Abstract
Pre-mRNA splicing is an essential step in gene expression that removes intron sequences efficiently and accurately to produce a mature mRNA for translation. It is the large and dynamic RNA-protein complex called the spliceosome that catalyzes intron removal. To carry out splicing the spliceosome not only needs to assemble correctly with the pre-mRNA but the spliceosome requires extensive remodelling of its RNA and protein components to execute the 2 steps of intron removal. Spliceosome remodelling is achieved through the action of ATPases that target both RNA and proteins to produce spliceosome conformations competent for each step of spliceosome activation, catalysis and disassembly. An increasing amount of research has pointed to the spliceosome associated NineTeen Complex (NTC) of proteins as targets for the action of a number of the spliceosomal ATPases during spliceosome remodelling. In this point-of-view article we present the latest findings on the changes in the NTC that occur following ATPase action that are required for spliceosome activation, catalysis and disassembly. We proposed that the NTC is one of the main targets of ATPase action during spliceosome remodelling required for pre-mRNA splicing.
Keywords: ATPase, Brr2, Cwc2, NineTeen Complex, PremRNA splicing, Prp19, Prp2, Prp16, Prp43, RNA helicase
Introduction
Splicing of pre-mRNA (pre-mRNA) is a complex mechanism where introns are removed, and exons are joined together to form a mature mRNA competent for translation. Pre-mRNA splicing is tightly regulated and its failure is linked to various tumors, pathologies of the endocrine system and neurodegenerative disorders.1 This intrinsic process of splicing is mediated by a multimegadalton ribonucleoprotein complex called the spliceosome. The spliceosome consists of 5 small nuclear RNAs (snRNAs), U1, U2, U4, U5 and U6, which are associated with proteins forming ribonucleoprotein particles (snRNPs).2,3 The snRNPs assemble with the pre-mRNA and are then remodelled into a number of specific complexes required for the 2 steps of splicing (Fig. 1). The process of splicing begins when the U1 and U2 snRNAs of the U1 and U2 snRNPs recognize, by base-pairing, the 5' splice site and the branch site of the pre-mRNA, respectively, to form the A complex. Then a preformed tri-snRNP, containing U4/U6 and U5 snRNPs, joins complex A to form complex B. At the same time a protein complex associated with Prp19, named the NineTeen Complex (NTC), also joins the spliceosome.4-6 Next, the spliceosome goes through dramatic rearrangements to form the Bact then B* complexes which involves dissociation of the U1 and U4 snRNPs as well as the removal and addition of certain proteins.7 The B* complex is now the catalytically activated spliceosome and is competent to carry out the first step of splicing, which is attack of the 5′ splice site phosphate by the 2′ hydroxyl of the branch site adenosine. Following the first step of splicing the B* complex is then rearranged to form the C complex through the removal of proteins and the addition of proteins that promote the second step of splicing.8 The C complex carries out the second step of splicing which is attack of the 3′ splice site phosphate by the 3′ hydroxyl of the 5′ exon to remove the intron and join the 2 exons. The resulting splicing complex is called the post-splicing complex and this complex must be disassembled to release the mRNA, leaving the intron associated with the snRNPs as the intron-lariat spliceosome (ILS). Finally, the ILS is disassembled allowing recycling of the snRNPs for subsequent rounds of splicing. The rearrangements and conformational changes required for spliceosome assembly, activation and disassembly are catalyzed by the spliceosomal ATPases.9 In this “Point of View” we will describe an increasing amount of experimental evidence that identifies the NTC, and NTC-associated proteins, as targets for a number of the spliceosomal ATPases required for remodelling the spliceosome during pre-mRNA splicing. We will concentrate on the yeast Saccharomyces cerevisiae system where there is the most evidence to date for this idea.
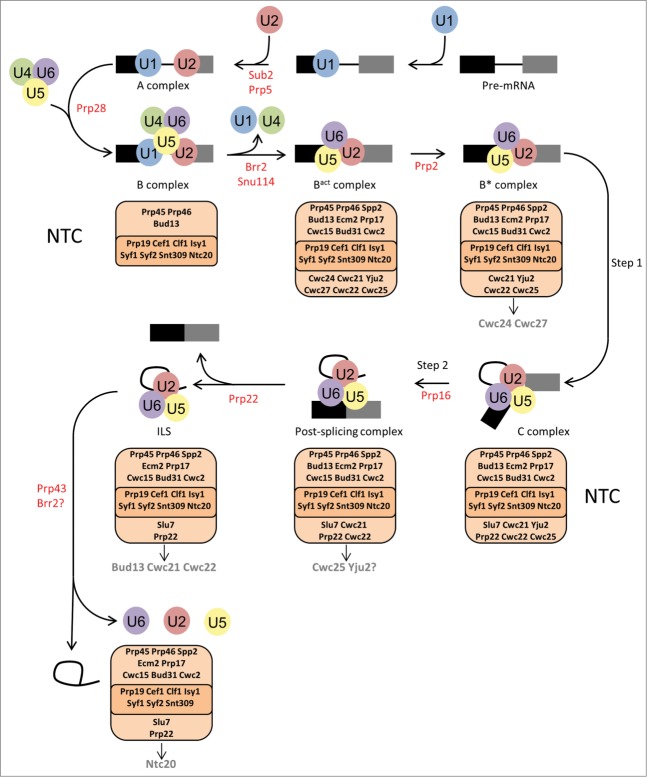
Figure 1.
NTC protein remodelling by ATPases during the spliceosome assembly, activation and disassembly process. The pathway of complexes formed on the pre-mRNA during spliceosome assembly, activation and disassembly are indicated with arrows and the names of each complex are given below the complex. The ATPases are shown in red under the arrow for the step that each ATPase promotes. The NTC core complex (dark orange) and NTC-associated proteins (light orange) are shown below each complex that they associate with. Arrows from the NTC complex are used to indicate the proteins that leave following ATPase action. Question marks are used to indicate that experimental evidence for Yju2 removal from the NTC and Brr2 action during spliceosome disassembly is not in agreement.
There are 8 ATPases involved in splicing (Table 1) and they share 8 conserved motifs divided into 2 domains, RecA1 and RecA2.9 The spliceosomal ATPases are used to modulate RNA-RNA, RNA-protein and protein-protein interactions during the splicing cycle.10 The ATPases promote correct conformations of the spliceosome for progression through the 2 steps of intron removal with accuracy and fidelity. It is the second role of some of the ATPases, that of proofreading certain steps of splicing, which provides the fidelity in the splicing process.11-15 Spliceosome conformations are monitored by the ATPases and can be rejected if a certain complex is not formed correctly or in a timely manner. The order of action of the ATPases during splicing is Sub2, Prp5, Prp28, Brr2, Prp2, Prp16, Prp22, Prp43 and Brr2 (Fig. 1).14 Of these ATPases, the action of Prp2, Prp16, Prp22, Prp43 and Brr2 appear to be related to changes in the interactions of the NTC and NTC-associated proteins with the spliceosome. The modulation of the NTC, therefore, is essential for the progression of the spliceosome through the splicing cycle.
Table 1.
Yeast Spliceosomal ATPases
| ATPase | Family | Function |
|---|---|---|
| Sub2/UAP56 | DEAD-box | Association of U2 snRNA with pre-mRNA |
| Prp5 | DEAD-box | Proofreads U2-branchsite interaction |
| Prp28 | DEAD-box | Release of U1 by disrupting the base-pairing between U1 and 5′splice site. |
| Prp2 | DEAH-box | Release of SF3a/SF3b |
| Prp16 | DEAH-box | Release of Yju2 and Cwc25 |
| U2/U6 helix I remodelling | ||
| Prp22 | DEAH-box | Release of mature mRNA |
| Prp43 | DEAH-box | Disassembly of the ILS |
| Brr2 | Ski2-like | Disrupt U4/U6 base-pairing |
| Disrupt U2/U6 base-pairing |
The NTC and NTC-associated proteins are found within the B, Bact, B*, C, post-splicing complex and ILS (Fig. 1).8,16,17 The core NTC complex nucleated by Prp19, is found in all these complexes, it is composed of 8 proteins, but an additional 18 NTC-associated proteins interact with, or co-purify with, these core proteins and can be found in one or more of the splicing complexes.5 At the core of the NTC, Prp19 forms tetramers via its central coiled-coil domain which are bound by Cef1.18 Prp19 also contains WD domains with 2 molecules of Cwc2 interacting with these WD domains in the Prp19 tetramer.19 The NTC is linked to the spliceosome active site through Cwc2 which interacts directly with both the U6 snRNA and the pre-mRNA.20,21 Recently, the S. pombe homolog of Cef1, called Cdc5, has been shown to bind double stranded RNA in vitro suggesting that Cef1/Cdc5 may also link the NTC to the active site RNAs of the spliceosome.22 The NTC, and NTC-associated proteins, are involved in a number of the spliceosome rearrangements and conformational changes during the splicing cycle. Associations and interactions of certain NTC proteins change following the action of certain ATPases. We now present the latest findings on the changes in the NTC that occur following ATPase action that are required for spliceosome activation, catalysis and disassembly.
Prp2 Remodelling of the Spliceosome for the First Step of Splicing Includes NTC Protein Remodelling
During the transition of the spliceosome B complex to the B* complex, which is now competent for the first step of splicing, it is the ATPase Prp2 that makes the final rearrangement of the spliceosome.23 The remodelling by Prp2 involves a number of NTC proteins. Prp2 action releases the SF3a/b complex associated with the U2 snRNP.24,25 The removal of the SF3a/b complex creates high affinity binding sites for the NTC proteins Cwc25 and Yju2. Both Cwc25 and Yju2 are required to enable the catalytically-activated spliceosome to carry out the first step of splicing.25,26 The action of Prp2 requires the NTC-associated proteins Spp2 and Cwc22.27-29 Prp2 remodelling also involves dissociation of the NTC proteins Cwc24 and Cwc27.7,25 Therefore, it is clear that the action of Prp2 not only requires NTC proteins but Prp2 action results in the association and dissociation of NTC proteins with the spliceosome required for the transition through the first step of splicing.
Prp16 Acts Through the NTC to Rearrange the Spliceosome for the Second Step of Splicing
Prp16 is an ATPase that proofreads the first step of splicing and promotes rearrangement of the spliceosome during the second step of splicing.11,12,30-32 During splicing a series of RNA-RNA interactions occur, and one crucial interaction is between the U2 and U6 snRNAs which base-pair to form helix I.33 The U2/U6 helix I is formed following the release of the U1 and U4 snRNPs, with helix I required for the first step of splicing then the second step and exon joining.30 It has been suggested that sequences encompassing helix I (U6 AGC triad) may form tertiary interactions with the U6 ACAGAGA box and the U6 internal stem loop (ISL) to bind a metal ion, enabling the spliceosome to have an active site resembling that found in group II introns.34-36 Evidence over time has pointed to a role for Prp16 in modulating a conformational change in the spliceosome involving the U2/U6 helix I.30,31,37,38 The U2/U6 helix I is destabilised between the 2 steps of splicing by Prp16 before helix I is reformed for the second step.30 As Prp16 interacts only transiently with the spliceosome, its influence on U2/U6 helix I must be applied through other spliceosome proteins. The first clue to how Prp16 could exert its action was found when deletion of the NTC protein Isy1 was shown to suppress the cold sensitive prp16–302 allele.38 This was the first link between Prp16 action and the NTC complex. Recently, we have found that another NTC protein, Cwc2, stabilizes U2/U6 helix I and appears to antagonize Prp16 action.39 The interactions of Cwc2 with the U6 snRNA and the pre-mRNA are influenced by Prp16 mutation.39 The prp16–302 allele stabilizes Cwc2 interactions with the U6 snRNA and destabilizes Cwc2 interactions with the pre-mRNA indicating that Cwc2 is one target for Prp16 action during splicing.39 Additionally, we have found that Cwc2 and Isy1 functionally cooperate during splicing.39 All together, these data point to the NTC proteins Isy1 and Cwc2, either directly or indirectly, as targets for Prp16 action in helix I remodelling during splicing.
In addition to modulating RNA-RNA and RNA-protein interactions between the 2 steps of splicing, there is evidence from both immunoprecipitation experiments and a purified yeast splicing system that Prp16 can also modulate the interactions of NTC proteins with the spliceosome prior to the second step of splicing.8,32 The binding of NTC proteins Cwc25 and Yju2 to the spliceosome, catalyzed by Prp2, is required to promote the first step of splicing.25,26 Following the first step of splicing Cwc25, and possibly Yju2, are removed to most likely allow new factors to bind to the spliceosome and promote the second step of splicing. It is the action of Prp16 that removes Cwc25, and potentially Yju2, after the first step of splicing. Using immunoprecipitation it was first shown that the action of Prp16 resulted in the release of Cwc25 and Yju2 from the spliceosome.32 However, recent work utilizing a purified yeast splicing system combined with mass spectrometry and dual-color fluorescence cross-correlation spectroscopy has found that Prp16 action causes a structural change in the spliceosome that reduces the binding affinity of Cwc25 allowing subsequent dissociation of Cwc25, but Prp16 action was not observed with this system to dissociate Yju2.8 Despite the conflicting data on Yju2 dissociation from the spliceosome, it is clear that the action of Prp16 influences the affinity of NTC proteins for the spliceosome to allow the second step of splicing.
Prp22 Dissociates the NTC Proteins Cwc21 and Cwc22 During Spliceosome Disassembly Along with the RES Complex
Following the second step of splicing the post-splicing complex must be disassembled to release the mRNA and recycle the snRNPs. The first step of the disassembly process is carried out through the action of the ATPase Prp22.40,41 During splicing, the U5 snRNA interacts with the 5’ exon and 3’ exon sequences to align the 2 exons for joining during the second step of splicing.42-47 After the second step of splicing, the interactions of the U5 snRNA with the 2 exon sequences are disrupted by the ATPase activity of Prp22 promoting release of the mature mRNA.48 Recent use of the purified yeast splicing system combined with mass spectrometry to follow the spliceosome disassembly process has revealed how the protein composition of the post-splicing complex changes following Prp22 action. It was found that the NTC-associated proteins Cwc21 and Cwc22 are significantly reduced in the ILS produced by Prp22 action.17 In addition, the RES (REtention and Splicing) complex proteins were also found to be significantly less abundant, or absent, from the ILS following Prp22 action.17 The RES complex associates with the B complex along with the NTC and is required for enhancing the splicing of certain pre-mRNAs and retention of unspliced pre-mRNAs.49-51 Significantly, the RES complex protein Bud13/Cwc26 is an NTC protein found to associate with Cef1.52 Therefore, it appears that the action of Prp22 targets proteins of the NTC to induce spliceosome disassembly.
Prp43 Disassembles the ILS to Allow Recycling of the snRNPs and the NTC for Further Rounds of Splicing
The second phase of spliceosome disassembly involves the removal of the snRNPs from the intron lariat RNA, but also dissociation of the snRNPs from each other, allowing the snRNPs to be recycled for subsequent rounds of splicing. The ATPase Prp43 is recruited for this disassembly step of the spliceosome. Prp43 associates with Ntr1 and Ntr2 (NTC-related proteins) and forms the NTR complex.53,54 It is Ntr1 that activates the ATPase activity of Prp43 to trigger release of the snRNPs from the intron lariat.55 The use of the purified yeast splicing system combined with mass spectrometry has also revealed how the protein composition of the snRNPs changes following Prp43 action to release the intron lariat and the snRNPs from each other. It has been found that the action of Prp43 completely dissociates the NTC protein Ntc20 from the snRNPs and intron-lariat.17 The other NTC proteins in the ILS appear to remain associated with the U2 and U5 snRNPs as well as the intron-lariat, but it is not clear how the NTC proteins are then further recycled from the released snRNPs and intron-lariat following Prp43 action.17 Nevertheless, it is apparent that the action of Prp43 influences NTC proteins during disassembly of the ILS.
Brr2 is Linked to the ATPases that Remodel the NTC During Splicing
The ATPase Brr2 is an essential U5 snRNP protein involved in remodeling RNA-RNA interactions during spliceosomal activation and disassembly.56 Brr2 disrupts the base-pairing of the U4/U6 snRNAs to promote the release of U4, but once the catalytic steps of splicing are completed, Brr2 again disrupts the base-pairing of U2/U6 marking the start of the spliceosome disassembly process.57-59 Brr2 activity is regulated by the GTPase Snu114.60 Brr2 contains 2 helicase cassettes, with the N-terminal cassette able to hydrolyse ATP whereas the C-terminal cassette has evolved into a protein binding module.56 While it does not appear that Brr2 ATPase action directly influences the NTC, Brr2 is known to interact with a number of ATPases that do remodel the NTC during splicing. Brr2 has been shown to interact with Prp2 by the 2-hybrid assay but also directly by pull-down assays.61,62 It has been proposed that Prp2 is recruited to the spliceosome by its interaction with Brr2.61 Brr2 has also been shown to interact with Prp16 which may be the way in which Prp16 associates with the spliceosome.62 Prp43 interacts with Ntr1 and Ntr2, with Prp43 being recruited to the spliceosome through Ntr2 interaction with Brr2.63 Ntr2 binding to Brr2 may be prevented by Prp16 and Slu7 binding to Brr2 providing a mechanism by which Prp43 action is regulated.64 Overall, Brr2 appears to be a binding platform for a number of the ATPases that modulate the NTC during splicing, indirectly linking Brr2 to NTC dynamics during splicing.
Conclusions and Future Directions
It is clear that the action of the ATPases Prp2, Prp16, Prp22 and Prp43 are related to the modulation of the NTC and NTC-associated proteins with the spliceosome during the splicing cycle. These changes in the NTC brought about by ATPase action are essential for providing the spliceosome conformations required for the first and second steps of splicing. Additionally, ATPase action is also required to modulate NTC interactions during spliceosome disassembly. Once the NTC is assimilated into the spliceosome it may not operate as a discrete complex as it appears only certain NTC proteins are modulated by ATPase action. Alternatively, it may be that the whole NTC is modulated by ATPase action but evidence is now only available for a few of the NTC proteins. In many cases it is not known whether the ATPases act directly or indirectly on the NTC proteins. In future, it will be important to determine the interaction network within the spliceosome by which the actions of the ATPases are transmitted to the NTC. There is no evidence to date for the action of the ATPases Sub2, Prp5 and Prp28 influencing the NTC as they act before the association of the NTC with the spliceosome. In humans a number of other DExD box ATPases like DDX5 (p68) and DDX17 (p72) are associated with the spliceosome.65 It is conceivable that the action of other ATPases may induce conformations during spliceosome assembly that allows incorporation of the NTC and NTC-associated proteins with the spliceosome. Nevertheless, the NTC appears to be a major target for ATPase remodelling of the spliceosome and the NTC is therefore intimately associated with the essential remodelling steps required for pre-mRNA splicing.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Due to space limitations we apologize to authors whose relevant primary publications were not cited. We would like to thank an anonymous reviewer for excellent comments and suggestions for improving the manuscript.
Funding
The work was supported by the Biotechnology and Biological Sciences Research Council.
References
- 1. Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med 2012; 18:472-82; PMID:22819011; http://dx.doi.org/ 10.1016/j.molmed.2012.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen HC, Cheng SC. Functional roles of protein splicing factors. Biosci Rep 2012; 32:345-59; PMID:22762203; http://dx.doi.org/ 10.1042/BSR20120007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009; 136:701-18; PMID:19239890; http://dx.doi.org/ 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 4. Chan SP, Kao DI, Tsai WY, Cheng SC. The Prp19p-associated complex in spliceosome activation. Science 2003; 302:279-82; PMID:12970570; http://dx.doi.org/ 10.1126/science.1086602 [DOI] [PubMed] [Google Scholar]
- 5. Hogg R, McGrail JC, O'Keefe RT. The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing. Biochem Soc Trans 2010; 38:1110-5; PMID:20659013; http://dx.doi.org/ 10.1042/BST0381110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saha D, Khandelia P, O'Keefe RT, Vijayraghavan U. Saccharomyces cerevisiae NineTeen complex (NTC)-associated factor Bud31/Ycr063w assembles on precatalytic spliceosomes and improves first and second step pre-mRNA splicing efficiency. J Biol Chem 2012; 287:5390-9; PMID:22215661; http://dx.doi.org/ 10.1074/jbc.M111.298547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ohrt T, Prior M, Dannenberg J, Odenwalder P, Dybkov O, Rasche N, Schmitzova J, Gregor I, Fabrizio P, Enderlein J, et al. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA 2012; 18:1244-56; PMID:22535589; http://dx.doi.org/ 10.1261/rna.033316.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ohrt T, Odenwalder P, Dannenberg J, Prior M, Warkocki Z, Schmitzova J, Karaduman R, Gregor I, Enderlein J, Fabrizio P, et al. Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in a purified system. RNA 2013; 19:902-15; PMID:23685439; http://dx.doi.org/ 10.1261/rna.039024.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cordin O, Beggs JD. RNA helicases in splicing. RNA Biol 2013; 10:83-95; PMID:23229095; http://dx.doi.org/ 10.4161/rna.22547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci 2011; 36:19-29; PMID:20813532; http://dx.doi.org/ 10.1016/j.tibs.2010.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burgess SM, Guthrie C. A mechanism to enhance mRNA splicing fidelity: The RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 1993; 73:1377-91; PMID:8324826; http://dx.doi.org/ 10.1016/0092-8674(93)90363-U [DOI] [PubMed] [Google Scholar]
- 12. Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5' splice site cleavage during pre-mRNA splicing. Mol Cell 2010; 39:385-95; PMID:20705241; http://dx.doi.org/ 10.1016/j.molcel.2010.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koodathingal P, Staley JP. Splicing fidelity: DEAD/H-box ATPases as molecular clocks. RNA Biol 2013; 10:1073-9; PMID:23770752; http://dx.doi.org/ 10.4161/rna.25245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Smith DJ, Query CC, Konarska MM. "Nought may endure but mutability": Spliceosome dynamics and the regulation of splicing. Mol Cell 2008; 30:657-66; PMID:18570869; http://dx.doi.org/ 10.1016/j.molcel.2008.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wlodaver AM, Staley JP. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome. RNA 2014; 20:282-94; PMID:24442613; http://dx.doi.org/ 10.1261/rna.042598.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 2009; 36:593-608; PMID:19941820; http://dx.doi.org/ 10.1016/j.molcel.2009.09.040 [DOI] [PubMed] [Google Scholar]
- 17. Fourmann JB, Schmitzova J, Christian H, Urlaub H, Ficner R, Boon KL, Fabrizio P, Luhrmann R. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev 2013; 27:413-28; PMID:23431055; http://dx.doi.org/ 10.1101/gad.207779.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ohi MD, Vander Kooi CW, Rosenberg JA, Ren L, Hirsch JP, Chazin WJ, Walz T, Gould KL. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol Cell Biol 2005; 25:451-60; PMID:15601865; http://dx.doi.org/ 10.1128/MCB.25.1.451-460.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vander Kooi CW, Ren L, Xu P, Ohi MD, Gould KL, Chazin WJ. The Prp19 WD40 domain contains a conserved protein interaction region essential for its function. Structure 2010; 18:584-93; PMID:20462492; http://dx.doi.org/ 10.1016/j.str.2010.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McGrail JC, Krause A, O'Keefe RT. The RNA binding protein Cwc2 interacts directly with the U6 snRNA to link the nineteen complex to the spliceosome during pre-mRNA splicing. Nucleic Acids Res 2009; 37:4205-17; PMID:19435883; http://dx.doi.org/ 10.1093/nar/gkp341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rasche N, Dybkov O, Schmitzova J, Akyildiz B, Fabrizio P, Lührmann R. Cwc2 and its human homologue RBM22 promote an active conformation of the spliceosome catalytic centre. EMBO J 2012; 31:1591-604; PMID:22246180; http://dx.doi.org/ 10.1038/emboj.2011.502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collier SE, Voehler M, Peng D, Ohi R, Gould KL, Reiter NJ, Ohi MD. Structural and Functional Insights into the N-Terminus of Schizosaccharomyces pombe Cdc5. Biochemistry 2014; 53:6439-51; PMID:25263959; http://dx.doi.org/ 10.1021/bi5008639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim SH, Lin RJ. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol Cell Biol 1996; 16:6810-9; PMID:8943336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lardelli RM, Thompson JX, Yates JR, 3rd, Stevens SW. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 2010; 16:516-28; PMID:20089683; http://dx.doi.org/ 10.1261/rna.2030510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warkocki Z, Odenwalder P, Schmitzova J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 2009; 16:1237-43; PMID:19935684; http://dx.doi.org/ 10.1038/nsmb.1729 [DOI] [PubMed] [Google Scholar]
- 26. Chiu YF, Liu YC, Chiang TW, Yeh TC, Tseng CK, Wu NY, Cheng SC. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol Cell Biol 2009; 29:5671-8; PMID:19704000; http://dx.doi.org/ 10.1128/MCB.00773-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Roy J, Kim K, Maddock JR, Anthony JG, Woolford JL, Jr. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1995; 1:375-90; PMID:7493316 [PMC free article] [PubMed] [Google Scholar]
- 28. Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol 2004; 24:10101-10; PMID:15542821; http://dx.doi.org/ 10.1128/MCB.24.23.10101-10110.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeh TC, Liu HL, Chung CS, Wu NY, Liu YC, Cheng SC. Splicing factor Cwc22 is required for the function of Prp2 and for the spliceosome to escape from a futile pathway. Mol Cell Biol 2011; 31:43-53; PMID:20956557; http://dx.doi.org/ 10.1128/MCB.00801-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mefford MA, Staley JP. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA 2009; 15:1386-97; PMID:19458033; http://dx.doi.org/ 10.1261/rna.1582609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J 1992; 11:5033-9; PMID:1464325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tseng CK, Liu HL, Cheng SC. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 2011; 17:145-54; PMID:21098140; http://dx.doi.org/ 10.1261/rna.2459611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 1992; 71:803-17; PMID:1423631; http://dx.doi.org/ 10.1016/0092-8674(92)90556-R [DOI] [PubMed] [Google Scholar]
- 34. Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature 2013; 503:229-34; PMID:24196718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Keating KS, Toor N, Perlman PS, Pyle AM. A structural analysis of the group II intron active site and implications for the spliceosome. RNA 2010; 16:1-9; PMID:19948765; http://dx.doi.org/ 10.1261/rna.1791310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee C, Jaladat Y, Mohammadi A, Sharifi A, Geisler S, Valadkhan S. Metal binding and substrate positioning by evolutionarily invariant U6 sequences in catalytically active protein-free snRNAs. RNA 2010; 16:2226-38; PMID:20826700; http://dx.doi.org/ 10.1261/rna.2170910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Madhani HD, Guthrie C. Genetic interactions between the yeast RNA helicase homolog Prp16 and spliceosomal snRNAs identify candidate ligands for the Prp16 RNA-dependent ATPase. Genetics 1994; 137:677-87; PMID:8088513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev 2005; 19:1894-904; PMID:16103217; http://dx.doi.org/ 10.1101/gad.1336305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hogg R, de Almeida RA, Ruckshanthi JP, O'Keefe RT. Remodeling of U2-U6 snRNA helix I during pre-mRNA splicing by Prp16 and the NineTeen Complex protein Cwc2. Nucleic Acids Res 2014; 42:8008-23; PMID:24848011; http://dx.doi.org/ 10.1093/nar/gku431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 1991; 349:487-93; PMID:1992352; http://dx.doi.org/ 10.1038/349487a0 [DOI] [PubMed] [Google Scholar]
- 41. Schwer B, Gross CH. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J 1998; 17:2086-94; PMID:9524130; http://dx.doi.org/ 10.1093/emboj/17.7.2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McGrail JC, O'Keefe RT. The U1, U2 and U5 snRNAs crosslink to the 5′ exon during yeast pre-mRNA splicing. Nucleic Acids Res 2008; 36:814-25; PMID:18084028; http://dx.doi.org/ 10.1093/nar/gkm1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McGrail JC, Tatum EM, O′Keefe RT. Mutation in the U2 snRNA influences exon interactions of U5 snRNA loop 1 during pre-mRNA splicing. EMBO J 2006; 25:3813-22; PMID:16888626; http://dx.doi.org/ 10.1038/sj.emboj.7601258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Newman A, Norman C. Mutations in yeast U5 snRNA alter the specificity of 5′ splice-site cleavage. Cell 1991; 65:115-23; PMID:2013092; http://dx.doi.org/ 10.1016/0092-8674(91)90413-S [DOI] [PubMed] [Google Scholar]
- 45. Newman AJ, Norman C. U5 snRNA interacts with exon sequences at 5′ and 3′ splice sites. Cell 1992; 68:743-54; PMID:1739979; http://dx.doi.org/ 10.1016/0092-8674(92)90149-7 [DOI] [PubMed] [Google Scholar]
- 46. O'Keefe RT, Newman AJ. Functional analysis of the U5 snRNA loop 1 in the second catalytic step of yeast pre-mRNA splicing. EMBO J 1998; 17:565-74; PMID:9430647; http://dx.doi.org/ 10.1093/emboj/17.2.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. O'Keefe RT, Norman C, Newman AJ. The invariant U5 snRNA loop 1 sequence is dispensable for the first catalytic step of pre-mRNA splicing in yeast. Cell 1996; 86:679-89; PMID:8752221; http://dx.doi.org/ 10.1016/S0092-8674(00)80140-3 [DOI] [PubMed] [Google Scholar]
- 48. Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell 2008; 30:743-54; PMID:18570877; http://dx.doi.org/ 10.1016/j.molcel.2008.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dreumont N, Seraphin B. Rapid screening of yeast mutants with reporters identifies new splicing phenotypes. FEBS J 2013; 280:2712-26; PMID:23560879; http://dx.doi.org/ 10.1111/febs.12277 [DOI] [PubMed] [Google Scholar]
- 50. Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J 2004; 23:4847-56; PMID:15565172; http://dx.doi.org/ 10.1038/sj.emboj.7600482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scherrer FW, Jr., Spingola M. A subset of Mer1p-dependent introns requires Bud13p for splicing activation and nuclear retention. RNA 2006; 12:1361-72; PMID:16738408; http://dx.doi.org/ 10.1261/rna.2276806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ohi MD, Link AJ, Ren LP, Jennings JL, McDonald WH, Gould KL. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol Cell Biol 2002; 22:2011-24; PMID:11884590; http://dx.doi.org/ 10.1128/MCB.22.7.2011-2024.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boon KL, Auchynnikava T, Edwalds-Gilbert G, Barrass JD, Droop AP, Dez C, Beggs JD. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol Cell Biol 2006; 26:6016-23; PMID:16880513; http://dx.doi.org/ 10.1128/MCB.02347-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 2005; 19:2991-3003; PMID:16357217; http://dx.doi.org/ 10.1101/gad.1377405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev 2007; 21:2312-25; PMID:17875666; http://dx.doi.org/ 10.1101/gad.1580507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hahn D, Beggs JD. Brr2p RNA helicase with a split personality: insights into structure and function. Biochem Soc Trans 2010; 38:1105-9; PMID:20659012; http://dx.doi.org/ 10.1042/BST0381105 [DOI] [PubMed] [Google Scholar]
- 57. Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA 1999; 5:959-71; PMID:10411139; http://dx.doi.org/ 10.1017/S135583829999012X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 1998; 8:847-55; PMID:9705931; http://dx.doi.org/ 10.1016/S0960-9822(07)00345-4 [DOI] [PubMed] [Google Scholar]
- 59. Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 2006; 23:389-99; PMID:16885028; http://dx.doi.org/ 10.1016/j.molcel.2006.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Frazer LN, Nancollis V, O'Keefe RT. The role of Snu114p during pre-mRNA splicing. Biochem Soc Trans 2008; 36:551-3; PMID:18482006; http://dx.doi.org/ 10.1042/BST0360551 [DOI] [PubMed] [Google Scholar]
- 61. Liu HL, Cheng SC. The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol Cell Biol 2012; 32:5056-66; PMID:23071087; http://dx.doi.org/ 10.1128/MCB.01109-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 2001; 157:1451-67; PMID:11290703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tsai RT, Tseng CK, Lee PJ, Chen HC, Fu RH, Chang KJ, Yeh FL, Cheng SC. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol Cell Biol 2007; 27:8027-37; PMID:17893323; http://dx.doi.org/ 10.1128/MCB.01213-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen HC, Tseng CK, Tsai RT, Chung CS, Cheng SC. Link of NTR-mediated spliceosome disassembly with DEAH-box ATPases Prp2, Prp16, and Prp22. Mol Cell Biol 2013; 33:514-25; PMID:23166295; http://dx.doi.org/ 10.1128/MCB.01093-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature 2002; 419:182-5; PMID:12226669; http://dx.doi.org/ 10.1038/nature01031 [DOI] [PubMed] [Google Scholar]