Abstract
Messenger ribonucleoprotein (mRNP) granules are dynamic, self-assembling structures that harbor non-translating mRNAs bound by various proteins that regulate mRNA translation, localization, and turnover. Their importance in gene expression regulation is far reaching, ranging from precise spatial-temporal control of mRNAs that drive developmental programs in oocytes and embryos, to similarly exquisite control of mRNAs in neurons that underpin synaptic plasticity, and thus, memory formation. Analysis of mRNP granules in their various contexts has revealed common themes of assembly, disassembly, and modes of mRNA regulation, yet new studies continue to reveal unexpected and important findings, such as links between aberrant mRNP granule assembly and neurodegenerative disease. Continued study of these enigmatic structures thus promises fascinating new insights into cellular function, and may also suggest novel therapeutic strategies in various disease states.
Keywords: stress granules, P-bodies, neuronal transport granules, germ granules, mRNA, translation, repression, decay, neurodegenerative disease
Abbreviations
- mRNP
messenger ribonucleoprotein
- IMC
inter-mitochondrial cement
- HSP
heat shock protein
- 4E-BP
eIF4E binding protein
- RISC
RNA induced silencing complex
- FRAP
fluorescence recovery after photobleaching
- MSP
multi-system proteinopathy
- ALS
amyotropic lateral sclerosis
- FTLD
frontotemporal lobar degeneration
Introduction
mRNA regulation is a crucial means by which cells control gene expression, as it enables both rapid and local changes in synthesis of specific proteins to occur. Three key aspects of mRNA regulation include control of mRNA localization, mRNA translation, and mRNA stability, all of which typically depend on specific regulatory proteins binding to an mRNA. In eukaryotes, non-translating mRNPs often assemble together into visible cytoplasmic structures that lack a limiting membrane, termed mRNP granules. Specific examples of these include P-bodies,1,2 stress granules,2,3 germ granules,4 and neuronal transport granules5 (Fig. 1). Recent work has uncovered much about how mRNAs are regulated within these structures, as well as how granules assemble, disassemble, and transit through cells. Importantly, mRNP granules are also strongly implicated in a variety of diseases, especially degenerative disorders, thus it is an exciting time for the field.
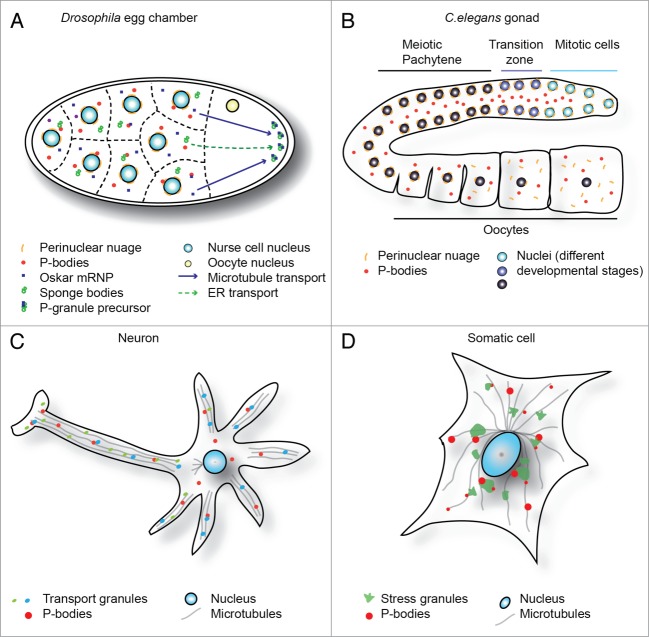
Figure 1.
mRNP granules across biology. (A) Graphic of Drosophila egg chamber indicates interaction of multiple mRNP granule types in the cytoplasm of nurse cells, and modes of transport to the germ plasm. (B) Gonad of a C.elegans hermaphrodite indicates “assembly line” maturation, from mitotic stem cells at distal end (top right) to oocytes at proximal end (bottom right). P-granules remain docked with nucleus until diplotene, then re-localize to the cytoplasm with nuclear pore components. P-bodies are distributed throughout the shared cytoplasm. (Panels A and B adapted from Voronina et al., 2011 and reproduced with permission). (C) Simplified neuronal cell schematic demonstrating specific localization of transport granules in dendrites and the axon. Interactions with P-bodies and transport along microtubules are highlighted. (D) Stressed somatic cell, indicating typical distribution of stress granules (often peri-nuclear) and P-bodies, which often dock with stress granules. Interactions of both granules with microtubules also highlighted.
Diversity and Similarity Among Different Mrnp Granule Types
Specific classification of mRNP granules depends upon their cellular context, presumed function, and the presence of particular protein markers. For instance, germ granules (subtypes of which include P-granules, nuage, chromatoid bodies, inter-mitochondrial cement, and sponge bodies) are usually defined as cytoplasmic mRNP foci present in germ cells, which often contain the RNA helicase Vasa,4 and are implicated in storage and localization of mRNAs. The developmental stage, cellular localization, composition, and model organism studied further dictates their sub-classification.4 In neuronal cells, transport granules are defined as cytoplasmic mRNP foci that transit along microtubules in axons and dendrites, with presumed functions in mRNA localization, particularly in growth cones and at synapses. Subtypes are mainly defined by the presence of RNA binding proteins such as Staufen. Despite the absence of translating mRNPs, many neuronal granules also harbor ribosomes.5 P-bodies, which are generally seen in all cell types, are defined by enrichment for mRNA decay proteins,1 whereas stress granules normally only form during cellular stress, and contain numerous initiation factors, including small ribosomal subunits.3 Both types are thought to contribute to regulation of translation, whereas P-bodies are also thought to function in mRNA decay.
Despite this diversity, all mRNP granules have features in common. First, they all contain repressed mRNAs that are capable of (re)-entering translation in response to appropriate signals.6-9 Second, they share many RNA binding proteins and mRNA species in common,10 and indeed factors concentrated in one granule often re-localize to another granule type with time or changes in cellular conditions.11-13 Third, mRNP granules exhibit dynamic interactions with one another such as docking, fusion, or apparent maturation from one granule type to the next. Examples include P-body-stress granule docking, fusion11,14 and apparent maturation,11,12 P-body-neuronal transport granule docking,15 P-granule-P-body docking16 and nuage-P-body fusion, and apparent maturation into sponge bodies.16,17 The simplest interpretation of such observations is that mRNPs are exchanged between different granules, though this has not been directly demonstrated,13 and remains an important unresolved issue.
mRNP Granules Assemble via Common Mechanisms
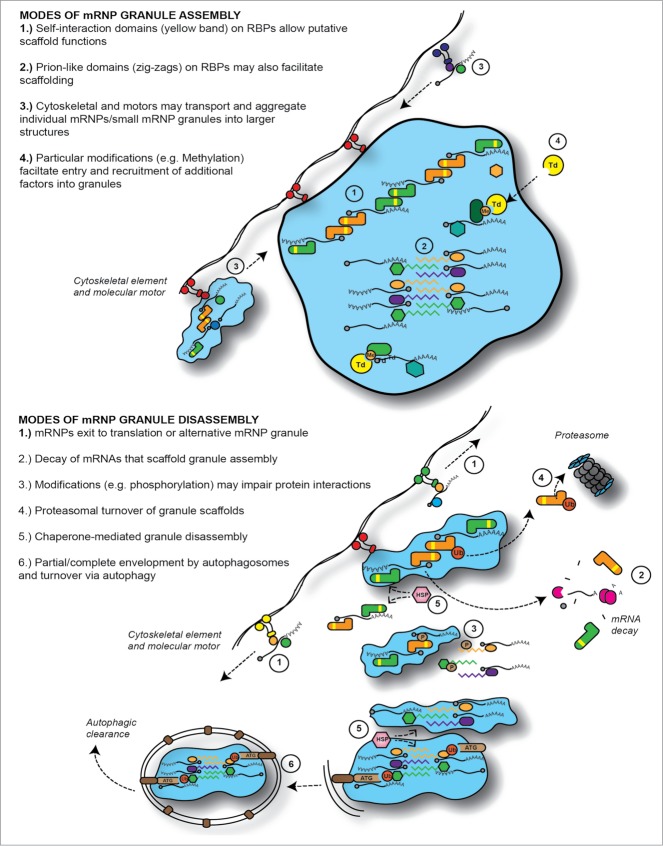
Studies of various mRNP granules types, particularly P-bodies and stress granules, have revealed common themes of assembly (Fig. 2). For example, non-translating mRNA is an essential component for assembly of all mRNP granules. Supporting this, P-bodies and stress granules cannot form in the presence of cycloheximide or emetine,19,20 which traps mRNAs in polysomes. In addition, perinuclear P-granules in C. elegans disassemble when transcription or mRNA export is inhibited.21 Semi-purified preparations of P-bodies and neuronal transport granules are also disassembled upon RNase treatment.21,22 Conversely, increasing the pool of non-translating mRNAs stimulates stress granule and P-body assembly, as shown by inhibition of translation initiation or mRNA decay,20,22,24 expression of decay resistant mRNA,22 or drugs that promote ribosome–mRNA dissociation.11,24
Figure 2.
Modes of mRNP granule assembly and disassembly. Circled numbers refer to text in figures describing putative assembly/disassembly mechanisms of mRNP granules. Note, the use and relative importance of these likely vary depending on granule type and context. Abbreviations: RBP, RNA binding protein; Td, tudor domain protein; Me, methylation; Ub, ubiquitination; P, phosphorylation; HSP, heat shock protein; ATG, autophagy factor.
Another common assembly mechanism relies on granule proteins that harbor self-interaction domains, and thus, may potentially act as scaffolds. Clear examples of deletion of self-interaction domains impairing granule assembly include the P-granule component PGL-3,26 the neuronal transport granule factor FMRP,27 the P-body factor Edc3,28 and the stress granule factor G3BP.29 Several other granule components that contain self-interaction motifs, including Staufen30 and Pat1,31 also facilitate granule assembly. Consistent with a scaffold function, many of these factors also exhibit multiple direct protein interactions with other granule components.28,31,32
Prion-like or low-complexity domains are often found in proteins affecting granule assembly. These domains are unusually common in proteins involved in RNA metabolism, and examples of proteins whose prion-like domain contributes to granule assembly include TIA-1/Pub1 in stress granules,12,33 and Lsm4 in yeast P-bodies.28,34 Many more components of various mRNP granules have prion-like domains, including TIAR/Ngr1,12,24 TDP-43,35 FUS36 hnRNPA2B1/A1,37 Dhh1, Ccr4, and Pop2.34 Recently, in vitro studies demonstrated that at sufficient concentrations, proteins containing prion-like domains such as FUS can assemble “hydrogel” structures, capable of interacting with other prion-like domains from a wide range of known mRNP granule proteins.38 Taken together, prion-like domains may therefore be important in localization of factors to mRNP granules, as well as granule assembly. An unresolved issue is how prion interactions in mRNP granules remain apparently dynamic, reversible, and heterotypic, in contrast to the stable, homotypic prion-interactions that underpin amyloid formation in several neurodegenerative diseases (see below).
The cytoskeleton and associated motor proteins also contribute to the assembly and disassembly of mRNP granules, although effects are specific to different conditions and different granule types. For instance, microtubule depolymerizing drugs prevent assembly of large stress granules39,40 and germ granules in early zebrafish embryos,41 whereas P-bodies become larger, and less mobile.42,43 Specific dynein and kinesin motor proteins also localize in stress granules, and appear to facilitate assembly and disassembly of stress granules, respectively.40 Dynein proteins also increase P-body assembly under stress.40 Contrasting results have been observed upon stress granule assembly following disruption of actin,40,44 while P-body disassembly in yeast is slowed by mutation of a myosin type V protein, Myo2.45 It is additionally very clear that localization of various germ and transport granules makes use of cytoskeletal elements for translocation and anchoring at specific sites; this subject has been well covered elsewhere.46,47
Protein modifications are also important in granule assembly, and recruitment of proteins to granules. For example, components of germ granules, such as Piwi-family argonautes, harbor methylated arginines that help recruit tudor domain-containing proteins. Interfering with this interaction can impair both localization of tudor-domain proteins, and the methylated proteins themselves,48,49 as well as germ granule assembly.50 Such interactions also underpin recruitment of tudor-domain proteins to stress granules.52,53 O-linked N-acetyl glucosamine,53 poly(ADP) ribosylation,54 acetylation, and ubiqutination39 are also modifications present on specific stress granule components, which have been implicated in stress granule assembly, albeit the mechanisms are not entirely clear. Finally, the phosphorylation of several proteins, including G3BP,29 TTP,55 Dcp1,42,56 Dcp2,57 and 4E-T58 alters their localization within, and/or the assembly of their respective granules. In addition, phosphorylation in the prion-like domain of FUS impairs hydrogel formation in vitro.59 Indeed, modifications have equal potential to promote disassembly, or prevent factor localization to mRNP granules29,55 (Fig. 2). Modification of key mRNP granule components thus offers an appealing means to explain the dynamic assembly and disassembly of granules in response to cellular stress, developmental, or synaptic signals.
Sporadic cases of assembly mechanisms described for other RNP granules warrant more widespread investigation. For example, a long non-coding RNA termed NEAT1 underpins the assembly of nuclear paraspeckles, which are RNP granules of unknown function.60 Interestingly, this lncRNA also binds and co-localizes in paraspeckles with TDP-43 and FUS, which are known stress granule and neuronal transport granule components. In principle, lncRNAs could help scaffold assembly of cytoplasmic mRNP granules just as mRNAs do, though examples of this are currently lacking.
Finally, intermitochondrial cement (IMC) in mouse spermatocytes may utilize organelle/molecular seeding to drive its assembly. This depends on the presence of phosphatidic acid on the surface of mitochondria. Depletion of phosphatidic acid impairs IMC formation, whereas increases in phospharidic acid drive IMC hyper-aggregation around mitochondria. Interaction of IMC components with phosphatidic acid was suggested as a means to concentrate and nucleate granule formation.61,62 It will be interesting to determine if similar mechanisms are used by other mRNP granules.
Principles of mRNP Granule Disassembly
mRNP granule disassembly (Fig. 2) is thought to most commonly occur by entry of the granule mRNAs into translation. Translation typically shows an inverse relationship with stress granule and P-body numbers.19,22 For instance, during cellular stress, bulk translation is inhibited, leading to increased granules, whereas during stress recovery, translation levels increase as granule numbers fall. This later phenomenon also holds true for specific mRNAs, which exit stress granules and enter translation during the recovery phase.62,63 Dendritic P-bodies also disassemble upon synaptic activation,15 which may reflect entry of mRNAs into translation.
mRNP granules could also be disassembled via mRNA decay, which is most likely relevant to P-bodies, given their enrichment for the decay machinery, the detection of mRNA decay intermediates within, and a correlative increase in P-body numbers when hard to degrade substrates are present.20 However, failure to degrade mRNAs due to blocks in mRNA decay also causes increases in stress granule numbers, at least in yeast,12 which may be due to exchange of mRNPs between the two granule subtypes. Thus, mRNA decay blocks could also affect germ granules and neuronal transport granules, with which P-bodies also physically interact.14,15 Notably, various mRNA degradative enzymes are found in mRNP granules besides P-bodies, such as Xrn1 in stress granules11 and Drosophila/mouse spermatid nuage,65 Dcp1/Dcp2 in various germ granules,66-68 and several Argonaute proteins with endonuclease activity in stress granules,69 and various germ granules.70-73 Whether mRNA degradation occurs in mRNP granules besides P-bodies remains poorly studied.
Turnover of protein components, particularly scaffolding factors, could also be an efficient way to disassemble mRNP granules. Indeed, inhibition of the Ubiquitin Proteasome system induces stress granules, although this appears to be caused by activation of a GCN2-mediated stress response rather than failure to degrade stress granule assembly factors.74 In contrast, proteasomal turnover of the helicase GLH-1 in C. elegans P-granules appears to be necessary to prevent excessive P-granule assembly that leads to sterility.75 Factors associated with proteasomal function were also identified in screens for factors affecting P-granule,76 P-body, and stress granule assembly.77
Chaperone proteins also affect disassembly of stress granules. Various heat shock proteins (HSPs) localize to, and affect, disassembly of stress granules in humans, flies, and yeast.24,33,74,78 An appealing model78 is that when cellular stress leads to accumulation of unfolded proteins, titration of HSPs would cause slower stress granule disassembly and trapping of mRNAs in non-translating states, thus helping cells conserve resources. As HSPs become available again either via increased synthesis, or from having dealt with other unfolded proteins, stress granules could once more be disassembled. However, chaperones are also implicated in P-body and stress granule assembly in inhibitor studies,79 and not all stresses that induce P-bodies and stress granules likely lead to significant levels of protein unfolding. Therefore, the role of chaperones in mRNP granule disassembly (or assembly) is probably complex. Whether HSPs regulate germ granules or neuronal transport granules is currently unclear.
Finally, two studies have recently shown that mRNP granules can be cleared by autophagy. The first demonstrated that in C. elegans autophagy mutants, P-granule components erroneously formed foci in somatic cells.80 A selective autophagy mechanism, depending on interaction of P-granule factors with the protein SEPA-1, was identified that helps clear formation of these aberrant foci.81 Similarly, stress granules can accumulate in yeast and mammalian cell autophagy mutants, and are targeted by autophagy under various growth and stress conditions.77 Defining the mechanism of such targeting, how mRNPs are affected, and whether autophagy contributes to the clearance of other mRNP granule types remain key issues to be addressed.
Functions of mRNP Granules
Given the conservation and utilization of mRNP granules in so many biological contexts, it is hard to imagine their assembly and form is without functional relevance. Nonetheless, identifying a role for mRNP granule assembly is not always straightforward, given the need to separate effects upon granule assembly per se from those arising from loss of a protein-specific function that directly regulates mRNAs. For instance, TIA-1 is a stress granule assembly factor,33 but also functions in splicing regulation82 and translation repression,83 thus asserting that changes in mRNA translation, decay, or localization are specifically due to changes in granule assembly must be carefully scrutinized and controlled for.
With this caveat in mind, there are two fundamental reasons why granule assembly may be beneficial. First, by virtue of a higher local concentration of proteins and mRNAs, certain processes may be increased in their efficiency. To illustrate this principle, formation of Cajal bodies, which are nuclear assembly sites for small nuclear ribonuclear protein particles (snRNPs), are thought to increase snRNP assembly rates by about 10-fold.84 Second, sequestration of proteins or mRNAs within granules may facilitate separation of processes that could interfere with one another (e.g., translation and translation repression/decay), or alter regulation of a particular process outside of the granule. Intriguing examples of the latter, unrelated to direct regulation of mRNPs, concerns localization of signaling factors in stress granules. Specifically, localization of factors such as RACK1,85 TRAF2,86 and TORC187,88 in stress granules affects signaling outcomes in response to cellular stress or extracellular signaling. However, since mRNPs form the primary component of these granules, most described or hypothesized roles have focused on possible effects on mRNA, examples of which are discussed below.
Role of mRNP Granules in mRNA Localization
Localization of mRNAs often occurs in mRNP granules. Oocytes, embryos, and neurons have proven excellent systems for study, given their highly polarized nature, although important insights have also been gained in yeast through study of the Ash1 “locasome,”89 an mRNP granule in which Ash1 is transported to the bud tip of daughter cells during cell division. The importance of mRNA localization has been illustrated in many contexts, and a striking example of its pervasiveness is the fact that 71% of 3370 mRNAs whose localization was studied in Drosophila embryos showed distinct localization patterns, often mirrored by the localization of their encoded proteins.90
A recurring theme of mRNA localization is that mRNAs, usually by virtue of several cis sequences or “zipcodes,” are bound by proteins that package the mRNA into translationally repressed mRNPs, often prior to nuclear export. Next, repressed mRNPs are typically localized to and anchored at particular cellular location via interactions with molecular motors and cytoskeletal elements, although sponge bodies are thought to move along ER membranes.18 A signaling event often precedes the switch of localized mRNAs from repression to translation, followed by granule disassembly.9,15,91 This topic has been well covered elsewhere,92 thus the focus here will be on how granule formation may contribute to localization.
First, it is important to note that a broad range exists in terms of how many mRNPs are actually packaged and transported in a given granule. Some of the earliest attempts at purifying nuclear transport granules identified large complexes (>1000S), with presumably multiple mRNAs.23 Staining with in situ probes against common leader sequence or poly-adenylated mRNAs also suggest a large population of mRNAs reside in germ granules,92,93 as well as P-bodies and stress granules.24,25 Approaches using specific mRNA labeling also shows co-migration of several mRNA species in neuronal granule subtypes,95 and the Ash1 locasome in yeast,96,97 and many mRNA species individually localize in stress granules or P-bodies under identical conditions.98,99 In contrast, recent quantitative imaging approaches in neurons indicates that some mRNA species can transit in granules or “particles” harboring just a single mRNA molecule.100-102 Why such variation in mRNP content occurs is unclear, but presumably reflects a differential requirement for the specificity of localizing individual mRNPs to particular cellular compartments under various conditions.
Localizing multiple mRNAs together in a single granule could be beneficial in allowing coordinated regulation of multiple mRNAs with related function. Supporting this, several mRNAs harboring identical cis regulatory elements can co-localize in particular transport granules.95,103 This might allow bursts of synthesis of several proteins with related function to locally respond to rapid signals that occurs at neuronal synapses. Grouping mRNPs in a single particle might also be more energy efficient in that fewer interactions with factors mediating transport (e.g., motor proteins) may be necessary, and maintenance of mRNPs in a repressed state might be facilitated by a higher local concentration of repressor proteins. However, neither of these ideas has been rigorously examined. In contrast, an obvious advantage of localizing mRNPs individually is greater specificity of control of mRNA function.
In principle, a hybrid of these approaches may be utilized, where mRNPs remodel between shared granules or individual mRNP particles, dependent upon the nature of the mRNP, cellular conditions or the localization status of the mRNP. Indeed, a recent study in live mouse neurons shows evidence of Beta-actin mRNPs being transported in granules harboring progressively fewer individual mRNPs, as distance from the soma increased.101 mRNP density in transport granules also decreased in this101 and another study following neuronal stimulation.100 Such factors might explain the large diversity in data observed in neurons, where it is also possible that neuronal granule mRNPs are exchanged with P-bodies under various conditions.15 Continued analysis at the single molecule level, ideally with multiple mRNPs in parallel, may help better understand this issue.
A key principle put forth for mRNP granules that have a primary function in localization (e.g., germ granules, neuronal transport granules) is that their mRNAs remain stable and non-translated until correctly localized. However, ribosomes are a component of many neuronal transport, and decay factors are also often found in neuronal and germ granules,104 and sometimes are even required for their localization.67 Thus, one assumes that the mRNP state within these granules is carefully regulated. Why transport factors involved in processes that are normally suppressed during localization? Regarding translation factors and ribosomes, one possibility is that their co-transport may aid rapid entry into translation upon arrival at their correct destination and following appropriate signals. As for decay factors, their presence may ensure that incorrectly localized mRNAs, which could be deleterious, are rapidly degraded as part of a recently theorized mis-localization quality control process.105
Role of mRNP Granules in Translational Control
Translation of mRNAs is highly regulated, with many mechanisms described that affect the ability of ribosomes to bind mRNAs, assemble productive initiation complexes,106 elongate through open reading frames and recycle efficiently.106,107 Broad translational control mechanisms that likely act upon most mRNAs include phosphorylation of the initiation factor eIF2α by various stress-responsive kinases,109 which limits levels of ternary complex, and thus, translation initiation rates. This event facilitates assembly of stress granules under most (but not all) circumstances.3 Another broad control mechanism is sequestration of the mRNA cap binding protein eIF4E by 4E-binding proteins (4E-BPs), a process largely regulated by the TOR signaling pathway, which integrates many growth factor and nutrient signals in order to regulate general protein synthesis rate.106 4E-BPs localize in P-bodies and stress granules, as does TORC1 itself (the main TOR kinase complex).87,88
More specific regulation of mRNAs usually involves proteins that bind cis elements in particular mRNAs, which then directly, or via recruitment of additional factors, disrupt the formation of productive initiation complexes. A common mechanism is impaired assembly of the eIF4E-eIF4G cap-binding complex, which facilitates recruitment of small ribosomal subunits to mRNAs.106 Another target of translational control is poly(A) tail length. mRNAs containing short poly(A) tails are translational impaired, which may reflect an inability to bind poly(A) binding protein, which facilitates eIF4G recruitment.110 Cytoplasmic Polyadenylation Element Binding Protein (CPEB) is a protein which can target both these steps, and which localizes in P-bodies, stress granules,111 germ granules,112 and neuronal transport granules.5 Specifically, CPEB binds to mRNA 3′UTRs harboring Cytoplasmic Polyadenylation Element (CPE) cis sequences and recruits Maskin, which binds and sequesters eIF4E. CPEB also recruits deadenylase enzymes to maintain short poly(A) tails. Further details of this mechanism, and the means by which CPEB also governs translational activation in response to appropriate signaling events has been excellently described elsewhere.113
Another prevalent and emerging mode of translational control is that enacted by miRNAs. These short oligonucleotides, in association with Ago proteins, form imperfect duplexes with target mRNAs, and recruit additional factors such as GW182 to form an RNA-induced silencing complex (RISC). While controversial as to which mechanism predominates, studies suggest that RISC can inhibit translation initiation, elongation, and promote deadenylation, leading to mRNA turnover.114 Importantly, RISC components also localize in P-bodies, stress granules, germ granules, and neuronal transport granules.115 It is likely therefore that multiple types of translationally repressed mRNPs can reside simultaneously in mRNP granules.
How might mRNP granule formation facilitate translation repression? One idea is that they may sterically restrict ribosome access; indeed the oligomerization of oskar mRNAs into high molecular-weight complexes by the translational repressor Bruno is thought to partially function in this manner.116 This may apply in some cases, but cannot explain why neuronal transport granule mRNAs appear silenced, as they often contain ribosomes. A second idea is that concentration of mRNAs and translational repressors might facilitate efficient assembly or maintenance of a translationally repressed mRNP state. However, there is clear data that suggests visible granule assembly is not always important for translational repression. For example, stress granule or P-body assembly is not required to inhibit translation globally during stress responses.12,38,39,118 Similar studies with germ granule or neuronal transport granules are currently lacking as assembly mutants that don't perturb mRNP interactions with the localization machinery, or affect translational repression directly, have not been well characterized. It thus remains unclear as to whether granule assembly is important for repression of only specific mRNAs, or whether particular granule types may function differentially with regards to translation repression.
The opposite notion, that some mRNP granules could promote translation, has also been proposed.4,10 Just as concentration of translational repressor proteins could in principle facilitate repression, concentration of translational components, as occurs in stress granules and neuronal transport granules, might facilitate assembly of productive translation complexes, such that mRNAs rapidly enter translation in response to appropriate cues. Supporting this, polysomes can be seen via electron microscopy on the surface of polar granules in Drosophila germ plasm at particular developmental stages,118 and interestingly, near the surface of P-bodies.119 Additionally, overexpression or mutation of FUS in cell lines leads to the formation of foci resembling stress granules, which surprisingly exhibit active protein synthesis within them.120 Such observations suggest that mRNP granules may both negatively and positively regulate translation depending on context. Therefore, careful analysis of the translational state of an mRNP granule should be verified, especially under diverse genetic or environmental conditions.
Role of mRNP Granules in mRNA Stability
Regulation of mRNA stability is a third role suggested for mRNP granules. Germ granules probably represent the clearest evidence of long-term storage or stabilization of mRNAs. For example, some mRNAs required for spermiogenesis are stored for several days in chromatoid bodies, until exiting and entering translation at appropriate developmental timepoints.121,122 Similarly, a subtype of germ granule referred to as “storage bodies” in the C. elegans gonad rely on the presence of CGH-1 (homolog of the P-body factor Dhh1/RCK) for stabilization of several maternal mRNAs.123 Stress granules and P-bodies may also harbor stable mRNAs, given that they contain mRNA stabilizing proteins such as HuR and Pab1. In addition, during stress, which induces P-body and stress granule numbers, many mRNAs are stabilized, in part by inhibition of deadenylation.125,126 Furthermore, repressed mRNAs can localize in P-bodies for short periods of time, and return to translation upon appropriate cues or following exit from stress,5,6 suggesting a transient storage function.
Despite this, studies utilizing fluorescence recovery after photobleaching (FRAP) indicate that most mRNP granule components shuttle in and out of mRNP granules, often quite rapidly. This is particularly true of most P-body and stress granule factors10 (with exceptions e.g., Dcp2 in P-bodies). Germ granule components (e.g., Vasa in Drosophila nuage; residency time of ≈60s;126 PGL-1 in C. elegans perinuclear P-granules ≈20s21), and neuronal granule components (e.g., Staufen in Drosophila neuronal granules; residency > 10 min;104 TDP-43 in Drosophila neuronal granules ≈ 10s127) also exhibit a wide range of shuttling rates. More telling are FRAP studies on fluorescently labeled mRNAs, which have indicated that both rapidly exchanging and immobile fractions of a single mRNA species can exist within stress granules simultaneously.12,128,129 Therefore, stress granules, and perhaps other granules, may indeed store a subpopulation of mRNAs, although at any moment, much of that mRNA species may switch to shuttling behavior, which could indicate a return to translation, or targeting to other mRNP granules. This is consistent with the previously proposed idea of stress granules as sites of mRNA triage, rather than just solely mRNA storage sites.3
In contrast to storage, mRNP granules may also promote decay, an idea that has only been seriously examined with P-bodies. Evidence that P-bodies can act as site of mRNA decay include the presence of all components of the 5′-3′ mRNA decay pathway, the detection of mRNA decay intermediates within P-body foci, and increases in P-body numbers when mRNA decay is inhibited at or following the mRNA decapping stage.1,20 In addition, in C. elegans embryos, mature P-body assembly within somatic blastomeres correlates spatially and temporally with the degradation of maternal mRNAs.16
However, studies in which assembly of P-bodies or stress granules is impaired typically reveal no significant change in the stability of the mRNAs examined.12,28,34,130 It is therefore likely that mRNA decay can proceed on individual mRNPs outside of granules. However, the studies referred to above examined only a handful of mRNAs, thus it remains possible that specific mRNAs are reliant on assembly into P-bodies, or related structures, for efficient decay. Alternatively, granule assembly may facilitate mRNA decay only under certain conditions. Indeed, the yeast P-body assembly factor Edc328 was identified as a decapping enhancer in strains where decapping activity was compromised. This suggests that examining decay under conditions when decapping is limiting, which can arise for many reasons,131 may reveal a clearer insight into the role of P-bodies in mRNA stability. The possibility of mRNA decay in other mRNP granule subtypes remains possible, but currently under investigated.
mRNP Granules and Disease
Given the wide-ranging effects mRNP granules may have on mRNA function and cell signaling, it is no surprise that mRNP granules are implicated in many diseases. For example, stress granules, P-bodies, and their components often affect, or are hijacked by RNA viruses during infection and replication (for review, see ref. 132). Stress granules are also upregulated in some cancers,133 and may facilitate cancer cell survival.88,134 Impaired localization of neuronal transport granules and translational control of their mRNPs are some of the mechanisms proposed to explain conditions such as spinal muscular atrophy and Fragile X syndrome (for review, see ref. 135). An area of intense recent focus however, is the emerging connection between mRNP granules and degenerative disease, which is now highlighted.
Toxic Stress Granules as a Cause of Degenerative Disease?
Recent work suggests that aberrant formation or persistence of stress granules may underpin a variety of (neuro) degenerative diseases. These include Multi-System Proteinopathy (MSP), Pagets disease, Amyotropic Lateral Sclerosis (ALS), and Frontotemporal Lobar Degeneration (FTLD). Several observations support this hypothesis. First, cells afflicted in these disease states (e.g., motor neurons in ALS) exhibit cytoplasmic foci or “inclusion bodies,” which compositionally resemble stress granules. Second, many mutated proteins associated with degenerative disease are RNA binding proteins (e.g., TDP-43, FUS, SMN1, hnRNPA1, hnRNPA2, Ataxin-2, TIA-1) that localize in stress granules,137,138 and other mRNP granule types. Third, mutated forms of these proteins often exhibit hyper-aggregative behavior in vitro and drive aberrant stress granule assembly in vivo.37,138 Fourth, in various model systems, toxicity of exogenously expressed disease proteins is suppressed by deletion or knockdown of factors that promote stress granule assembly.139-141 Finally, a second class of mutated proteins associated with degenerative disease are autophagy-promoting factors (VCP, optineurin, p62, ubiquilin-2142), which is notable given that stress granules are cleared via autophagy,77 and cellular autophagy dysfunction is commonly observed in degenerative disease.
This data has led to a working model136,143,144 in which aberrant formation of stress granules through granule-promoting factors (e.g., hyper-aggregating RNA-binding proteins) or failure to clear such aggregates via autophagy, leads to the persistent formation of stress granules in cells, which for some reason cannot be disassembled via other means, such as returning mRNAs to translation. One idea proposed to explain this is that the abundance of prion-like domains in stress granule components, coupled with prolonged concentration of such proteins together, eventually leads to the formation of hyper-stable amyloid-like structures that may result in stress granule being refractory to remodeling and disassembly, leaving clearance via autophagy as a last resort.136 Supporting these ideas, ALS pathology is suppressed in model systems and patients where autophagy is upregulated,145,146 or when stress granule assembly is impaired.147 In addition, pathogenic forms of TDP-43 expressed in C. elegans or Drosophila show slower exchange rates from aggregates or neuronal granules by FRAP.127,148
Several reasons have been proposed as to why persistent stress granules or related mRNP aggregates could be toxic to cells.136,144 Sequestration of protein factors in granules could lead to a reduced ability of granules to appropriately remodel mRNPs, and may limit the available pool of a protein that also functions elsewhere in the cell. For instance, many stress granule proteins shuttle between the nucleus and cytoplasm, and regulate processes such as splicing and export. mRNAs, and other associated mRNP factors, including miRNAs, may also be trapped in such granules, and thus, mis-regulated. Related to this, it is noteworthy that the RNA binding activity of mutant forms of TDP-43 and FUS are typically required for toxicity in model systems, even when their ability to induce aggregation is not strongly affected.144 Specific functions of mRNP granules themselves could also be impaired. For example, disease alleles of TDP-43 impair transport along axons of particular neuronal transport granules.149 Finally, aberrant sequestration of signaling factors in stress granules, which can alter growth and apoptotic decisions in several cases85-88 could also be critical to disease progression. Determining the causes of cellular toxicity, and why cell types such as neurons appear especially susceptible to mutations that affect mRNP granules, are outstanding questions that now lie before the field.
Future Challenges Ahead
Several key questions regarding mRNP granules remain to be addressed. First, what is the complete RNA and protein composition of mRNP granules in their various contexts? Such knowledge would lead to insight as to both the mechanism and specificity of regulation of mRNAs within granules, as well as illuminate links to other areas of cell biology (e.g., signaling, autophagy). Second, what are the mRNP remodeling events that govern entry and exit of mRNAs to, from, and between mRNP granules? This could shed light on a number of gene expression control mechanisms at the mRNA level. Third, how do granules assemble and disassemble, and how are these processes affected in disease? Although much has been learned here, new mechanisms continue to arise, understanding of which could reveal novel therapeutic targets in diseases involving aberrant granule behavior. Finally, can clear direct functions be assigned to the aggregation of various mRNP granules? Identifying assembly defects that do not impinge directly on other mRNP regulatory functions, coupled with the realization that mRNA regulatory processes are often interdependent, mean this will be a challenging goal. Nonetheless, new ideas coupled with new technologies promise answers and exciting new questions in the years ahead.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I thank Daniela Zarnescu and Allison Buchanan for critical feedback. I apologize for not being able to cite all relevant works due to space constraints.
References
- 1. Jain S, Parker R. The discovery and analysis of P Bodies. Adv Exp Med Biol 2013; 768:23-43; PMID:23224963; http://dx.doi.org/ 10.1007/978-1-4614-5107-5_3 [DOI] [PubMed] [Google Scholar]
- 2. Erickson SL, Lykke-Andersen J. Cytoplasmic mRNP granules at a glance. J Cell Sci 2011; 124:293-7; PMID:21242308; http://dx.doi.org/ 10.1242/jcs.072140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci 2008; 33:141-50; PMID:18291657; .http://dx.doi.org/ 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- 4. Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. RNA granules in germ cells. Cold Spring Harb Perspect Biol 2011; 3:3; PMID:21768607; http://dx.doi.org/ 10.1101/cshperspect.a002774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron 2006; 51:685-90; PMID:16982415; http://dx.doi.org/ 10.1016/j.neuron.2006.08.021 [DOI] [PubMed] [Google Scholar]
- 6. Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 2005; 310:486-9; PMID:16141371; http://dx.doi.org/ 10.1126/science.1115791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol 2006; 71:513-21; PMID:17381334; http://dx.doi.org/ 10.1101/sqb.2006.71.038 [DOI] [PubMed] [Google Scholar]
- 8. Nagamori I, Cruickshank VA, Sassone-Corsi P. Regulation of an RNA granule during spermatogenesis: acetylation of MVH in the chromatoid body of germ cells. J Cell Sci 2011; 124:4346-55; PMID:22223882; http://dx.doi.org/ 10.1242/jcs.096461 [DOI] [PubMed] [Google Scholar]
- 9. Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 2005; 438:512-5; PMID:16306994; http://dx.doi.org/ 10.1038/nature04115 [DOI] [PubMed] [Google Scholar]
- 10. Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009; 36:932-41; PMID:20064460; http://dx.doi.org/ 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 2005; 169:871-84; PMID:15967811; http://dx.doi.org/ 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 2008; 183:441-55; PMID:18981231; http://dx.doi.org/ 10.1083/jcb.200807043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol Biol Cell 2008; 19:4469-79; PMID:18632980; http://dx.doi.org/ 10.1091/mbc.E08-05-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoyle NP, Castelli LM, Campbell SG, Holmes LEA, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol 2007; 179:65-74; PMID:17908917; http://dx.doi.org/ 10.1083/jcb.200707010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zeitelhofer M, Karra D, Macchi P, Tolino M, Thomas S, Schwarz M, Kiebler M, Dahm R. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci 2008; 28:7555-62; PMID:18650333; http://dx.doi.org/ 10.1523/JNEUROSCI.0104-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev Biol 2008; 323:76-87; PMID:18692039; http://dx.doi.org/ 10.1016/j.ydbio.2008.07.008 [DOI] [PubMed] [Google Scholar]
- 17. Wilsch-Bräuninger M, Schwarz H, Nüsslein-Volhard C. A sponge-like structure involved in the association and transport of maternal products during Drosophila oogenesis. J Cell Biol 1997; 139:817-29; PMID:9348297; http://dx.doi.org/ 10.1083/jcb.139.3.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jaglarz MK, Kloc M, Jankowska W, Szymanska B, Bilinski SM. Nuage morphogenesis becomes more complex: two translocation pathways and two forms of nuage coexist in Drosophila germline syncytia. Cell Tissue Res 2011; 344:169-81; PMID:21365220; http://dx.doi.org/ 10.1007/s00441-011-1145-2 [DOI] [PubMed] [Google Scholar]
- 19. Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 2000; 151:1257-68; PMID:11121440; http://dx.doi.org/ 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 2003; 300:805-8; PMID:12730603; http://dx.doi.org/ 10.1126/science.1082320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sheth U, Pitt J, Dennis S, Priess JR. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 2010; 137:1305-14; PMID:20223759; http://dx.doi.org/ 10.1242/dev.044255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 2005; 11:371-82; PMID:15703442; http://dx.doi.org/ 10.1261/rna.7258505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 2004; 43:513-25; PMID:15312650; http://dx.doi.org/ 10.1016/j.neuron.2004.07.022 [DOI] [PubMed] [Google Scholar]
- 24. Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol 1999; 147:1431-42; PMID:10613902; http://dx.doi.org/ 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cougot N, Babajko S, Séraphin B. Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 2004; 165:31-40; PMID:15067023; http://dx.doi.org/ 10.1083/jcb.200309008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanazawa M, Yonetani M, Sugimoto A. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J Cell Biol 2011; 192:929-37; PMID:21402787; http://dx.doi.org/ 10.1083/jcb.201010106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gareau C, Martel D, Coudert L, Mellaoui S, Mazroui R. Characterization of Fragile X Mental Retardation Protein granules formation and dynamics in Drosophila. Biol Open 2013; 2:68-81; PMID:23336078; http://dx.doi.org/ 10.1242/bio.20123012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 2007; 179:437-49; PMID:17984320; http://dx.doi.org/ 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol 2003; 160:823-31; PMID:12642610; http://dx.doi.org/ 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Martel C, Dugré-Brisson S, Boulay K, Breton B, Lapointe G, Armando S, Trépanier V, Duchaîne T, Bouvier M, Desgroseillers L. Multimerization of Staufen1 in live cells. RNA 2010; 16:585-97; PMID:20075165; http://dx.doi.org/ 10.1261/rna.1664210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol 2010; 30:4308-23; PMID:20584987; http://dx.doi.org/ 10.1128/MCB.00429-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell 2010; 39:773-83; PMID:20832728; http://dx.doi.org/ 10.1016/j.molcel.2010.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 2004; 15:5383-98; PMID:15371533; http://dx.doi.org/ 10.1091/mbc.E04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reijns MAM, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci 2008; 121:2463-72; PMID:18611963; http://dx.doi.org/ 10.1242/jcs.024976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu-Yesucevitz L, Bilgutay A, Zhang Y-J, Vanderweyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One 2010; 5:e13250; PMID:20948999; http://dx.doi.org/ 10.1371/journal.pone.0013250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Jr., Sapp P, McKenna-Yasek D, Brown RH, Jr., Hayward LJ. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 2010; 19:4160-75; PMID:20699327; http://dx.doi.org/ 10.1093/hmg/ddq335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim HJ, Kim NC, Wang Y-D, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 2013; 495:467-73; PMID:23455423; http://dx.doi.org/ 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753-67; PMID:22579281; http://dx.doi.org/ 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev 2007; 21:3381-94; PMID:18079183; http://dx.doi.org/ 10.1101/gad.461107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loschi M, Leishman CC, Berardone N, Boccaccio GL. Dynein and kinesin regulate stress-granule and P-body dynamics. J Cell Sci 2009; 122:3973-82; PMID:19825938; http://dx.doi.org/ 10.1242/jcs.051383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Theusch EV, Brown KJ, Pelegri F. Separate pathways of RNA recruitment lead to the compartmentalization of the zebrafish germ plasm. Dev Biol 2006; 292:129-41; PMID:16457796; http://dx.doi.org/ 10.1016/j.ydbio.2005.12.045 [DOI] [PubMed] [Google Scholar]
- 42. Aizer A, Kafri P, Kalo A, Shav-Tal Y. The P body protein Dcp1a is hyper-phosphorylated during mitosis. PLoS One 2013; 8:e49783; PMID:23300942; http://dx.doi.org/ 10.1371/journal.pone.0049783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sweet TJ, Boyer B, Hu W, Baker KE, Coller J. Microtubule disruption stimulates P-body formation. RNA 2007; 13:493-502; PMID:17307817; http://dx.doi.org/ 10.1261/rna.355807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ivanov PA, Chudinova EM, Nadezhdina ES. Disruption of microtubules inhibits cytoplasmic ribonucleoprotein stress granule formation. Exp Cell Res 2003; 290:227-33; PMID:14567982; http://dx.doi.org/ 10.1016/S0014-4827(03)00290-8 [DOI] [PubMed] [Google Scholar]
- 45. Chang W, Zaarour RF, Reck-Peterson S, Rinn J, Singer RH, Snyder M, Novick P, Mooseker MS. Myo2p, a class V myosin in budding yeast, associates with a large ribonucleic acid-protein complex that contains mRNAs and subunits of the RNA-processing body. RNA 2008; 14:491-502; PMID:18218704; http://dx.doi.org/ 10.1261/rna.665008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal 2011; 23:324-34; PMID:20813183; http://dx.doi.org/ 10.1016/j.cellsig.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gaspar I. Microtubule-based motor-mediated mRNA localization in Drosophila oocytes and embryos. Biochem Soc Trans 2011; 39:1197-201; PMID:21936788; http://dx.doi.org/ 10.1042/BST0391197 [DOI] [PubMed] [Google Scholar]
- 48. Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 2010; 16:70-8; PMID:19926723; http://dx.doi.org/ 10.1261/rna.1869710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goulet I, Boisvenue S, Mokas S, Mazroui R, Côté J. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum Mol Genet 2008; 17:3055-74; PMID:18632687; http://dx.doi.org/ 10.1093/hmg/ddn203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis 2004; 40:164-70; PMID:15495201; http://dx.doi.org/ 10.1002/gene.20079 [DOI] [PubMed] [Google Scholar]
- 51. Linder B, Plöttner O, Kroiss M, Hartmann E, Laggerbauer B, Meister G, Keidel E, Fischer U. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum Mol Genet 2008; 17:3236-46; PMID:18664458; http://dx.doi.org/ 10.1093/hmg/ddn219 [DOI] [PubMed] [Google Scholar]
- 52. De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res 2007; 313:4130-44; PMID:17967451; http://dx.doi.org/ 10.1016/j.yexcr.2007.09.017 [DOI] [PubMed] [Google Scholar]
- 53. Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 2008; 10:1224-31; PMID:18794846; http://dx.doi.org/ 10.1038/ncb1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Leung AKL, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 2011; 42:489-99; PMID:21596313; http://dx.doi.org/ 10.1016/j.molcel.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WFC, Blackwell TK, Anderson P. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 2004; 23:1313-24; PMID:15014438; http://dx.doi.org/ 10.1038/sj.emboj.7600163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rzeczkowski K, Beuerlein K, Müller H, Dittrich-Breiholz O, Schneider H, Kettner-Buhrow D, Holtmann H, Kracht M. c-Jun N-terminal kinase phosphorylates DCP1a to control formation of P bodies. J Cell Biol 2011; 194:581-96; PMID:21859862; http://dx.doi.org/ 10.1083/jcb.201006089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yoon J-H, Choi E-J, Parker R. Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J Cell Biol 2010; 189:813-27; PMID:20513766; http://dx.doi.org/ 10.1083/jcb.200912019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cargnello M, Tcherkezian J, Dorn JF, Huttlin EL, Maddox PS, Gygi SP, Roux PP. Phosphorylation of the eukaryotic translation initiation factor 4E-transporter (4E-T) by c-Jun N-terminal kinase promotes stress-dependent P-body assembly. Mol Cell Biol 2012; 32:4572-84; PMID:22966201; http://dx.doi.org/ 10.1128/MCB.00544-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 2012; 149:768-79; PMID:22579282; http://dx.doi.org/ 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 60. Nakagawa S, Hirose T. Paraspeckle nuclear bodies–useful uselessness? Cell Mol Life Sci 2012; 69:3027-36; PMID:22476590; http://dx.doi.org/ 10.1007/s00018-012-0973-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 2011; 20:364-75; PMID:21397847; http://dx.doi.org/ 10.1016/j.devcel.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang H, Gao Q, Peng X, Choi S-Y, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 2011; 20:376-87; PMID:21397848; http://dx.doi.org/ 10.1016/j.devcel.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsai N-P, Ho P-C, Wei L-N. Regulation of stress granule dynamics by Grb7 and FAK signalling pathway. EMBO J 2008; 27:715-26; PMID:18273060; http://dx.doi.org/ 10.1038/emboj.2008.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lian XJ, Gallouzi I-E. Oxidative Stress Increases the Number of Stress Granules in Senescent Cells and Triggers a Rapid Decrease in p21waf1/cip1 Translation. J Biol Chem 2009; 284:8877-87; PMID:19176530; http://dx.doi.org/ 10.1074/jbc.M806372200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci USA 2006; 103:2647-52; PMID:16477042; http://dx.doi.org/ 10.1073/pnas.0509333103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lim AK, Tao L, Kai T. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol 2009; 186:333-42; PMID:19651888; http://dx.doi.org/ 10.1083/jcb.200904063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Aravin AA, van der Heijden GW, Castañeda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 2009; 5:e1000764; PMID:20011505; http://dx.doi.org/ 10.1371/journal.pgen.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin M-D, Fan S-J, Hsu W-S, Chou T-B. Drosophila decapping protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev Cell 2006; 10:601-13; PMID:16678775; http://dx.doi.org/ 10.1016/j.devcel.2006.02.021 [DOI] [PubMed] [Google Scholar]
- 69. Leung AKL, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A 2006; 103:18125-30; PMID:17116888; http://dx.doi.org/ 10.1073/pnas.0608845103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol 2006; 16:1884-94; PMID:16949822; http://dx.doi.org/ 10.1016/j.cub.2006.08.051 [DOI] [PubMed] [Google Scholar]
- 71. Kotaja N, Lin H, Parvinen M, Sassone-Corsi P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci 2006; 119:2819-25; PMID:16787948; http://dx.doi.org/ 10.1242/jcs.03022 [DOI] [PubMed] [Google Scholar]
- 72. Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 2007; 129:69-82; PMID:17418787; http://dx.doi.org/ 10.1016/j.cell.2007.03.026 [DOI] [PubMed] [Google Scholar]
- 73. Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 2008; 31:67-78; PMID:18571452; http://dx.doi.org/ 10.1016/j.molcel.2008.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mazroui R, Di Marco S, Kaufman RJ, Gallouzi I-E. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol Biol Cell 2007; 18:2603-18; PMID:17475769; http://dx.doi.org/ 10.1091/mbc.E06-12-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Orsborn AM, Li W, McEwen TJ, Mizuno T, Kuzmin E, Matsumoto K, Bennett KL. GLH-1, the C. elegans P granule protein, is controlled by the JNK KGB-1 and by the COP9 subunit CSN-5. Development 2007; 134:3383-92; PMID:17699606; http://dx.doi.org/ 10.1242/dev.005181 [DOI] [PubMed] [Google Scholar]
- 76. Updike DL, Strome S. A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics 2009; 183:1397-419; PMID:19805813; http://dx.doi.org/ 10.1534/genetics.109.110171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Buchan JR, Kolaitis R-M, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 2013; 153:1461-74; PMID:23791177; http://dx.doi.org/ 10.1016/j.cell.2013.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol 2013; 23:2452-62; PMID:24291094; http://dx.doi.org/ 10.1016/j.cub.2013.09.058 [DOI] [PubMed] [Google Scholar]
- 79. Matsumoto K, Minami M, Shinozaki F, Suzuki Y, Abe K, Zenno S, Matsumoto S, Minami Y. Hsp90 is involved in the formation of P-bodies and stress granules. Biochem Biophys Res Commun 2011; 407:720-4; PMID:21439943; http://dx.doi.org/ 10.1016/j.bbrc.2011.03.088 [DOI] [PubMed] [Google Scholar]
- 80. Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 2009; 136:308-21; PMID:19167332; http://dx.doi.org/ 10.1016/j.cell.2008.12.022 [DOI] [PubMed] [Google Scholar]
- 81. Zhao Y, Tian E, Zhang H. Selective autophagic degradation of maternally-loaded germline P granule components in somatic cells during C. elegans embryogenesis. Autophagy 2009; 5:717-9; PMID:19372764; http://dx.doi.org/ 10.4161/auto.5.5.8552 [DOI] [PubMed] [Google Scholar]
- 82. Del Gatto-Konczak F, Bourgeois CF, Le Guiner C, Kister L, Gesnel MC, Stévenin J, Breathnach R. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5′ splice site. Mol Cell Biol 2000; 20:6287-99; PMID:10938105; http://dx.doi.org/ 10.1128/MCB.20.17.6287-6299.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J 2000; 19:4154-63; PMID:10921895; http://dx.doi.org/ 10.1093/emboj/19.15.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell 2006; 17:4972-81; PMID:16987958; http://dx.doi.org/ 10.1091/mbc.E06-06-0513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol 2008; 10:1324-32; PMID:18836437; http://dx.doi.org/ 10.1038/ncb1791 [DOI] [PubMed] [Google Scholar]
- 86. Kim WJ, Back SH, Kim V, Ryu I, Jang SK. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol Cell Biol 2005; 25:2450-62; PMID:15743837; http://dx.doi.org/ 10.1128/MCB.25.6.2450-2462.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Takahara T, Maeda T. Transient sequestration of TORC1 into stress granules during heat stress. Mol Cell 2012; 47:242-52; PMID:22727621; http://dx.doi.org/ 10.1016/j.molcel.2012.05.019 [DOI] [PubMed] [Google Scholar]
- 88. Thedieck K, Holzwarth B, Prentzell MT, Boehlke C, Kläsener K, Ruf S, Sonntag AG, Maerz L, Grellscheid S-N, Kremmer E, et al. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 2013; 154:859-74; PMID:23953116; http://dx.doi.org/ 10.1016/j.cell.2013.07.031 [DOI] [PubMed] [Google Scholar]
- 89. Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell 1998; 2:437-45; PMID:9809065; http://dx.doi.org/ 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- 90. Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007; 131:174-87; PMID:17923096; http://dx.doi.org/ 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 91. Paquin N, Ménade M, Poirier G, Donato D, Drouet E, Chartrand P. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol Cell 2007; 26:795-809; PMID:17588515; http://dx.doi.org/ 10.1016/j.molcel.2007.05.016 [DOI] [PubMed] [Google Scholar]
- 92. Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell 2009; 136:719-30; PMID:19239891; http://dx.doi.org/ 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Seydoux G, Fire A. Soma-germline asymmetry in the distributions of embryonic RNAs in Caenorhabditis elegans. Development 1994; 120:2823-34; PMID:7607073 [DOI] [PubMed] [Google Scholar]
- 94. Pitt JN, Schisa JA, Priess JR. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol 2000; 219:315-33; PMID:10694425; http://dx.doi.org/ 10.1006/dbio.2000.9607 [DOI] [PubMed] [Google Scholar]
- 95. Gao Y, Tatavarty V, Korza G, Levin MK, Carson JH. Multiplexed dendritic targeting of alpha calcium calmodulin-dependent protein kinase II, neurogranin, and activity-regulated cytoskeleton-associated protein RNAs by the A2 pathway. Mol Biol Cell 2008; 19:2311-27; PMID:18305102; http://dx.doi.org/ 10.1091/mbc.E07-09-0914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci U S A 2003; 100:11429-34; PMID:13679573; http://dx.doi.org/ 10.1073/pnas.2033246100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Lange S, Katayama Y, Schmid M, Burkacky O, Bräuchle C, Lamb DC, Jansen R-P. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic 2008; 9:1256-67; PMID:18485054; http://dx.doi.org/ 10.1111/j.1600-0854.2008.00763.x [DOI] [PubMed] [Google Scholar]
- 98. Stöhr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Hüttelmaier S. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol 2006; 175:527-34; PMID:17101699; http://dx.doi.org/ 10.1083/jcb.200608071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Simpson CE, Lui J, Kershaw CJ, Sims PFG, Ashe MP. mRNA localization to P-bodies in yeast is bi-phasic with many mRNAs captured in a late Bfr1p-dependent wave. J Cell Sci 2014; 127:1254-62; PMID:24424022; http://dx.doi.org/ 10.1242/jcs.139055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Mikl M, Vendra G, Kiebler MA. Independent localization of MAP2, CaMKIIα and β-actin RNAs in low copy numbers. EMBO Rep 2011; 12:1077-84; PMID:21869818; http://dx.doi.org/ 10.1038/embor.2011.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, Lopez-Jones M, Meng X, Singer RH. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 2014; 343:422-4; PMID:24458643; http://dx.doi.org/ 10.1126/science.1239200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Batish M, van den Bogaard P, Kramer FR, Tyagi S. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci U S A 2012; 109:4645-50; PMID:22392993; http://dx.doi.org/ 10.1073/pnas.1111226109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Tübing F, Vendra G, Mikl M, Macchi P, Thomas S, Kiebler MA. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J Neurosci 2010; 30:4160-70; PMID:20237286; http://dx.doi.org/ 10.1523/JNEUROSCI.3537-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Barbee SA, Estes PS, Cziko A-M, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 2006; 52:997-1009; PMID:17178403; http://dx.doi.org/ 10.1016/j.neuron.2006.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Walters R, Parker R. Quality control: Is there quality control of localized mRNAs? J Cell Biol 2014; 204:863-8; PMID:24637320; http://dx.doi.org/ 10.1083/jcb.201401059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; http://dx.doi.org/ 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Groppo R, Richter JD. Translational control from head to tail. Curr Opin Cell Biol 2009; 21:444-51; PMID:19285851; http://dx.doi.org/ 10.1016/j.ceb.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Inada T. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim Biophys Acta 2013; 1829:634-42; PMID:23416749; http://dx.doi.org/ 10.1016/j.bbagrm.2013.02.004 [DOI] [PubMed] [Google Scholar]
- 109. Donnelly N, Gorman AM, Gupta S, Samali A. The eIF2α kinases: their structures and functions. Cell Mol Life Sci 2013; 70:3493-511; PMID:23354059; http://dx.doi.org/ 10.1007/s00018-012-1252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Charlesworth A, Meijer HA, de Moor CH. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip Rev RNA 2013; 4:437-61; PMID:23776146; http://dx.doi.org/ 10.1002/wrna.1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Wilczynska A, Aigueperse C, Kress M, Dautry F, Weil D. The translational regulator CPEB1 provides a link between dcp1 bodies and stress granules. J Cell Sci 2005; 118:981-92; PMID:15731006; http://dx.doi.org/ 10.1242/jcs.01692 [DOI] [PubMed] [Google Scholar]
- 112. Standart N, Minshall N. Translational control in early development: CPEB, P-bodies and germinal granules. Biochem Soc Trans 2008; 36:671-6; PMID:18631138; http://dx.doi.org/ 10.1042/BST0360671 [DOI] [PubMed] [Google Scholar]
- 113. Richter JD. CPEB: a life in translation. Trends Biochem Sci 2007; 32:279-85; PMID:17481902; http://dx.doi.org/ 10.1016/j.tibs.2007.04.004 [DOI] [PubMed] [Google Scholar]
- 114. Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys 2013; 42:217-39; PMID:23654304; http://dx.doi.org/ 10.1146/annurev-biophys-083012-130404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Moser JJ, Fritzler MJ. Relationship of other cytoplasmic ribonucleoprotein bodies (cRNPB) to GW/P bodies. Adv Exp Med Biol 2013; 768:213-42; PMID:23224973; http://dx.doi.org/ 10.1007/978-1-4614-5107-5_13 [DOI] [PubMed] [Google Scholar]
- 116. Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 2006; 124:521-33; PMID:16469699; http://dx.doi.org/ 10.1016/j.cell.2006.01.031 [DOI] [PubMed] [Google Scholar]
- 117. Mokas S, Mills JR, Garreau C, Fournier M-J, Robert F, Arya P, Kaufman RJ, Pelletier J, Mazroui R. Uncoupling stress granule assembly and translation initiation inhibition. Mol Biol Cell 2009; 20:2673-83; PMID:19369421; http://dx.doi.org/ 10.1091/mbc.E08-10-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Amikura R, Kashikawa M, Nakamura A, Kobayashi S. Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc Natl Acad Sci U S A 2001; 98:9133-8; PMID:11470924; http://dx.doi.org/ 10.1073/pnas.171286998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Cougot N, Molza A-E, Giudice E, Cavalier A, Thomas D, Gillet R. Structural organization of the polysomes adjacent to mammalian processing bodies (P-bodies). RNA Biol 2013; 10:314-20; PMID:23324601; http://dx.doi.org/ 10.4161/rna.23342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Yasuda K, Zhang H, Loiselle D, Haystead T, Macara IG, Mili S. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J Cell Biol 2013; 203:737-46; PMID:24297750; http://dx.doi.org/ 10.1083/jcb.201306058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Kleene KC. Multiple controls over the efficiency of translation of the mRNAs encoding transition proteins, protamines, and the mitochondrial capsule selenoprotein in late spermatids in mice. Dev Biol 1993; 159:720-31; PMID:8405691; http://dx.doi.org/ 10.1006/dbio.1993.1277 [DOI] [PubMed] [Google Scholar]
- 122. Nguyen Chi M, Chalmel F, Agius E, Vanzo N, Khabar KSA, Jégou B, Morello D. Temporally regulated traffic of HuR and its associated ARE-containing mRNAs from the chromatoid body to polysomes during mouse spermatogenesis. PLoS One 2009; 4:e4900; PMID:19333380; http://dx.doi.org/ 10.1371/journal.pone.0004900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Boag PR, Atalay A, Robida S, Reinke V, Blackwell TK. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol 2008; 182:543-57; PMID:18695045; http://dx.doi.org/ 10.1083/jcb.200801183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hilgers V, Teixeira D, Parker R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA 2006; 12:1835-45; PMID:16940550; http://dx.doi.org/ 10.1261/rna.241006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Gowrishankar G, Winzen R, Dittrich-Breiholz O, Redich N, Kracht M, Holtmann H. Inhibition of mRNA deadenylation and degradation by different types of cell stress. Biol Chem 2006; 387:323-7; PMID:16542155; http://dx.doi.org/ 10.1515/BC.2006.043 [DOI] [PubMed] [Google Scholar]
- 126. Snee MJ, Macdonald PM. Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J Cell Sci 2004; 117:2109-20; PMID:15090597; http://dx.doi.org/ 10.1242/jcs.01059 [DOI] [PubMed] [Google Scholar]
- 127. Estes PS, Daniel SG, McCallum AP, Boehringer AV, Sukhina AS, Zwick RA, Zarnescu DC. Motor neurons and glia exhibit specific individualized responses to TDP-43 expression in a Drosophila model of amyotrophic lateral sclerosis. Dis Model Mech 2013; 6:721-33; PMID:23471911; http://dx.doi.org/ 10.1242/dmm.010710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zhang J, Okabe K, Tani T, Funatsu T. Dynamic association-dissociation and harboring of endogenous mRNAs in stress granules. J Cell Sci 2011; 124:4087-95; PMID:22135363; http://dx.doi.org/ 10.1242/jcs.090951 [DOI] [PubMed] [Google Scholar]
- 129. van der Laan AMA, van Gemert AMC, Dirks RW, Noordermeer JN, Fradkin LG, Tanke HJ, Jost CR. mRNA cycles through hypoxia-induced stress granules in live Drosophila embryonic muscles. Int J Dev Biol 2012; 56:701-9; PMID:23319346; http://dx.doi.org/ 10.1387/ijdb.103172al [DOI] [PubMed] [Google Scholar]
- 130. Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol 2007; 27:3970-81; PMID:17403906; http://dx.doi.org/ 10.1128/MCB.00128-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip Rev RNA 2010; 1:253-65; PMID:21935889; http://dx.doi.org/ 10.1002/wrna.15 [DOI] [PubMed] [Google Scholar]
- 132. Lloyd RE. Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdiscip Rev RNA 2013; 4:317-31; PMID:23554219; http://dx.doi.org/ 10.1002/wrna.1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Baguet A, Degot S, Cougot N, Bertrand E, Chenard M-P, Wendling C, Kessler P, Le Hir H, Rio M-C, Tomasetto C. The exon-junction-complex-component metastatic lymph node 51 functions in stress-granule assembly. J Cell Sci 2007; 120:2774-84; PMID:17652158; http://dx.doi.org/ 10.1242/jcs.009225 [DOI] [PubMed] [Google Scholar]
- 134. Fournier M-J, Gareau C, Mazroui R. The chemotherapeutic agent bortezomib induces the formation of stress granules. Cancer Cell Int 2010; 10:12; PMID:20429927; http://dx.doi.org/ 10.1186/1475-2867-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci 2011; 31:16086-93; PMID:22072660; http://dx.doi.org/ 10.1523/JNEUROSCI.4105-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 2013; 154:727-36; PMID:23953108; http://dx.doi.org/ 10.1016/j.cell.2013.07.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener 2012; 7:56; PMID:23164372; http://dx.doi.org/ 10.1186/1750-1326-7-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem 2009; 284:20329-39; PMID:19465477; http://dx.doi.org/ 10.1074/jbc.M109.010264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol 2011; 9:e1000614; PMID:21541367; http://dx.doi.org/ 10.1371/journal.pbio.1000614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Elden AC, Kim H-J, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010; 466:1069-75; PMID:20740007; http://dx.doi.org/ 10.1038/nature09320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ, et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci 2010; 30:7729-39; PMID:20519548; http://dx.doi.org/ 10.1523/JNEUROSCI.5894-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Thomas M, Alegre-Abarrategui J, Wade-Martins R. RNA dysfunction and aggrephagy at the centre of an amyotrophic lateral sclerosis/frontotemporal dementia disease continuum. Brain 2013; 136:1345-60; PMID:23474849; http://dx.doi.org/ 10.1093/brain/awt030 [DOI] [PubMed] [Google Scholar]
- 143. Dewey CM, Cenik B, Sephton CF, Johnson BA, Herz J, Yu G. TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res 2012; 1462:16-25; PMID:22405725; http://dx.doi.org/ 10.1016/j.brainres.2012.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol 2013; 201:361-72; PMID:23629963; http://dx.doi.org/ 10.1083/jcb.201302044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Zhang X, Chen S, Song L, Tang Y, Shen Y, Jia L, Le W. MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy 2014; 10:588-602; PMID:24441414; http://dx.doi.org/ 10.4161/auto.27710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Fornai F, Longone P, Cafaro L, Kastsiuchenka O, Ferrucci M, Manca ML, Lazzeri G, Spalloni A, Bellio N, Lenzi P, et al. Lithium delays progression of amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A 2008; 105:2052-7; PMID:18250315; http://dx.doi.org/ 10.1073/pnas.0708022105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Kim H-J, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ, Lee VM-Y, Finkbeiner S, Gitler AD, Bonini NM. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet 2014; 46:152-60; PMID:24336168; http://dx.doi.org/ 10.1038/ng.2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Zhang T, Mullane PC, Periz G, Wang J. TDP-43 neurotoxicity and protein aggregation modulated by heat shock factor and insulin/IGF-1 signaling. Hum Mol Genet 2011; 20:1952-65; PMID:21355045; http://dx.doi.org/ 10.1093/hmg/ddr076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SSW, Kiskinis E, Winborn B, Freibaum BD, Kanagaraj A, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron 2014; 81:536-43; PMID:24507191; http://dx.doi.org/ 10.1016/j.neuron.2013.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]