stem.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y, et al. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. Journal of hepatology. 2014;60:127–134. doi: 10.1016/j.jhep.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 57.Chang CJ, Hung MC. The role of EZH2 in tumour progression. British journal of cancer. 2012;106:243–247. doi: 10.1038/bjc.2011.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 59.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 60.R Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2014. http://www.R-project.org/ [Google Scholar]
- 61.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 62.Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant methods. 2007;3:11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.