Abstract
There is an increasing recognition of the role that cancer cell derived exosomes play in intercellular signaling upon fusion or uptake with a target cell, including immune system evasion, tumor growth and metastasis. To date, however, although exosomal membrane and cargo lipids are expected to play a pivotal role in exosome biogenesis and secretion, as well as in fusion or uptake and target cell functional response, the detailed characterization of cancer cell derived exosome lipids across a range of different cancers has not yet been broadly explored. Here, a simple and straightforward lipidome analysis strategy consisting of optimized sample extraction and novel sample derivatization techniques, coupled with high-resolution ‘shotgun’ mass spectrometry and ‘targeted’ tandem mass spectrometry methods, is demonstrated for the rapid identification of >520 individual lipids in 36 lipid classes and sub classes from exosomes secreted by the colorectal cancer cell line, LIM1215. Relative quantification and comparison of exosome versus cellular lipid profiles reveals significant enrichment of certain lipid classes, as well as substantial lipid subclass remodeling and changes in abundance of individual lipids, including sphingolipids, sterol lipids, glycerolipids and glycerophospholipids, and particularly plasmalogen- and alkyl ether-containing glycerophospholipids. This analysis strategy therefore provides a platform for comprehensive lipidome profiling across a wide range of cancer cell or tissue derived exosomes, that will facilitate subsequent functional studies aimed at elucidating the role of specific cellular or exosome lipids in the onset and progression of colorectal cancer, or to identify specific lipid(s) that could serve as effective diagnostic or prognostic disease biomarkers.
Keywords: lipidome, mass spectrometry, colorectal cancer, exosome
2. Introduction
The disruption of lipid metabolism pathways involved in the dynamic regulation of membrane structure, energy homeostasis and signaling can affect critical cellular processes including cell proliferation, differentiation and mobility, and are known to be associated with the onset and progression of various diseases, including cancer [1-8]. For example, changes in the compositions and/or abundance of certain lipid classes including glycerolipids, glycerophospholipids, sphingolipids and sterol lipids, or their bioactive signaling lipid metabolites, have been shown to be correlated with malignancy and metastatic capacity in numerous human cancers [9-14]. Therefore, elucidating the molecular compositions and functions of these lipids is expected to significantly increase our understanding of colon cancer biology, as well as enable the identification of diagnostic or prognostic biomarkers [15-17], or novel therapeutic targets, for the disease.
There is also an increasing recognition of the role that cancer cell derived exosomes (40-100 nm vesicles formed within multivesicular bodies and secreted from parent cells upon fusion of late endosomes with the cellular membrane) can play in cell-cell communication and in effecting phenotypic changes upon uptake or fusion with a recipient cell [18-24]. For example, numerous reports indicate that exosome mediated intercellular transfer of protein, mRNA and miRNA can facilitate a range of processes including immune system evasion, the promotion of angiogenesis, tumor growth, migration and cancer metastasis [25-27]. This inter-cellular communication role of exosomes is especially important since cancer cells exhibit increased secretion of exosomes relative to non-malignant cells [23,24].
Exosomal membrane lipids have also been shown to play a pivotal role in exosome formation and secretion, fusion and uptake, and in target cell functional response [28-34]. Numerous studies have clearly demonstrated that exosome lipid compositions are different to those of the parent cells, with significant enrichment observed in multiple lipid classes including cholesterol, sphingomyelin, phosphatidylserine, ganglioside, and saturated phosphatidylcholine and phosphatidylethanolamine molecular lipid species. [28,29,35-39]. Exosomes have also been shown to contain the necessary functional components of active lipid metabolism, including enzymes (e.g., phospholipases) and bioactive signalling lipids (e.g., prostaglandins and leukotrienes), in addition to the above complex lipid precursors of lipid metabolism contained within the exosomal membrane [33,34].
To date, however, the functional (i.e., causative) roles of exosome derived lipids in malignant and metastatic cancer cell transformation, and exploration of their potential to serve as biomarkers of colorectal cancer malignancy and metastatic capacity, remains poorly understood. Indeed, only a few studies have been performed to date to systematically characterize the diverse range of hundreds, or potentially thousands, of individual molecular lipid species that may be present within cancer cell secreted exosomes (i.e., the lipidome), or to quantitatively monitor the global exosome lipid profile alterations occurring as a function of cancer cell or tissue molecular phenotype, or malignancy and metastatic potential. One notable example is the recent report by Llorente, et al. who used a combination of different lipid extraction strategies coupled with a series of ‘targeted’ positive and negative ionization mode precursor ion scanning and neutral loss ‘shotgun’ (i.e., direct infusion) tandem mass spectrometry (MS/MS) and liquid chromatography (LC)-MS/MS methods, to perform an in depth lipidomic study of exosome compositions from PC-3 prostate cancer cells. From this study, an overall 8.4-fold enrichment of lipids per mg of protein in exosomes was observed, along with significant enrichment and remodelling of phosphatidylserine, cholesterol, sphingomyelin and glycosphingolipids compared to their parent cells, and the identification and quantitative analysis of 280 individual lipid species [39], at the molecular lipid level [40].
The development of high-resolution / accurate mass spectrometry instrumentation [41-44], and optimized lipid extraction protocols [45], has facilitated additional recent improvements for direct ‘shotgun’ lipidomic analysis of a broad range of both non-polar and polar lipid classes and their individual lipids within crude lipid extracts, across at least three orders of magnitude ion abundance, without need for complex liquid-liquid extraction or chromatographic fractionation techniques prior to mass spectrometry analysis, and with limited sample consumption requirement. These strategies are further enhanced by the use of functional group selective derivatization reactions that result in the resolution of isobaric mass overlap of several common lipid classes and their subsequent unambiguous assignment, as reported previously by our group [44,46]. These benefits are particularly attractive for exosome lipidome analysis, where limited sample availability is an ever present challenge. Here, therefore, we have combined each of these different strategies, i.e., optimized sample extraction, sample derivatization and high-resolution ‘shotgun’ mass spectrometry and ‘targeted’ tandem mass spectrometry (MS/MS), for the rapid identification and determination of normalized lipid abundance changes for a wide variety of lipid classes, sub classes and individual lipid species from exosomes secreted by the colorectal cancer cell line, LIM1215 [47].
3. Materials and Methods
3.1 Materials
Ammonium formate was purchased from Alfa Aesar (Ward Hill, MA). Iodine, N,N-dimethylformamide (DMF), and triethylamine (TEA) were from Jade Scientific (Westland, MI). Internal standard lipids PC(14:0/14:0), PE(14:0/14:0), and PS(14:0/14:0) were purchased from Avanti Polar Lipids (Alabaster, AL). Ammonium bicarbonate, isopropanol, methanol, and water were from J.T. Baker (Phillipsburg, NJ). Chloroform was from EMD Chemicals (Billerica, MA). All solvents used were high performance liquid chromatography grade, and all lipid extraction and storage solvents contained 0.01% butylated hydroxytoluene (BHT) from Sigma Aldrich (St. Louis, MO). 13C1-S,S’-dimethylthiobutanoylhydroxysuccinimide ester (13C1-DMBNHS) was synthesized as previously described [44,46]. RPMI 1640 cell culture medium and Penicillin-Streptomycin were from GIBCO, Life Technologies (Grand Island, NY). Fetal calf serum (FCS) was from SAFC® Bioscience (St. Louis, MO).
3.2 LIM1215 cell culture
The human colorectal cancer LIM1215 cell line was cultured in RPMI 1640 medium supplemented with 10% FCS and 100 Units/mL of Penicillin-Streptomycin in 150 cm2 BD FalconTM tissue culture flasks at 37°C in 5% CO2. 1.48 × 107 cells, containing 2976 micrograms of protein, were collected then lyophilized and stored at −80°C prior to lipid analysis.
3.3 Isolation of secreted exosomes from LIM1215 cells
LIM1215 cells were grown to ~80% confluent (~5 × 108) in IntegridTM 150 mm cell culture dishes then washed three times to remove residual FCS. Subsequently, cells were cultured for 24 h in RPMI 1640 media supplemented with 10% exosome depleted FCS. The cell viability was more than 98% as determined by Tryphan Blue assay. The conditioned media from the plates was then collected and centrifuged at 500 g for 10 min. A subsequent spin at 2,000 g for 20 min was performed to remove floating cells and cell debris followed by 10,000 g for 30 min. The supernatant was further subjected to ultracentrifugation at 100,000 g in a Beckman SW-28 rotor for 1 h at 4°C. Finally, pellets were washed with PBS to remove residual traces of media and centrifuged again at 100,000 g for 1 h at 4°C to isolate exosomes. The exosome preparation containing 230 micrograms of protein was then lyophilized and stored at −80°C prior to lipid analysis.
3.4 Protocol for monophasic lipid extraction from LIM1215 cells and secreted exosomes
Lyophilized cell pellets containing 2.9 mg of cellular protein, or lyophilized exosome pellets containing 230 micrograms of exosomal protein, were combined with 1.0 mL of ice-cold 40% methanol and homogenized for five minutes using a Bullet Blender (Next Advance, Averill Park, NY) and 0.5 mm zirconium oxide beads.
Homogenates were transferred to glass tubes (13 × 100 mm) and 1.4 mL of water was added.
5.0 mL of methanol and 2.7 mL of chloroform was then added to each homogenate and the samples were vortexed thoroughly, then incubated with shaking for 30 minutes at room temperature.
Samples were centrifuged for 30 minutes at 2000 × g at room temperature.
Supernatants were collected to a new glass tube. To the remaining protein pellets, 1.0 mL water was added. Samples were vortexed vigorously, followed by addition of 4.0 mL chloroform : methanol (1 : 2, v : v), then incubated with shaking for 30 minutes at room temperature.
Samples were centrifuged as in step 4, then the supernatants collected and combined with the supernatant from the first extraction.
Extraction solvents were evaporated under nitrogen. The dried lipid extracts were then washed three times with 0.5 mL 10 mM ammonium bicarbonate, then taken to complete dryness again under nitrogen.
Lipids were resuspended in isopropanol : methanol : chloroform (4 : 2 : 1, v:v:v, containing 0.01% BHT) at a final concentration of 1 µL solvent / µg protein extracted.
Notes
We have recently reported that monophasic lipid extraction with methanol : chloroform : water (2 : 1 : 0.74, v:v:v) enables the enhanced recovery of both polar and nonpolar lipid classes compared to traditional biphasic lipid extraction methods [45]. Here the volumes used for monophasic lipid extraction allowed lipid extraction from the equivalent of several milligrams of protein. However, volumes for extracting and resuspending lipids may be scaled up or down as needed. Extraction volumes must be increased if the sample to be extracted is sufficiently large that a biphasic mixture results after the addition of all solvents to the sample homogenate, or the size of the protein pellet produced after centrifugation (step 4) is too large to allow sufficient recovery of the supernatant. In the present study, the volume of extraction solvent used relative to the amount of extracted material was sufficient to minimize any potential differences in recovery between the two sample quantities.
3.5 Protocol for sequential functional group derivatization of aminophospholipids and plasmalogen-containing lipids
20 µL of LIM1215 cell lipid extract, or 10 µL of exosome lipid extract were aliquoted in triplicate to individual wells of a Whatman multichem 96-well plate (Sigma Aldrich, St. Louis, MO).
To each lipid extract, 10 µL of a mixture of 2 µM each PC(14:0/14:0), PE(14:0/14:0) and PS(14:0/14:0) were added as internal standards for relative lipid quantification and confirmation of aminophospholipid derivatization reaction efficiency. Solvents were then evaporated under a blanket of nitrogen.
A 2.5 mM stock solution of TEA in chloroform was freshly prepared by adding 3.4 µL TEA to 10 mL of chloroform.
A 2.5 mM stock solution of 13C1-DMBNHS was freshly prepared by dissolving 3.26 mg 13C1-DMBNHS in 3.35 mL of DMF.
40 µL of a solution of 39 : 1.1 : 1 (v:v:v) chloroform : 2.5 mM TEA : 2.5 mM 13C1-DMBNHS was added to each dried lipid extract.
The 96-well plate was sealed with Teflon Ultra Thin Sealing Tape (Analytical Sales and Services, Pompton Plains, NJ) and incubated at room temperature with gentle shaking for 30 minutes.
The sealing tape was removed from the 96-well plate and the derivatization solvent was evaporated under nitrogen. Each lipid extract was washed twice by adding 40 µL of 10 mM aq. ammonium bicarbonate and carefully aspirating the wash solution. Remaining traces of water were then removed by drying under nitrogen.
While keeping the 96-well plate under nitrogen, the plate was chilled in an ice bath for 10 minutes.
A stock solution of 3.94 mM iodine was freshly prepared by dissolving 10 mg iodine in 10 mL chloroform.
A stock solution of 15 mM ammonium bicarbonate was freshly prepared by dissolving 12 mg ammonium bicarbonate in 10 mL of HPLC methanol.
A solution of 2 : 1 (v:v) chloroform : methanol containing 266 µM iodine and 2mM ammonium bicarbonate was prepared by adding 400 µL of 3.94 mM iodine in chloroform to 3.6 mL chloroform, and 800 µL of 15 mM ammonium bicarbonate in HPLC methanol to 1.2 mL HPLC methanol, then combined and placed in an ice bath.
40 µL of the 2 : 1 (v:v) chloroform : methanol containing 266 µM iodine and 2mM ammonium bicarbonate was then added to each dried lipid extract, and the plate was sealed with sealing tape and reacted for 5 minutes in the ice bath. The reaction solvents were then completely removed by evaporation under nitrogen, while maintaining the 96-well plate in the ice bath.
Each lipid extract was washed twice by adding 40 µL of 10 mM aq. ammonium bicarbonate and carefully aspirating the wash solution. Remaining traces of water were then removed by drying under nitrogen.
The derivatized lipid extracts were then resuspended in 40 µL of isopropanol : methanol : chloroform (4 : 2 : 1, v : v : v) containing 20 mM ammonium formate, and the 96-well plate was sealed with sealing tape.
Notes
Prior to sequential functional group derivatization and mass spectrometry analysis, the cell and exosome lipid extracts were analyzed in positive ionization mode over a range of sample dilutions to determine the analytical conditions in which the cell and exosome lipid concentrations were approximately equal, and were within the linear range of detector response, as previously described [45].
3.6 High resolution accurate mass spectrometry and tandem mass spectrometry analysis
10 µL of each derivatized lipid sample was aspirated and introduced via nanoESI to a high resolution / accurate mass Thermo Scientific model LTQ Orbitrap Velos mass spectrometer (San Jose, CA) using an Advion Triversa Nanomate nESI source (Advion, Ithaca, NY) operating with a spray voltage of 1.4 kV and a gas pressure of 0.3 psi. The ion source interface settings (inlet temperature of 100°C and S-Lens value of 50%) were optimized to maximize the sensitivity of the precursor ions while minimizing ‘in-source’ fragmentation. High resolution mass spectra were acquired over the range of 200-2000 m/z in positive ionization mode using the FT analyzer operating at 100,000 mass resolving power (at m/z 400), and signal averaged for 2 minutes. Following initial lipid assignment by database analysis of the high resolution mass spectra (see below), a series of ‘targeted’ Higher-Energy Collision Induced Dissociation (HCD-MS/MS) product ion spectra were acquired on selected precursor ions using the FT analyzer operating at 100,000 mass resolving power and default activation times in positive ionization mode using the derivatized lipid extracts to confirm the identities of lipid headgroups, or in negative ionization mode using underivatized lipid extracts for fatty acid chain identification. HCD-MS/MS collision energies were individually optimized for each lipid class of interest using commercially available lipid standards whenever possible.
3.7 Peak finding, lipid identification, and relative quantification
Accurate mass-based ‘sum composition’ [40] lipid assignments were achieved using the Lipid Mass Spectrum Analysis (LIMSA) v.1.0 software linear fit algorithm [48], in conjunction with a user-defined database of hypothetical lipid compounds for automated peak finding and correction of 13C isotope effects. Lipids were labelled using the nomenclature proposed by the LIPID MAPS consortium [49]. Quantification of the abundances of lipid species in the cell and exosome samples was performed by comparison of target lipid ion peak areas to the PC(14:0/14:0) internal standard, with subsequent normalization of quantitated ion abundances to the protein equivalents utilized for analysis of each cell or exosome lipid extract. Mean normalized abundances for each lipid species were determined from triplicate analyses of each cell or exosome lipid extract. As a single internal standard was used for relative quantification of all lipid species detected in positive ionization mode mass spectra [44,45], and no attempts were made to correct for differences in ionization efficiency as a function of lipid headgroup identity and/or fatty acid length and degree of unsaturation, the normalized lipid ion abundances presented here are not intended to represent ‘absolute’ lipid abundance. However, this semi-quantitative measure of lipid ion abundances allows the rapid determination of differences in lipid abundances between cell and exosome lipid extracts per unit of protein. All data will be available through Vesiclepedia [50].
3.8 Statistical analysis
Differences in the mean abundance of lipid classes and individual assigned lipid species between LIM1215 cells and secreted exosomes were determined by ANOVA followed by Sidak’s multiple comparisons test using GraphPad Prism 6.0 software. Differences in mean fatty acid ratios were evaluated by two-tailed Student’s t-test. Statistical significance was set at P<0.05.
4. Results and Discussion
4.1 Workflow for comprehensive lipidome analysis of LIM1215 cells and their secreted exosomes
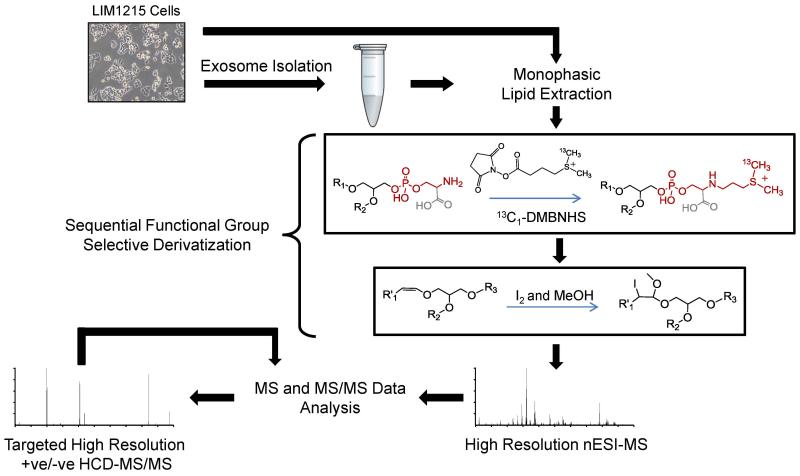
The overall workflow employed here for the comprehensive lipidome analysis of LIM1215 cells and exosomes is shown in Scheme 1, and consists of (i) cell culture, (ii) exosome isolation, (iii) monophasic lipid extraction, (iv) sequential functional group derivatization reactions, (v) positive ionization mode ESI-MS followed by database analysis of the resultant high resolution/accurate mass spectra for initial lipid assignment at the ‘sum composition’ level, and finally (vi) ‘targeted’ high resolution positive and/or negative ionization mode HCD-MS/MS for confirmation of lipid assignments and further structural characterization at the ‘molecular lipid’ level (albeit without assignment of fatty acyl/alkyl/alkenyl chain sn-linkage position). Specific details of each step associated with this workflow are described above in the Materials and Methods section.
Scheme 1.
Workflow for lipidome analysis of secreted exosomes and their parent cells. After exosome isolation by ultracentrifugation, exosomes and their parent cells are subjected to monophasic lipid extraction, followed by sequential functional group selective derivatization of aminophospholipids with 13C1-DMBNHS, and plasmalogen 1-alk-enyl double bonds with iodine and methanol. Derivatized lipid extracts are then analyzed in positive ionization mode by direct infusion nESI-high resolution / accurate mass Orbitrap MS, and assigned lipid identities are confirmed by ‘targeted’ higher-energy collisional dissociation - tandem mass spectrometry (HCD-MS/MS).
4.2 Characterization of exosomes secreted by LIM1215 colorectal cancer cells
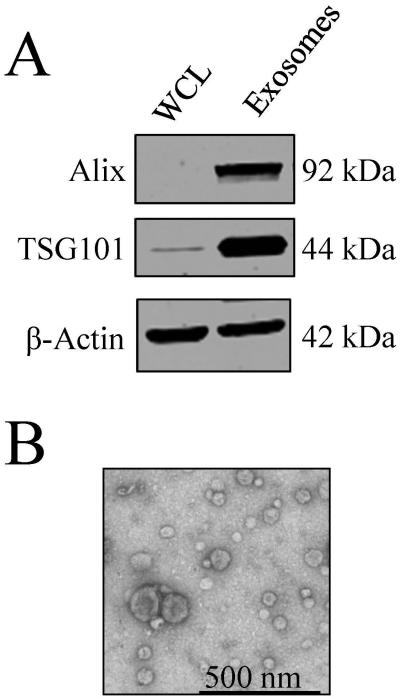
In order to characterize the isolated exosomes from the LIM1215 cells prior to comprehensive lipidome analysis, Western blotting analysis was performed to confirm the presence of the exosome markers Alix and TSG101. As shown in Figure 1A, Alix and TSG101 were found to be enriched in exosomes compared to the whole cell lysate samples. Additionally, TEM analysis confirmed the presence of membranous vesicles in the range of 40-100 nm in diameter (Figure 1B).
Figure 1.
Characterization of exosomes secreted by LIM1215 colorectal cancer cells. A) Western blotting analysis of the exosome markers Alix and TSG101. B) TEM analysis to confirm the presence of membranous vesicles in the range of 40-100 nm in diameter.
4.3 nanoESI - high resolution accurate mass spectrometry of LIM1215 cells and secreted exosome lipid extracts
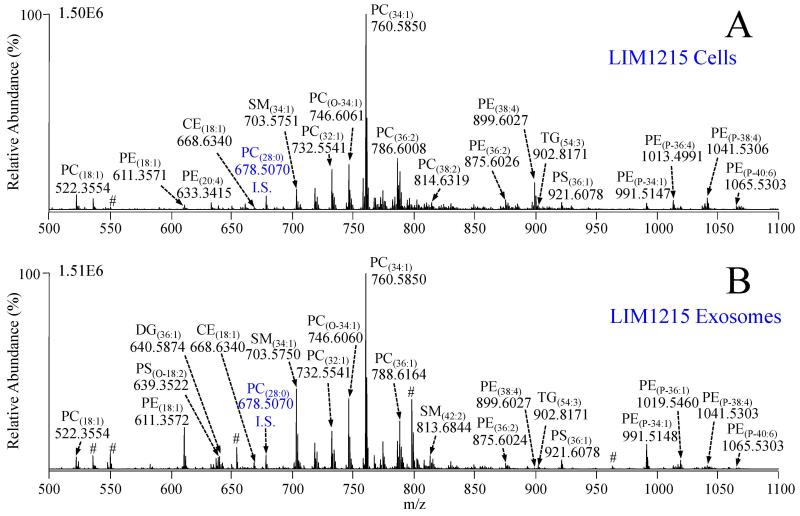
Figure 2A shows a positive ionization mode high resolution accurate mass spectrum acquired from a crude lipid extract of the LIM1215 cells, following sample preparation using the strategy outlined in Scheme 1 and described in the Materials and Methods section above. Accurate mass measurements enabled initial lipid assignment at the sum composition level (i.e., assignment of the headgroup identity, the nature of the fatty acyl/alkyl/alkenyl chain linkage, and the total number of carbons : total double bonds (TC : TDB) within the fatty acyl/alkyl/alkenyl chains) for a broad range of glycerolipid, glycerophospholipid, sphingolipid and sterol lipid species based on agreement of the measured accurate masses and isotope distributions with the calculated theoretical masses and isotope distributions of lipid species contained within the LIMSA database. As previously reported by us, 13C1-DMBNHS labeling of aminophospholipids prevents any isobaric overlap between even numbered carbon chain length phosphatidylethanolamine (PE) lipids with certain odd numbered carbon chain length molecular species of phosphatidylcholine (PC) and ammonium adducts of phosphatidic acid (PA) lipids (and vice versa), and the prevention of isobaric overlap between phosphatidylserine (PS) lipids with the ammonium adducts of phosphatidylglycerol (PG) lipids, as well as simultaneously increasing the sensitivity of detection for aminophospholipids due to the introduction of a fixed positive charge [44,46]. Additionally, derivatization of plasmalogen alk-1-enyl double bonds with iodine and methanol as described in section 2.5 results in an ability to discern plasmalogen (P-) lipids from potentially isobaric alkyl ether (O-) lipid species, based solely on their accurate mass measurements [46]. Importantly, these two derivatization reactions can be performed sequentially on crude lipid extracts in individual wells of a 96-well plate, with only minimal additional sample handling.
Figure 2.
Positive ionization mode high resolution / accurate mass spectra of lipid extracts from A) LIM1215 cells and B) LIM1215 secreted exosomes. Cell and exosome total lipid extracts were derivatized with 13C1-DMBNHS and iodine/methanol, then diluted to achieve approximately equal lipid concentrations prior to analysis. High resolution (R=100,000) accurate mass spectra were acquired and averaged for a period of 2 minutes each. PC(28:0) (500 nM) was include as an internal standard (I.S.) # Non-lipid variable background ion.
Figure 2B shows a positive ionization mode high resolution mass spectrum from the LIM1215 secreted exosomes, which were subjected to the same workflow used for analysis of the LIM1215 parent cells. Importantly, to minimize both absolute and relative ion suppression effects, and to enable direct comparison of relative quantitative differences in individual lipids between the complex cell and exosome lipid extracts, both the LIM1215 cell and exosome lipid extracts were initially diluted such that their lipid concentrations were approximately equal, and that each lipid extract was within the empirically determined linear range of detector response [45]. In practice, normalization of lipid levels between the cell and exosome samples required the exosome lipid extracts studied here to be diluted two-fold more than the LIM1215 cell lipid extracts, due to a higher lipid-to-protein ratio in the exosomes relative to their parent cells, similar to that reported by Llorente et al [39]. Numerous differences in the abundances of individual lipid species relative to the PC(28:0) internal standard (m/z 678.5070) in each sample were readily observed by visual comparison of the spectra in Figure 2A and Figure 2B. For example, m/z 611.3572 assigned as lyso lipid PE(18:1), m/z 703.5370 assigned as sphingomyelin lipid SM(34:1), m/z 746.6060 assigned as alkyl ether lipid PC(0-34:1), and m/z 991.5147 assigned as plasmalogen lipid PE(P-34:1) in Figure 2B from the LIM1215 exosomes were all increased in abundance compared to the relative abundances of the respective ions in the mass spectrum from the LIM1215 cells in Figure 2A. Conversely, m/z 899.6027 assigned as diacyl PE(38:4) was drastically reduced in the exosomes compared to the parent cell lipid extract. The observed differences between the mass spectra in Figure 2A and 2B clearly indicate that the distribution of lipid abundances within the LIM1215 cells and their secreted exosomes are divergent in numerous lipid classes and subclasses.
4.4 Global lipid profiles of LIM1215 cells and their secreted exosomes
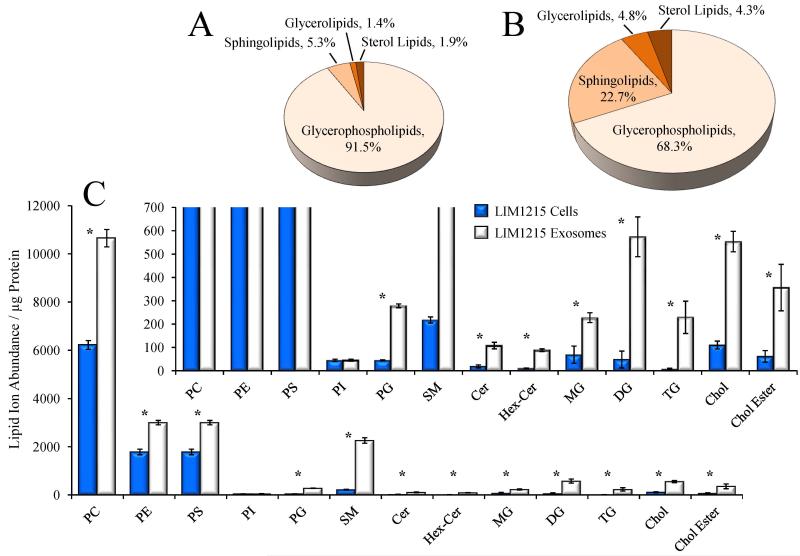
To enable a direct quantitative comparison of LIM1215 cell and exosome lipid class distributions and individual lipid ion abundances, the ions observed in the mass spectra shown in Figure 2A and Figure 2B were subjected to isotope correction and normalization against the PC(28:0) internal standard, then each normalized lipid ion abundance was normalized against the equivalent amount of cell or exosome protein utilized for each mass spectrometry analysis. The sums of the protein-adjusted normalized lipid abundances for all assigned lipid species within each lipid class then enabled a direct comparison of the global lipid profiles obtained for the LIM1215 cells and exosomes across multiple lipid classes. Figure 3A and 3B show the % distributions of the four broadly classified lipid classes defined by the Lipid MAPS consortium (www.lipidmaps.org), i.e. glycerolipids, glycerophospholipids, sphingolipids, and sterol lipids, within the LIM1215 cells and exosomes, respectively. Notably, an approximate 2.3-fold increase in total protein-adjusted normalized lipid abundance was observed in the LIM1215 exosomes relative to the cellular control, expressed here by the proportional increase in the area of Figure 3B compared to that in Figure 3A. Glycerophospholipids were found to comprise 91.5% of the total cellular lipid ion abundances, followed by sphingolipids (5.3%), sterol lipids (1.9%) and glycerolipids (1.4%), whereas the LIM1215 exosomes were greatly enriched in sphingolipids (22.7%), sterol lipids (4.3%), and glycerolipids (4.8%), with a concomitant decrease in the relative fraction of glycerophospholipid ion abundances. This overall increase in protein-adjusted normalized total lipid levels and general redistribution of lipid classes in secreted exosomes relative to their parent cell lines is consistent with numerous previous analyses of exosomal lipid content [28,35-39].
Figure 3.
Global lipidome analysis of LIM1215 cells and secreted exosomes. A: Pie chart representing the distribution of summed ion abundances of glycerolipid, glycerophospholipid, sphingolipid and sterol lipid classes, for all lipids detected in A) LIM1215 cells and B) LIM1215 secreted exosomes. The area of each pie chart is proportional to the total lipid ion abundance, normalized to protein concentration. C) Comparison of the total normalized summed ion abundances for each lipid class from the data in Figure 1. LIM1215 cells are shown in dark bars and LIM1215 secreted exosomes are shown in white bars. The inset to panel C shows an expanded vertical axis to allow comparison of low abundance lipid classes. Error bars show the standard deviations determined by triplicate analytical replicate measurement. * P < 0.05.
A comparison of the total normalized abundances for specific lipid sub-classes determined from triplicate analyses of the LIM1215 cells and exosome crude lipid extracts is shown as a bar graph in Figure 3C. With the exception of phosphatidylinositol (PI) lipids, the total normalized ion abundances (per microgram of protein) for all other lipid sub-classes were observed to increase in the exosomes compared to LIM1215 cells. The inset to Figure 3C shows an expanded region of the bar graph to highlight the increases in abundance of low abundance lipid sub-classes in the exosomes compared to LIM1215 cells. The observed increase in protein-adjusted normalized total lipid ion abundances in the exosome lipid extract, particularly for the sphingomyelin (SM), glycerophosphatidylserine (PS) and cholesterol (Cho) lipids, is also very consistent with previous observations of altered exosomal lipid compositions relative to their parent cell lines [28,35-39].
Importantly, the data shown in Figures 2 and 3 clearly demonstrate the range of lipid species, and depth of lipidome coverage across more than three orders of magnitude of ion abundance that can be obtained from a single positive ion mode ESI-MS spectrum acquired for each sample using the workflow described in Scheme 1 and in the Methods sections above, with minimal sample handling (i.e., without the need for chromatographic separation of lipid classes prior to MS), and with only a minimal amount (i.e., µg quantities) of sample required.
Following database analysis of the ions observed in the high resolution accurate mass spectra shown in Figure 2A and 2B, a total of 535 individual lipid species were assigned at the sum composition level from the LIM1215 cellular lipid extracts, and 527 individual lipid species from the LIM1215 secreted exosomes, across a broad range of glycerolipid, glycerophospholipid, sphingolipid and sterol lipid classes and sub-classes (Table 1). Importantly, this represents the most detailed analysis of exosome lipid content reported to date. In general the types and abundances of lipid classes and subclasses in the LIM1215 exosomes are similar to those recently reported from an analysis of exosomes secreted from PC-3 prostate cancer cells [39], as well as other studies involving the characterization of cancer cell derived exosomes [28,29,38]. In the present study, however, we did not observe any significant levels of ganglioside lipids, such as GM1 or GM3, that were reported in the PC-3 exosome study. The monophasic lipid extraction and high resolution nESI-MS analysis strategy employed here has previously been successfully utilized for determination of ganglioside content in neural retina tissue [45]. Thus, it is likely that ganglioside lipids (if present) in the non-neural LIM1215 cells and secreted exosomes examined here are below the limit of detection.
Table 1.
Summary of LIM1215 cell and exosome lipids assigned via sequential functional group derivatization and direct infusion nano electrospray ionization - high resolution accurate mass spectrometry (nESI-MS).
| Lipid Class | Lipid Sub-Class | LIM1215 Cells | LIM1215 Exosomes |
|---|---|---|---|
| Total Lipids Assigned (Sum Composition) | |||
| Glycerophospholipids | PC | 61 | 51 |
| P-PC | 37 | 18 | |
| O-PC | 69 | 53 | |
| LPC | 14 | 13 | |
| O-LPC | 7 | 7 | |
| PE | 48 | 30 | |
| P-PE | 46 | 41 | |
| O-PE | 34 | 27 | |
| LPE | 16 | 23 | |
| P-LPE | 3 | 5 | |
| O-LPE | 6 | 6 | |
| PS | 24 | 18 | |
| P-PS | 0 | 1 | |
| O-PS | 19 | 6 | |
| LPS | 0 | 3 | |
| P-LPS | 4 | 2 | |
| O-LPS | 12 | 9 | |
| PI | 15 | 9 | |
| O-PI | 2 | 0 | |
| PG | 16 | 28 | |
| P-PG | 0 | 1 | |
| O-PG | 0 | 5 | |
| LPG | 2 | 4 | |
| Sphingolipids | SM | 18 | 36 |
| Cer | 11 | 16 | |
| Hex-Cer | 4 | 17 | |
| Glycerolipids | MG | 6 | 11 |
| O-MG | 10 | 7 | |
| DG | 19 | 25 | |
| O-DG | 8 | 8 | |
| TG | 14 | 26 | |
| P-TG | 0 | 1 | |
| O-TG | 0 | 10 | |
| Sterol Lipids | Chol | 1 | 1 |
| Chol Ester | 9 | 9 | |
| Total | 535 | 527 | |
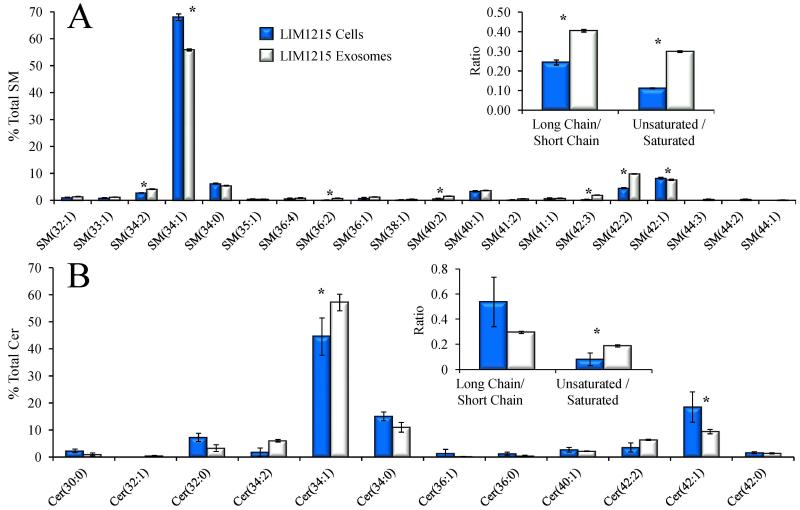
4.5 Comparison of sphingomyelin and ceramide lipid species in LIM1215 exosomes and cells
Figure 3 demonstrates that the LIM1215 exosomes contain more lipid per microgram of protein than their parent cells for the majority of detected lipid sub-classes. Given the overall higher lipid abundance in exosomes, we next determined that the exosome lipid profiles also underwent a significant relative redistribution of individual lipid species within a given lipid sub-class. For example, Figures 4A and 4B show clear differences in the relative abundances of individual SM and ceramide (Cer) lipid species, expressed as the percent total SM and Cer lipid abundances, respectively, in the SM and Cer pools from the LIM1215 exosomes cells compared to the cellular SM and Cer pools. Notably, abundant SM(34:1), SM(42:1) and SM(42:2), and Cer(34:1), Cer(42:1) and Cer(42:2) species were observed in both the cell and exosome lipid extracts, presumably containing C16-, C24- and C24:1- amide-linked fatty acid chains with a sphingosine backbone. The high relative abundance of these lipids in the LIM1215 colorectal cancer cells, originally derived from an Omental metastatic tumor [47], are consistent with prior observations that the levels of C16-, C24- and C24:1- amide-linked fatty acid chain containing ceramides are significantly increased in malignant breast cancer tissue compared to benign tissue, from which it was suggested that increases in certain long-chain ceramides may contribute to or result from the carcinogenesis process [9].
Figure 4.
Comparison of the % total individual A) sphingomyelin (SM) and B) ceramide (Cer) lipid species in LIM1215 cells (dark bars) and LIM1215 secreted exosomes (white bars) assigned by positive ionization mode high resolution / accurate mass spectrometry from the data in Figure 1. Only the most abundant lipids are shown for clarity. The insets to each panel show the ratio of the total lipid ion abundances (normalized to protein) of species that contain at least one long chain fatty acid (≥ 20 carbons) to species that contain only short chain fatty acids, and the ratio of the abundances of species containing at least one unsaturated fatty acid to species containing only saturated fatty acids. Error bars show the standard deviations determined by triplicate measurement. * P < 0.05.
Interestingly, the LIM1215 exosome SM and Cer lipids also exhibited significant changes in the distribution of these abundant lipids compared to the parent cells. For example, a significantly decreased proportion of SM(34:1) was observed relative to the LIM1215 cells, with a concomitant increase in long chain SM(42:2) and SM(42:1) species, while a significant increase in Cer(34:1) and Cer(42:2) and a concomitant decrease in Cer(42:1) was observed for the Cer lipids. This later observation was also reported in the PC-3 exosome study [39]. The observed amide-linked fatty acid chain remodeling in the exosome derived SM lipids led to a significant increase in the ratio of the abundances of long chain versus short chain SM species (long chain amide-linked fatty acids are defined here as greater than or equal to C20, and short chain amide-linked fatty acids defined as less than C20), as well as a significant increase in the ratio of unsaturated versus saturated amide-linked fatty acid containing SM species (Figure 4A, inset). In contrast, the ratio of long chain versus short chain Cer species (Figure 4B, inset) decreased in the exosomes compared to the cells, while the ratio of unsaturated versus saturated amide-linked fatty acid containing Cer species (Figure 4B, inset) increased similar to the SM lipids.
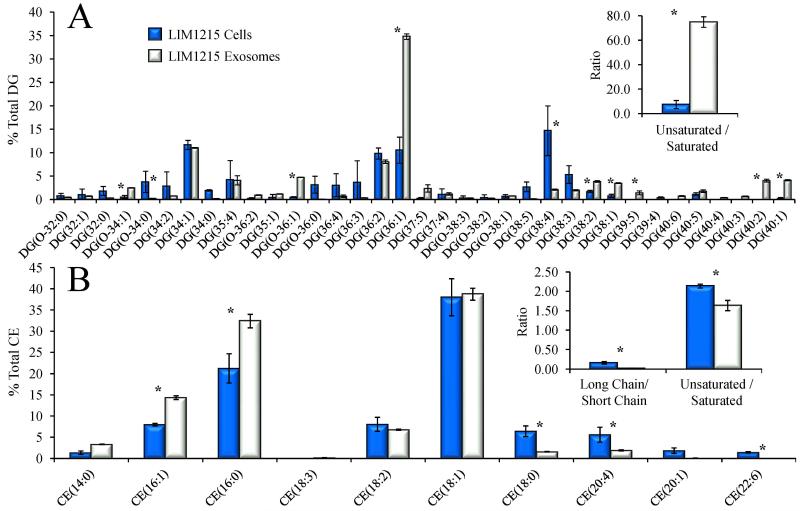
4.6 Comparison of diglyceride and cholesterol ester lipid species in LIM1215 exosomes and cells
A comparison of the relative distributions of diglycerides in the LIM1215 cell and exosome lipid extracts is shown in Figure 5A. Interestingly, this data clearly illustrates that exosomes contain a relatively larger proportion of the mono-unsaturated species DG(36:1), as well as other abundant mono- and poly-unsaturated species including DG(38:2), DG(38:1), DG(40:2) and DG(40:1) compared to the LIM1215 cellular diglycerides. The net effect of the disparate fatty acid content in cell and exosome diglycerides, seen in the inset of Figure 5A, is an approximate 12-fold increase in the ratio of the total abundances of unsaturated versus saturated fatty acids in exosomes compared to LIM1215 cells, similar to that observed for the exosome sphingomyelin lipids (compare the insets to Figure 4A and 5A).
Figure 5.
Comparison of the % total individual A) diglyceride (DG) and B) cholesterol ester (CE) molecular species in LIM1215 cells (dark bars) and LIM1215 secreted exosomes (white bars) detected as ammonium adducts by positive ionization mode high resolution / accurate mass spectrometry from the data in Figure 1. Only the most abundant lipids are shown for clarity. The inset to panel A shows the ratio of the total lipid ion abundances (normalized to protein) of DG species that contain at least one unsaturated fatty acid to species containing only saturated fatty acids. The inset to panel B shows the ratio of the total lipid ion abundances (normalized to protein) of CE species that contain at least one long chain fatty acid (≥ 20 carbons) to species that contain only short chain fatty acids, and the ratio of the abundances of species containing at least 1 unsaturated fatty acid to species containing only saturated fatty acids. Error bars show the standard deviations determined by triplicate measurement. * P < 0.05.
Figure 5B shows the changes observed in the percent total lipid abundances of the LIM1215 cell and exosome derived cholesterol ester lipids. These fatty acid-bearing sterol lipids are expected to reflect the pool of fatty acids generally available for complex lipid synthesis. As seen in the inset to Figure 5B, the ratios of the total abundances of long chain versus short chain fatty acids, and of unsaturated versus saturated fatty acids both show an overall decrease in the LIM1215 exosomes compared to their parent cells. Interestingly, the decrease in exosome cholesterol ester fatty acid length and unsaturation ratios is opposite to that observed for the sphingomyelin and diglyceride lipid sub-classes. The decrease in both these ratios in the cholesterol esters is principally due to relative decreases in the content of the polyunsaturated fatty acids 20:4 and 22:6 in the exosomes, while saturated and monounsaturated C14-C16 fatty acids are relatively increased compared to their levels in LIM1215 cells. The pool of fatty acids esterified to cholesterol therefore does not necessarily reflect the fatty acids preferentially incorporated into more complex exosome lipids, such as diglycerides and sphingomyelin, suggesting that exosomal lipids may arise from distinct cellular subdomains or via selective inclusion of specific lipids into exosomal vesicles.
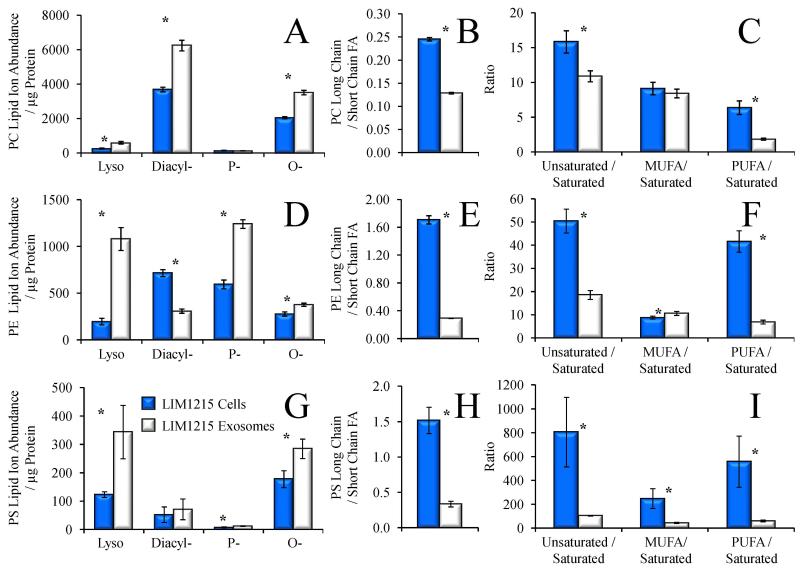
4.7 Comparison of phospholipid subclasses and fatty acid ratios in LIM1215 exosomes and cells
In addition to the information obtained regarding the global lipid profiles and changes in relative ion distributions of specific lipid classes and the length and degree of unsaturation of acyl chain compositions within specific lipids between the LIM1215 cells and their secreted exosomes, the lipid analysis workflow employed here also enabled rapid assignment of the distribution of lyso-, diacyl-, plasmalogen (P-) and alkyl ether (O-) sub-classes within the lipid classes shown in Figure 3C. For example, Figure 6A shows the total protein-adjusted normalized ion abundances for each glycerophosphatidylcholine (PC) subclass in the LIM1215 cells and their secreted exosomes. From this data it can be seen that in addition to the overall increase in total PC lipid ion abundance shown in Figure 3C, the distribution of PC lipids within the exosomes is significantly shifted toward lyso-, diacyl-, and O-PC lipids compared to the parent LIM1215 cells. Figure 6B depicts the ratio of total normalized abundances of long chain versus short chain containing fatty acids of the PC species, which decreased approximately 50% in the LIM1215 exosomes compared to their parent cells, while Figure 6C shows the ratio of the total normalized abundances of unsaturated versus saturated PC species, similar to those shown in Figure 4 and Figure 5 for other lipid classes. Here, however, the unsaturated PC lipids were further subdivided into species that contain monounsaturated fatty acids (MUFA) or at least one polyunsaturated fatty acid (PUFA), then their total normalized abundances were expressed relative to the total normalized abundances of PC species containing only saturated fatty acids. Overall, a marked decrease was observed in the ratio of exosome PC unsaturated to saturated fatty acids, which was exclusively due to a drastic decrease in PUFA-containing PC species, as the PC species containing only MUFA did not change significantly between the LIM1215 cells and exosomes.
Figure 6.
Characterization of phospholipid subclasses and fatty acid ratios from the data in Figure 1. Comparison of A) the total lipid ion abundances (normalized to protein) of PC lyso-, diacyl-, plasmalogen (P-), and alkyl ether (O-) subclasses, B) the ratio of the total abundance of PC species containing at least one long chain fatty acid (≥ 20 carbons) to PC species containing only short chain fatty acids, and C) the ratio of the abundance of PC species containing at least one unsaturated double bond to PC species containing only saturated fatty acids in LIM1215 cells (dark bars) and LIM1215 secreted exosomes (white bars). Comparison of D) the lipid ion abundance (normalized to protein) of PE lyso-, diacyl-, plasmalogen (P-), and alkyl ether (O-) subclasses, E), the ratio of the abundance of PE species containing at least one long chain fatty acid (≥ 20 carbons) to PE species containing only short chain fatty acids, and F) the ratio of the abundance of PE species containing at least one unsaturated double bond to PE species containing only saturated fatty acids in LIM1215 cells (dark bars) and LIM1215 secreted exosomes (white bars). MUFA, monounsaturated fatty acid. PUFA, polyunsaturated fatty acid. Error bars show the standard deviations determined by triplicate measurement. * P < 0.05.
Figure 6D-F and Figure 6G-I show an identical set of measurements obtained from the glycerophosphatidylethanolamine (PE) and glycerophosphatidylserine (PS) lipids, respectively. Similar to the results shown for the PC lipids in Figure 6A, the total normalized abundances of exosome derived lyso-, diacyl-, plasmalogen (P-) and alkyl ether (O-) sub-classes of PE and PS lipids (Figure 6D and 6G) both exhibited sharp divergence from their parent cells. However, in contrast to the results obtained for the PC subclasses, the exosome PE lipids were dominated by increases in lyso- and plasmalogen subclasses, and alkyl-ether lipids to a lesser extent, with a sharp decrease in diacyl species relative to the parent LIM1215 cells, whereas the PS lipids underwent an increase only in the lyso-lipid subclass, and a decrease in alkyl-ether linked species relative to their parent LIM1215 cells. Figures 6E and 6H demonstrate that the exosome PE and PS lipid pool both underwent significant decreases in the ratio of long chain versus short chain containing fatty acids, similar to the PC lipids, along with significantly lower ratios of unsaturated to saturated fatty acids, principally due to a decrease in exosome PUFA content compared to the LIM1215 parent cells for the PE lipids (Figure 6F), and a decrease in both MUFA and PUFA for the PS lipids (Figure 6I).
The existence of atypical ether-linked PS lipids, as well as ether-linked PI, PA and phosphatidylthreonine (PT) lipids), have been reported previously by several groups. For example, Ivanova et al. identified these species in macrophages [51], while Deeley et al. reported the presence of several ether-linked PS species in human lens tissue [52]. Other studies have also observed some of these ether lipid classes in differentiating leukemia cells [53], and in primary and metastatic colorectal cancer cell lines [44], e.g., ether linked-PI and ether-linked LPS, respectively. Increased levels of cellular plasmalogen lipids have previously been proposed to play a role as endogenous antioxidants [54], in facilitating membrane fusion [55], in the regulation of membrane cholesterol ester processing [56] and have also been correlated with the levels of several oncogenic signaling lipids involved in the regulation of cell survival, cancer aggressiveness, and tumor growth [12]. Interestingly, Phuyal et al. recently reported that the ether lipid precursor hexadecylglycerol added to PC-3 cells doubled the cellular levels of ether lipids in the cells, with increased levels of ether lipids also observed in their exosomes. Furthermore, cells containing high levels of ether lipids were found to release more exosomes than control cells [57]. To date however, the possible functional role of ether lipids in exosome mediated intercellular signaling have not yet been explored.
Taken together, these results demonstrate that exosome glycerophospholipid compositions are clearly distinct from the parent cells from which the exosomes were secreted, indicating that exosome formation and/or secretion requires unique partitioning of particular lipid classes and subclasses, while requiring fatty acids with differing degrees of length and unsaturation. Moreover, the decreases in exosome PC, PE and PS long chain to short chain fatty acid ratios, and unsaturated to saturated fatty acid ratios relative to the LIM1215 parent cells follow the same general patterns observed for cholesterol ester fatty acids. Cholesterol ester fatty acid content may therefore reflect exosome incorporation of fatty acids into phospholipids, but not neutral lipids or sphingolipids, further illustrating that exosomes utilize specific subsets of cellular lipids which may themselves arise from discreet pools of fatty acids or the preferential incorporation of specific types of fatty acids in a lipid class-specific manner.
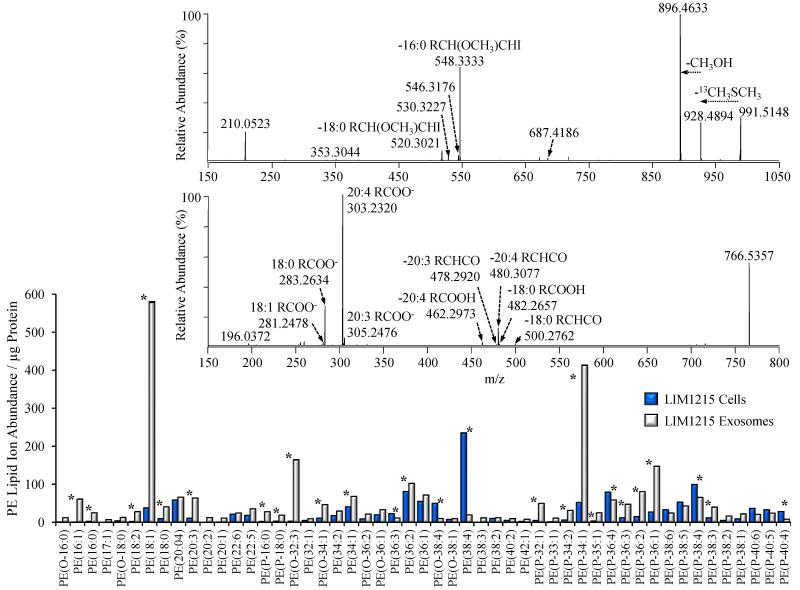
4.8 Comparison of LIM1215 cell and secreted exosome individual PE species and their confirmation and structural characterization by high resolution HCD-MS/MS
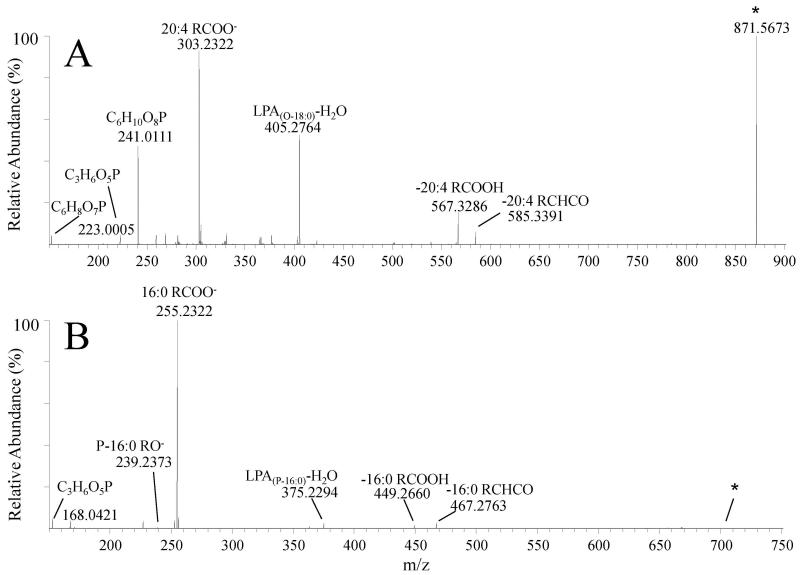
The bar graph in Figure 7 shows the fifty most abundant PE species identified in the LIM1215 cells and secreted exosomes, that were initially assigned at the sum composition level from the high resolution accurate mass spectra in Figure 2. Here, the PE species are expressed as their protein-adjusted normalized ion abundances in order to allow an examination of the absolute, rather than relative, changes in PE lipid distributions between the LIM1215 cells and exosomes. Numerous striking differences are apparent in the exosome PE species compared to the LIM1215 cells. While the LIM1215 cells clearly contain a higher abundance of the diacyl PE(38:4) species compared to the exosomes, the exosomes in general contain a greater abundance of most PE species (in agreement with Figure 3C), with particularly large increases in lyso PE(18:1), alkyl ether PE(O-32:3), and the plasmalogen lipids PE(P-34:1) and PE(P-36:1).
Figure 7.
Identification of LIM1215 cell and secreted exosome PE, P-PE, O-PE, LPE, P-LPE and O-LPE lipid species by high resolution / accurate mass HCD-MS/MS. Only the top 50 most abundant lipids are shown for clarity. Comparison of the total lipid ion abundances (normalized to protein) of PE molecular species in LIM1215 cells (dark bars) and LIM1215 secreted exosomes (white bars) assigned by positive ionization mode accurate mass spectrometry from the data in Figure 1. Error bars show the standard deviations determined by triplicate measurement. * P < 0.05. Top Inset: Positive ionization mode high resolution (R=100,000) HCD-MS/MS analysis to confirm the identity of the assigned PE(P-34:1) lipid, acquired from the LIM1215 exosome lipid extract derivatized with 13C1-DMBNHS and iodine/methanol. Bottom Inset: Negative ionization mode high resolution (R=100,000) HCD-MS/MS analysis to confirm the identity of the assigned PE(38:4) lipid, acquired from an underivatized LIM1215 cell lipid extract. Key product ions used for structural characterization are labeled.
High resolution accurate mass based lipid assignments afford a high degree of confidence in assigning lipid identities at the sum composition level, and their abundances, provided that the analytical conditions are properly controlled [45] and that sufficient instrument resolution exists to provide mass accuracy at the parts per million (ppm) or sub-ppm levels. However, high resolution tandem mass spectrometry (MS/MS) in both positive and negative ionization modes from derivatized and underivatized lipid extracts provide a valuable mechanism for confirmation of the initially assigned lipid species, and, when desired, further molecular characterization of lipids of interest from exosomes and their parent cells by generating characteristic product ions corresponding to the lipid headgroup and the fatty acid and/or backbone components of individual lipid species.
For example, in order to confirm the assignment of the 13C1-DMBNHS and iodine/methanol derivatized plasmalogen PE(P-34:1) species at m/z 991.5148 in Figure 2B, and to determine the precise identity of its acyl and alkenyl chain constituents, the ion was isolated and subjected to ‘targeted’ high resolution positive ionization mode higher energy collisional dissociation (HCD)-MS/MS analysis (top inset to Figure 7). From this spectrum, characteristic product ions indicative of the presence of an abundant plasmalogen PE(P-16:0_18:1) species, along with two other lower abundance PE(P-16:1_18:0) PE(P-18:0_16:1) lipids were observed. The key ions that enabled the identification of these lipids included m/z 928.4894, m/z 896.4633 and m/z 210.0523, indicative of the neutral loss of 13C dimethylsulfide, loss of 13C dimethylsulfide and methanol, and a protonated derivatized PE head group product ion, respectively, along with m/z 548.3333, m/z 546.3176 and m/z 520.3021 indicative of the neutral losses of 13C dimethylsulfide and a methanol/iodine derivatized P-16:0, P-16:1 and P-18:0 alcohols, respectively, as previously described [46]. The ability to readily identify and structurally characterize plasmalogen lipids by positive ion HCD-MS/MS from the derivatized lipid extracts is an additional significant benefit of the lipidomics workflow described in the present study.
The acquisition of negative ionization mode HCD-MS/MS data of underivatized lipids enables the elucidation of the fatty acid content of most diacyl lipid species, particularly for glycerophospholipid classes. As an example, the lower inset of Figure 7 shows the spectrum obtained by negative ionization mode HCD-MS/MS of m/z 766.5357 from an underivatized LIM1215 cell lipid extract (corresponding to the 13C1-DMBNHS derivatized diacyl PE(38:4) species at m/z 1041.5306 in Figure 2A). Identification of the PE(38:4) fatty acid substituents was performed via observation of characteristic product ions formed upon neutral losses of fatty acid (RCOOH) and ketene (RCHCO) species, indicative of the presence of an abundant PE(18:0_20:4) species, along with a low abundance PE(18:1_20:3) lipid (i.e., m/z 500.2762 (neutral loss of 18:0 ketene), m/z 482.2657 (neutral loss of 18:0 fatty acid), m/z 480.3077 (neutral loss of 20:4 ketene), m/z 478.2920 (neutral loss of 20:3 ketene) and m/z 462.2973 (loss of 20:4 fatty acid)). Given that positional isomers of the sn1 and sn2 fatty acids may be present, and the MS/MS data do not provide sufficient information for distinguishing one versus the other, we have utilized the descriptive nomenclature for lipid species suggested by Liebisch et. al. [40]. While abundant fatty acid anions are also present in the HCD-MS/MS spectrum (e.g., the 20:4 and 18:0 fatty acid anions at m/z 303.2320 and m/z 283.2634, respectively), the use of the product ions resulting from fatty acid or ketene neutral losses from the PE(38:4) parent ion are preferred for unequivocal identification of the fatty acid constituents, due to the possibility of other precursor ions also being present within the nominal m/z isolation window used for HCD-MS/MS that may also give rise to the fatty acid anion products. Additional examples of the application of negative ion mode MS/MS to confirm the sum composition assignments initially based on accurate mass measurements, and for further structural characterization of acyl, alkyl or alkenyl chain compositions, of two of the atypical ether-linked lipids observed in this study (PI(O-38:4) from the LIM1215 cells and PG(P-32:0) from the LIM1215 exosomes), are shown in Figure 8.
Figure 8.
Confirmation of accurate mass ‘sum’ composition assignments and structural characterization of LIM1215 cell and secreted exosome atypical ether-linked lipids (A) PI(O-38:4) acquired from an underivatized LIM1215 cell lipid extract (assigned as PI(O-18:0_20:4)) and (B) PG(P-32:0) acquired from an underivatized LIM1215 exosome lipid extract (assigned as PG(P-16:0_16:0)), by high resolution / accurate mass negative ion mode HCD-MS/MS. Key product ions used for structural characterization are labeled.
5. Conclusions
A comprehensive understanding of cancer cell secreted exosome lipid molecular compositions is expected to provide an improved understanding of the functional role that exosome derived lipids play in intercellular signaling processes associated with the onset, progression and metastatic transformation of cancer cells. However, the detailed identification and characterization of exosome lipids requires exceedingly sensitive analytical techniques capable of maximizing the depth of information obtainable from the limited sample quantities that are typically available. The workflow outlined herein provides a rapid and robust lipidome analysis platform that meets this requirement. Using this approach, and from a single positive ion high resolution mass spectrum, we have achieved the most comprehensive exosome lipid analysis reported to date, with lipid assignments at the sum composition level across a broad range of glycerophospholipid, glycerolipid, sphingolipid, and sterol lipid classes and subclasses, and across several orders of magnitude of ion abundance. Importantly, the results from this simple analysis procedure are in broad agreement with other studies on the lipid compositions of exosomes from other cancer types, and therefore may be used to provide insights into the role of exosomes in inter-cellular communication and the onset, progression and metastatic transformation of cancer cells.
Highlights.
Novel workflow for rapid and comprehensive cancer exosome lipid characterization.
Lipid content of LIM1215 exosomes is distinct from their parent colon cancer cells.
Most in-depth characterization of exosome lipid profiles to date.
6. Acknowledgements
Support for this work was provided by funding from an Australian Research Council Discovery Project grant to SM and GER (DP130100535), and from the National Institutes of Health to GER (GM103508). SM is supported by an Australian NH&MRC fellowship (1016599) and an Australian Research Council DECRA fellowship (DE150101777). The funding bodies had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
List of Abbreviations
- BHT
butylated hydroxytoluene
- CE
cholesterol ester
- Cer
ceramide
- Chol
cholesterol
- 13C1-DMBNHS
13C1-S,S’-dimethylthiobutanoylhydroxysuccinimide ester
- DG
diradylglycerol
- DMF
N,N-dimethylformamide
- FCS
fetal calf serum
- HCD
higher-energy collisional dissociation
- Hex-Cer
hexosylceramide (cerebroside)
- LC
Liquid chromatography
- LPC
lysoglycerophosphocholine
- LPE
lysoglycerophosphoethanolamine
- LPG
lysoglycerophosphoglycerol
- LPS
lysoglycerophosphoserine
- MG
monoradylglycerol
- MS
mass spectrometry
- MUFA
monounsaturated fatty acid
- nESI
nano-electrospray ionization
- O-
alkyl ether lipid
- P-
plasmalogen lipid
- PBS
phosphate buffered saline
- PC
glycerophosphocholine
- PE
glycerophosphoethanolamine
- PG
glycerophosphoglycerol
- PI
glycerophosphoinositol
- PS
glycerophosphoserine
- PUFA
polyunsaturated fatty acid
- SM
sphingomyelin
- Tandem mass spectrometry (MS/MS)
- TEA
triethylamine
- TEM
transmission electron microscopy
- TG
triradylglycerol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References
- 1.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 2.Currie E, Schulze A, Zechner R, Walther TC, Farese RV. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritz V, Fajas L. Metabolism and proliferation share common regulatory pathways in cancer cells. Oncogene. 2010;29:4369–4377. doi: 10.1038/onc.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruggieri S, Mugnai G, Mannini A, Calorini L, Fallani A, Barletta E, Mannori G, Cecconi O. Lipid Characteristics in Metastatic Cells. Clin. Exp. Metastasis. 1999;17:271–276. doi: 10.1023/a:1006662811948. [DOI] [PubMed] [Google Scholar]
- 5.Roos DS, Choppin PW. Tumorigenicity of cell lines with altered lipid composition. Proc. Natl. Acad. Sci. USA. 1984;81:7622–7626. doi: 10.1073/pnas.81.23.7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azordegan N, Fraser V, Le K, Hillyer LM, Ma DWL, Fischer G, Moghadasian MH. Carcinogenesis alters fatty acid profile in breast tissue. Mol. Cell. Biochem. 2013;374:223–232. doi: 10.1007/s11010-012-1523-4. [DOI] [PubMed] [Google Scholar]
- 7.Fernandis AZ, Wenk MR. Lipid-based biomarkers for cancer. J. Chromatogr. B. 2009;877:2830–2835. doi: 10.1016/j.jchromb.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 8.Louie SM, Roberts LS, Mulvihill MM, Luo K, Nomura DK. Cancer cells incorporate and remodel exogenous palmitate into structural and oncogenic signaling lipids. Biochim Biophys Acta. 2013;1831:1566–1572. doi: 10.1016/j.bbalip.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryland LK, Fox TE, Liu X, Loughran TP, Kester M. Dysregulation of sphingolipid metabolism in cancer. Cancer Biol Ther. 2011;11:138–149. doi: 10.4161/cbt.11.2.14624. [DOI] [PubMed] [Google Scholar]
- 10.Braverman NE, Moser AB. Functions of plasmalogen lipids in health and disease. Biochim. Biophys. Acta. 2012;1822:1442–1452. doi: 10.1016/j.bbadis.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Snyder F, Wood R. Alkyl and alk-1-enyl ethers of glycerol in lipids from normal and neoplastic human tissues. Cancer Res. 1969;29:251. [PubMed] [Google Scholar]
- 12.Benjamin DI, Cozzo A, Ji X, Roberts LS, Louie SM, Mulvihill MM, Luo K, Nomura DK. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc Natl Acad Sci U S A. 2013;110:14912–14917. doi: 10.1073/pnas.1310894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith RR, White HB. Neutral Lipid Patterns of Normal and Pathologic Nervous Tissue. Arch. Neurol. 1968;19:54–59. doi: 10.1001/archneur.1968.00480010072005. [DOI] [PubMed] [Google Scholar]
- 14.Dória ML, Cotrim Z, Macedo B, Simões C, Domingues P, Helguero L, Domingues MR. Lipidomic approach to identify patterns in phospholipid profiles and define class differences in mammary epithelial and breast cancer cells. Breast Cancer Res. Treat. 2012;133:635–648. doi: 10.1007/s10549-011-1823-5. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Z, Xiao Y, Elson P, Tan H, Plummer SJ, Berk M, Aung PP, Lavery IC, Achkar JP, Li L, Casey G, Xu Y. Plasma Lysophosphatidylcholine Levels: Potential Biomarker for Colorectal Cancer. J. Clin. Oncol. 2007;25:2696–2701. doi: 10.1200/JCO.2006.08.5571. [DOI] [PubMed] [Google Scholar]
- 16.Smith RE, Lespi P, Di Luca M, Busos C, Marra FA, de Alaniz MJT, Marra CA. A Reliable Biomarker Derived from Plasmalogens to Evaluate Malignancy and Metastatic Capacity of Human Cancers. Lipids. 2008;43:79–89. doi: 10.1007/s11745-007-3133-6. [DOI] [PubMed] [Google Scholar]
- 17.Min HK, Lim S, Chung BC, Moon MH. Shotgun lipidomics for candidate biomarkers of urinary phospholipids in prostate cancer. Anal. Bioanal. Chem. 2011;399:823–830. doi: 10.1007/s00216-010-4290-7. [DOI] [PubMed] [Google Scholar]
- 18.Mathivanan S, Ji H, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. Journal of Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immun. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 20.Simons M, Raposo G. Exosomes-vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Lee TH, D'Asti E, Magnus N, Al-Nedawi K, Meehan B, Rak J. Microvesicles as mediators of intercellular communication in cancer-the emerging science of cellular 'debris'. Semin Immunopathol. 2011;33:455–467. doi: 10.1007/s00281-011-0250-3. [DOI] [PubMed] [Google Scholar]
- 22.Urbanelli. A. Magini L, Buratta S, Brozzi A, Sagini K, Polchi A, Tancini B, Emiliani C. Signaling Pathways in Exosomes Biogenesis, Secretion and Fate. Genes. 2013;4:152–170. doi: 10.3390/genes4020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rabinowits C, Gercel-Taylor JM, Day DD, Taylor GH. Kloecker, Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. G. [DOI] [PubMed] [Google Scholar]
- 24.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers for ovarian cancer. Gynecol. Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 25.Yu S, Liu C, Su K, Wang J, Liu Y, Zhang L, Li C, Cong Y, Kimberly R, Grizzle WE, Falkson C, Zhang H. Tumor Exosomes Inhibit Differentiation of Bone Marrow Dendritic Cells. J Immunol. 2007;178:6867–6875. doi: 10.4049/jimmunol.178.11.6867. [DOI] [PubMed] [Google Scholar]
- 26.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer Exosomes Trigger Fibroblast to Myofibroblast Differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 27.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar CM, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma Exosomes Educate Bone Marrow Progenitor Cells Toward a Pro-Metastatic Phenotype Through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochimica et Biophysica Acta. 2014;1841:108–120. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Record M. Exosomal Lipids in Cell–Cell Communication. In: Zhang H-G, editor. Emerging Concepts of Tumor Exosome–Mediated Cell–Cell Communication. Springer; New York: 2013. pp. 47–68. [Google Scholar]
- 30.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 31.Beloribi S, Ristorcelli E, Breuzard G, Silvy F, Betrand-Michel J, Beraud E, Verine A, Lombardo D. Exosomal Lipids Impact Notch Signaling and Induce Death of Human Pancreatic Tumoral SOJ-6 Cells. PLoS ONE. 2012;7:e47480. doi: 10.1371/journal.pone.0047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, Colone M, Tatti M, Sargiacomo M, Fais S. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, Paillasse M, De Medina P, Monsarrat B, Perret B, Silvente-Poirot S, Poirot M, Record M. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010;51:2105–2120. doi: 10.1194/jlr.M003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laulagnier K, Grand D, Dujardin A, Hamdi S, Vincent-Schneider H, Lankar D, Salles JP, Bonnerot C, Perret B, Record M. PLD2 is enriched on exosomes and its activity is correlated to the release of exosomes. FEBS Lett. 2004;572:11–14. doi: 10.1016/j.febslet.2004.06.082. [DOI] [PubMed] [Google Scholar]
- 35.Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–212. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Carayon K, Chaoui K, Ronzier E, Lazar I, Bertrand-Michel J, Roques V, Balor S, Terce F, Lopez A, Salome L, Joly E. Proteolipidic Composition of Exosomes Changes during Reticulocyte Maturation. J Biol Chem. 2011;286:34426–34439. doi: 10.1074/jbc.M111.257444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laulagnier K, Motta C, Hamdi S, Roy S, Fauvelle F, Pageaux J, Kobayashi T, Salles J, Perret B, Bonnerot C, Record M. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004;380:161–171. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rappa G, Mercapide J, Anzanello F, Pope RM, Lorico A. Biochemical and biological characterization of exosomes containing prominin-1/CD133. Molecular Cancer. 2013;12:62. doi: 10.1186/1476-4598-12-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Llorente A, Skotland T, Sylvanne T, Kauhanen D, Rog T, Orlowski A, Vattulainen I, Ekroos K, Sandvig K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim Biophys Acta. 2013;1831:1302–1309. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 40.Liebisch G, Vizcaino JA, Kofeler H, Trotzmuller M, Griffiths WJ, Schmitz G, Spener F, Wakelam MJ. Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res. 2013;54:1523–1530. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwudke D, Schuhmann K, Herzog R, Bornstein SR, Shevchenko A. Shotgun Lipidomics on High Resolution Mass Spectrometers. Cold Spring Harb. Perspect. Biol. 2011;3:a004614. doi: 10.1101/cshperspect.a004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuhmann K, Herzog R, Schudke D, Metelmann-strupat W, Bornstein SR, Shevchenko A. Bottom-up shotgun lipidomics by higher energy collisional dissociation on LTQ orbitrap mass spectrometers. Anal. Chem. 2011;83:5480–5487. doi: 10.1021/ac102505f. [DOI] [PubMed] [Google Scholar]
- 43.Schuhmann K, Almeida R, Baumert M, Herzog R, Bornstein SR, Shevchenko A. Shotgun Lipidomics on a LTQ Orbitrap Mass Spectrometer by Successive Switching Between Acquisition Polarity Modes. J. Mass Spectrom. 2012;47:96–104. doi: 10.1002/jms.2031. [DOI] [PubMed] [Google Scholar]
- 44.Fhaner CJ, Liu S, Ji H, Simpson RJ, Reid GE. Comprehensive Lipidome Profiling of Isogenic Primary and Metastatic Colon Adenocarcinoma Cell Lines. Anal Chem. 2012;84:8917–8926. doi: 10.1021/ac302154g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lydic TA, Busik JV, Reid GE. A Monophasic Extraction Strategy for the Simultaneous Lipidome Analysis of Polar and Nonpolar Retina Lipids. J Lipid Res. 2014;55:1797–1809. doi: 10.1194/jlr.D050302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fhaner CJ, Liu S, Zhou X, Reid GE. Functional Group Selective Derivatization and Gas-Phase Fragmentation Reactions of Plasmalogen Glycerophospholipids. Mass Spectrom (Tokyo) 2013;2:S0015. doi: 10.5702/massspectrometry.S0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitehead RH, Macrae FA, St John DJ, Ma J. A colon cancer cell line (LIM1215) derived from a patient with inherited nonpolyposis colorectal cancer. J. Natl. Cancer Inst. 1985;74:759–765. [PubMed] [Google Scholar]
- 48.Haimi P, Uphoff A, Hermansson M, Somerharju P. Software tools for analysis of mass spectrometric lipidome data. Anal. Chem. 2006;78:8324–8331. doi: 10.1021/ac061390w. [DOI] [PubMed] [Google Scholar]
- 49.Fahy E, Cotter D, Sud M, Subramaniam S. Lipid classification, structures and tools. Biochim Biophys Acta. 2011;1811:637–647. doi: 10.1016/j.bbalip.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, Bond VC, Borràs FE, Breakefield X, Budnik V, Buzas E, Camussi G, Clayton A, Cocucci E, Falcon-Perez JM, Gabrielsson S, Gho YS, Gupta D, Harsha HC, Hendrix A, Hill AF, Inaal JM, Jenster G, Kiang LS, Krämer-Albers E-M, Llorente A, Lötvall J, Mincheva-Nilsson L, Nazarenko I, Nieuwland R, Nolte-'t Hoen ENM, Pandey A, Patel T, Piper MG, Pluchino S, Prasad TS, Rajendran L, Raposo G, Record M, Reid GE, Sánchez-Madrid F, Schiffelers RM, Siljander P, Stoorvogel W, Taylor D, Thery C, Valadi H, van Balkom BWM, Vázquez J, Vidal M, Yáñez-Mó M, Zoeller M, Mathivanan S. Vesiclepedia: A compendium for extracellular vesicles that enables continuous community annotation. PLoS Biol. 2012;10:e1001450. doi: 10.1371/journal.pbio.1001450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivanova PT, Milne SB, Brown H. A Identification of atypical ether-linked glycerophospholipid species in macrophages by mass spectrometry. J. Lipid Res. 2010;51:1581–1590. doi: 10.1194/jlr.D003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deeley JM, Thomas MC, Truscott RJW, Mitchell TW, Blanksby SJ. Identification of Abundant Alkyl Ether Glycerophospholipids in the Human Lens by Tandem Mass Spectrometry Techniques. Anal. Chem. 2009;81:1920–1930. doi: 10.1021/ac802395d. [DOI] [PubMed] [Google Scholar]
- 53.Naito M, Kudo I, Nakagawa Y, Waku K, Nojiri H, Saito M, Inoue K. Lipids of human promyelocytic leukemia cell HL-60: increasing levels of ether-linked phospholipids during retinoic acid-induced differentiation. J. Biochem. 1987;102:155–162. doi: 10.1093/oxfordjournals.jbchem.a122027. [DOI] [PubMed] [Google Scholar]
- 54.Lessig J, Fuchs B. Plasmalogens in biological systems: their role in oxidative processes in biological membranes, their contribution to pathological processes and aging and plasmalogen analysis. Curr. Med. Chem. 2009;16:2021–2041. doi: 10.2174/092986709788682164. [DOI] [PubMed] [Google Scholar]
- 55.Glaser PE, Gross RW. Plasmenylethanolamine facilitates rapid membrane fusion: a stopped-flow kinetic investigation correlating the propensity of a major plasma membrane constituent to adopt an HII phase with its ability to promote membrane fusion. Biochemistry. 1994;33:5805–5812. doi: 10.1021/bi00185a019. [DOI] [PubMed] [Google Scholar]
- 56.Mankidy R, Ahiahonu PWK, Ma H, Jayasinghe D, Ritchie SA, Khan MA, Su-Myat KK, Wood PL, Goodenowe DB. Membrane plasmalogen composition and cellular cholesterol regulation: a structure activity study. Lipids Health Dis. 2010;9:62. doi: 10.1186/1476-511X-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phuyal S, Skotland T, Hessvik NP, Simolin H, Øverbye A, Brech A, Parton RG, Ekroos K, Sandvig K, Llorente A. The Ether Lipid Precursor Hexadecylglycerol Stimulates the Release and Changes the Composition of Exosomes Derived from PC-3 Cells. J. Biol. Chem. 2015;290:4225–4237. doi: 10.1074/jbc.M114.593962. [DOI] [PMC free article] [PubMed] [Google Scholar]