Abstract
The process of mRNA decay and surveillance is considered to be one of the main posttranscriptional gene expression regulation platforms in eukaryotes. The degradation of stable, protein-coding transcripts is normally initiated by removal of the poly(A) tail followed by 5’-cap hydrolysis and degradation of the remaining mRNA body by Xrn1. Alternatively, the exosome complex degrades mRNA in the 3’>5’direction. The newly discovered uridinylation-dependent pathway, which is present in many different organisms, also seems to play a role in bulk mRNA degradation. Simultaneously, to avoid the synthesis of incorrect proteins, special cellular machinery is responsible for the removal of faulty transcripts via nonsense-mediated, no-go, non-stop or non-functional 18S rRNA decay. This review is focused on the major eukaryotic cytoplasmic mRNA degradation pathways showing many similarities and pointing out main differences between the main model-species: yeast, Drosophila, plants and mammals.
Keywords: Arabidopsis thaliana, Drosophila melanogaster, human, mRNA decay, mRNA surveillance, Saccharomyces cerevisiae, Schizosaccharomyces pombe
Introduction
mRNA molecules synthesized in the nucleus by RNA polymerase II are relatively unstable. In the cytoplasm, they are protected from the attack of exonucleases by the 5′-cap structure and the 3′-poly(A) tail. The stability of mRNAs depends on its innate features, but is predetermined by the nucleotide sequence and relates to the functions of the protein it encodes. Therefore, mRNA decay is considered the main posttranscriptional gene expression regulation platform. The cytoplasmic bulk mRNA degradation pathway in eukaryotic cells starts with shortening of the poly(A) tail. This process is performed by 3′>5′ exonucleases. To date, 8 different deadenylases have been characterized in metazoan, primarily including the CCR4-NOT complex, PAN2-PAN3 and PARN.1 Deadenylation is the first and therefore often rate-limiting step of mRNA decay; however, it is reversible to some extent, as transcripts can be readenylated and translated into functional proteins.2 Furthermore, the 5′-cap may be removed in the process known as decapping by the Dcp1-Dcp2 complex and mRNA may subsequently be subjected to degradation by Xrn1 exonuclease. Alternatively, the exosome complex degrades mRNA in the 3′>5′ direction. Deletions of the main enzymes of the 3′>5′ or 5′>3′ degradation pathways do not result in the total accumulation of aberrant mRNA, which suggests that those enzymes work in cooperation.2 In this article, we review and compare our knowledge regarding dominant cytoplasmic RNA decay pathways, excluding RNAi, in the major model systems: yeast, human, Drosophila and plants.
General mRNA Degradation Pathways
Deadenylation
The length of the poly(A) tail in a newly synthesized mRNA molecule depends on the organism. After entering the cytoplasm, it is either stabilized by poly(A) binding proteins (PABP), which facilitates translation, or shortened by cytoplasmic nucleases.3 Poly(A) tail removal triggers mRNA degradation in almost all eukaryotic decay pathways (Fig. 1). This includes the degradation of stable, protein-coding mRNAs, ARE-mediated decay (for transcripts containing AU-rich destabilizing elements (AREs) in their 3′ UTRs4), nonsense-mediated decay (for mRNAs with premature stop codons),5 miRNA-mediated decay6,7 and the degradation mediated by destabilizing elements in protein coding regions, as in the case of the proto-oncogene c-fos.8 Therefore, eukaryotic genomes encode a wide variety of deadenylases.
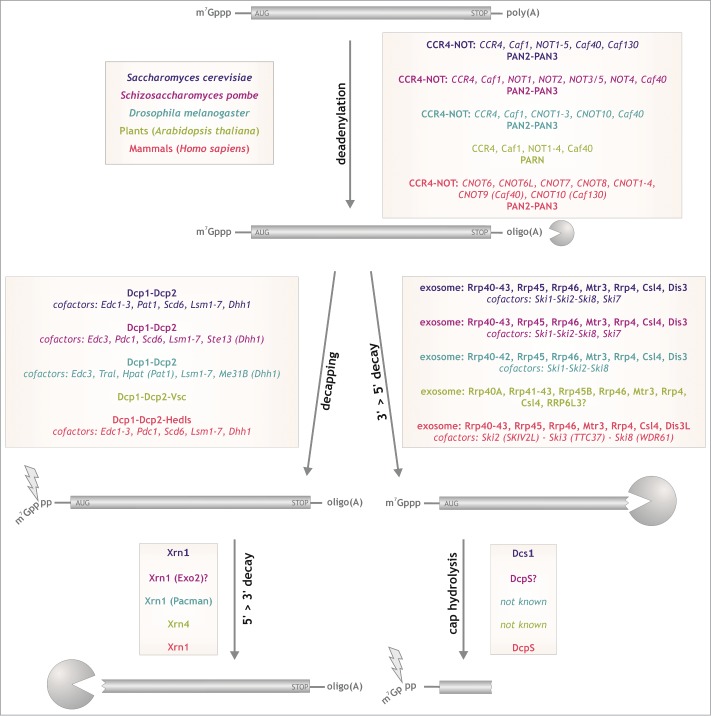
Figure 1.
Deadenylation-dependent degradation: pathways and enzymes. Degradation of properly synthesized mRNAs starts with shortening of the poly(A) tail (deadenylation), which is followed by decapping and subsequent 5’>3’ degradation by Xrn1 or exosome-mediated 3’>5’ degradation and cap hydrolysis. Color coding represents homologs from different organisms. Question mark indicates that relevant homolog is present in a database of a given species but its involvement in the process was not confirmed experimentally.
Based on biochemical and bioinformatic approaches, deadenylases can be divided into 2 main groups: DEDD or EEP nucleases.1,9 Enzymes from both groups are Mg2+-dependent exoribonucleases, which degrade RNA in a 3′>5′ direction, releasing 5′-AMP. The DEDD group is characterized by the presence of Asp and Glu residues in 3 active motifs that coordinate divalent metal ions.10 This group includes Caf1 (Pop2), Caf1Z, PARN and PAN2. Deadenylases from the EEP (exonuclease-endonuclease-phosphatase) superfamily contain conserved catalytic Asp and His residues in their active site and catalyze phosphate ester hydrolysis with the help of 2 magnesium ions.11 CCR4, NOCTURNIN, ANGEL and 2′PDE are the enzymes that have been assigned to this group.
Saccharomyces cerevisiae
In S. cerevisiae, all cytoplasmic mRNA degradation pathways are initiated by removal of the poly(A) tail. This process is performed by 2 main complexes, CCR4-NOT and PAN2-PAN3. The CCR4-NOT complex is a multifunctional protein assembly that has been extensively studied in yeast.12 It consists of 9 main subunits, 2 of which, CCR4 and Caf1 (Pop2), belong to the exonuclease families.13 CCR4, an EEP nuclease, is the main deadenylase in the S. cerevisiae CCR4-NOT complex: deleting ccr4 along with pan2 blocks deadenylation entirely.14 CCR4 is incorporated into the whole complex through Caf1 via a leucine-rich repeat motif (LRR)15 and this interaction is conserved across the entire eukaryotic domain. Caf1 (Pop2), the second exonuclease of the complex, is not required for the deadenylation of mRNA in vitro as it lacks the canonical catalytic residues in its active site.10 It might be active under certain conditions or able to enhance the activity of CCR4; however, biochemical evidence shows that CCR4 is mainly responsible for deadenylation in yeast cells.16,17 The eukaryotic CCR4-NOT complex, apart from 2 deadenylases, consists of 7 more subunits with distinct yet not fully characterized functions. NOT1 is the scaffold protein of the complex, without any enzymatic activity, NOT2, NOT3 and NOT5 form a NOT module interacting with the C-terminus of NOT118 and NOT4 (Mot2) is an active E3 ubiquitin ligase.19 Caf130 and Caf40, which are also incorporated into the yeast complex, possess no clear function.13 Their role in the deadenylation process is probably regulatory, as the deletion of either of them impairs poly(A) tail removal.16
The second deadenylation complex present in yeast, but also well conserved in higher eukaryotes, is a heterotrimer formed by the PAN2 protein interacting with the homodimer of PAN3. PAN2 is a distributive, hydrolytic 3′-exonuclease, belonging to the DEDD superfamily.11 Its activity depends on Pab1,20 and it is involved in the initial shortening of the poly(A) tail to a length of around 60–80 nt.21 As Pab1 promotes activity of the PAN2-PAN3 complex and inhibits CCR4-NOT,16,20 the model of cooperative cytoplasmic deadenylation was proposed. According to this model, PAN2-PAN3 nuclease is responsible for the rapid removal of the first exposed adenine residues,22 leaving the poly(A) tail of around 65 nucleotides. After this, the poly(A) tail is slowly and distributively degraded by the CCR4 nuclease.23 However, it seems that the activity of both deadenylating complexes is partially redundant. Deletion of ccr4 gene gives slower deadenylation rates, but only the ccr4Δ, pan2Δ double mutant accumulates poly(A) tailed mRNAs and has severe growth defects.16
Schizosaccharomyces pombe
In fission yeast, Schizosaccharomyces pombe, mRNA turnover has been less extensively studied; however, the main pathways and enzymes are well conserved. Orthologs of the components of CCR4-NOT and PAN2-PAN3 deadenylases are present,24,25 indicating a similar role in the initiation or mRNA degradation by poly(A) tail removal. The CCR4-NOT complex in S. pombe consists of 7 main subunits (CCR4, Caf1 (Pop2), NOT1, NOT2, NOT3/5, NOT4 (Mot2), Caf40 (Rcd1)). Unlike in S. cerevisiae, Caf1 has a fully conserved DEDD active site and has been shown to be a functional 3′>5′ exonuclease.26 Activity of PAN2-PAN3 complex has not been proven.
In the genome of S. pombe, an ortholog of PARN, a mainly nuclear deadenylase which is absent in S. cerevisiae, has been identified.25 Its physiological role in fission yeast is unclear, but it was suggested to be involved in the Dicer-independent RNAi pathway in the nucleus.27
Drosophila melanogaster
The deadenylation-dependent mRNA decay pathway has also been quite extensively studied in Drosophila melanogaster cells. The genome of the fruit fly encodes homologs of all yeast deadenylases: Caf1, CCR4, and PAN2-PAN3. However, no homolog of mammalian PARN has been identified.28 It was shown that Caf1, CCR4, NOT1, NOT2, NOT3, Caf40 and the fly ortholog of human CNOT10 form a stable complex, with the main deadenylation activity assigned to Caf1 subunit, rather than to CCR4, as was observed in S. cerevisiae.29 Interestingly, NOT4, a stable component of the complex in yeast, does not seem to be incorporated into the fly CCR4-NOT.29
Homologs of different exonucleases from EEP nuclease family can be found in the D. melanogaster genome: ANGEL, 2′PDE and Nocturnin, with the latter interacting with NOT1 protein and also probably affecting the deadenylation rates.29 The mechanism of deadenylation by PAN2-PAN3 complex in fly cells was not well studied. It was shown, however, that it interacts with the GW182 protein and is responsible for the deadenylation of miRNA targets.30
Plants
The main mRNA degradation pathways are also proposed to exist in plants, as most of the enzymes can be found in genomes of various plant species.31 Genes encoding deadenylating enzymes are often present in many copies. The Arabidopsis thaliana genome encodes 11 paralogs of Caf1; 16 can be found in rice Oryza sativa and 4 in grapes (Vitis vinifera). It was not shown whether all of those proteins are active deadenylases; however, all, as in S. pombe, contain the fully conserved DEDD motif of an active site.32 Two of the 11 A. thaliana Caf1 homologs were examined. Studies have shown their deadenylation activity in vitro and involvement in stress response.32,33 Other homologs of CCR4-NOT subunits were identified in the A. thaliana genome: CCR4, NOT1, NOT2, NOT3, NOT4, Caf40. However, their function and whether they form a stable complex, as in other organisms, remains unknown.34 Poly(A) ribonuclease (PARN), absent in budding yeast and fruit fly, plays an important role in A. thaliana embryogenesis as an active cytoplasmic deadenylase.35 Plant genomes encode also PAN homologs; nevertheless, their potential activity and function has not yet been examined.
Mammals
The majority of mammalian cytoplasmic mRNAs are degraded through a deadenylation-dependent pathway. The human genome encodes 12 deadenylases from both superfamilies.9 Nevertheless, removal of the poly(A) tail is performed by the cooperative work of 2 deadenylases: CCR4-NOT and PAN2-PAN3.36 The human CCR4-NOT complex consist of 10 subunits: CNOT1, CNOT2, CNOT3, CNOT4 (probably not a stable component), CNOT9/Caf40/Rcd1, CNOT10/Caf130, 2 copies of CCR4 homologs: CNOT6, CNOT6L, and 2 of Caf1: CNOT7, CNOT8.37 All 4 nucleases are enzymatically active; however, they demonstrate different substrate specificity.36-38
Human PAN2 is a distributive exonuclease that interacts with PAN3, which binds through its PAM2 motif with poly(A)-binding protein (PABP).39 Initially, poly(A) tails of a stable mammalian mRNAs are deadenylated by the PAN2-PAN3 complex to the length of ∼110 nt and subsequently degraded by CCR4-NOT complex until the length is ∼10 adenine residues.40,41 PARN, PARNL, ANGEL, ANGEL2, Nocturnin and 2′PDE deadenylases are encoded in the human genome. Among them, PARN has been shown to be involved in the deadenylation of specific mRNAs.9,42
Decapping and Xrn1-mediated 5′ to 3′ mRNA degradation
One of the 2 possible scenarios for mRNA degradation after initial deadenylation is removal of the 5′-cap structure (Fig. 1). This process is performed by the Dcp2 enzyme, which belongs to the Nudix hydrolase family and is conserved among eukaryotes.43 The reaction products are m7GDP and 5′ monophosphate RNA. Activity of the decapping enzyme depends on divalent cations and specific cap methylation on N7.44 The RNA body is also involved in catalysis and the enzyme has a preference for RNA substrates no shorter than 25 nucleotides.45 Dcp2 forms a complex with Dcp1,46,47 which is a small protein containing an EVH1 domain which is a common platform, bridging protein-protein interactions.48 Moreover, Dcp1 is the main activator of Dcp2.49 The decapping process is regulated by a plethora of activators and inhibitors, but their composition varies between the organisms.43,50,51 The best defined decapping activator is a conserved in all eukaryotes. The Lsm1-7-Pat1 complex, which preferentially binds the 3′-end of oligoadenylated mRNA,52,53 enhances decapping and inhibits exosome attachment.54 Decapped (5′ monophosphorylated) RNA is exposed to attack and complete degradation by Xrn1, a processive 5′>3′ exonuclease.44 Decapping and the 5′>3′ degradation are coupled, as Xrn1 nuclease interacts directly with one of the components of the decapping machinery, Dcp1, Pat1 or Edc4, depending on the organism.55,56 This Xrn1-dependent decapping mechanism supports a model in which Xrn1 is a global transcript level buffering protein.57
Saccharomyces cerevisiae
In yeast, the main mRNA degradation pathway starts with removal of the poly(A) tail followed by decapping and rapid degradation of the RNA body from the 5′ end by Xrn1 nuclease.46,58 Decapping is performed by the Dcp2 protein in a complex with Dcp1.45,46 The conserved region of Dcp2 (residues 248-300) interacts with the decapping activator Edc3 and possibly also mediates interactions with some other factors.58 The Saccharomyces cerevisiae genome encodes a range of decapping regulators: Edc1-3 (enhancers of decapping 1, 2, 3), Pat1, Scd6, Lsm1-7 and Dhh1. All of these have been demonstrated to bind RNA and activate decapping in vivo.55 Except for Edc1 and Edc2, which seem to be specific to S. cerevisiae, all of the activators are conserved in evolution. Among them, the Lsm1-7-Pat1 complex seems to be crucial for the activation of decapping. The crystal structure of the yeast Lsm1-7-Pat1 complex shows that Lsm2-Lsm3 conserved helices bind the C-terminus of Pat153,59 and the latter interacts with Xrn1 nuclease.55
Schizosaccharomyces pombe
Fission yeast Dcp2 also forms a complex with its activator, Dcp1, and is responsible for the decapping of mRNA molecules.48,60,61 Its activity is also stimulated by the set of activators: Edc3, Pdc1 (functional homolog of Helds), Lsm1-7, Ste13 (Dhh1) and Scd6.62,63 Moreover, in Schizosaccharomyces pombe, decapping can be stimulated by 3′ uridylation, which is a novel pathway of mRNA degradation.64 The S. pombe genome encodes a homolog of Xrn1 exonuclease, Exo2. However, direct evidence for its role in 5′>3′ mRNA degradation has not yet been shown.65
Drosophila melanogaster
In Drosophila melanogaster, the decapping machinery includes the decapping enzyme Dcp2, which directly interacts with its activators: Dcp1, Tral, Edc3, Lsm1-7, helicase Me31B (homolog of Dhh1)66 and HPat homologs to the yeast Pat1.67 For the efficient decapping of mRNA, the interaction between Dcp1 and Xrn1/Pacman is also required.56
Plants
Removal of the 5′-cap structure in plants is performed by the complex consisting of Dcp1, Dcp2 and Varicose (Vsc), a homolog of mammalian Hedls/Ge-1. The Dcp2 Hudix domain displays enzymatic activity, which is stimulated by Dcp1 and Vsc.68 While decapping in yeast is regulated by a variety of decapping activators, in plants, the Dcp5 protein has been found to associate with Dcp1, Dcp2 and influence translation repression, P-body formation and postembryonic development.69 Arabidopsis thaliana has 3 orthologs of yeast Xrn1: Xrn2, Xrn3, Xrn4.34 Xrn2 and Xrn3 are involved in the processing of rRNA and snoRNA in the nucleus, while Xrn4 is located in the cytoplasm70 and is responsible for the 5′>3′ degradation of specific transcripts.71
Mammals
The 5′>3′ degradation machinery has been best studied in yeast and mammals. Dcp2 is the main decapping enzyme, with RNA binding properties.72 Unique to mammalian cells, the Hedls protein (also called Edc4, Ge-1), bridges the interaction between Dcp2 and its activator Dcp1 and also influences its activity.73 A range of other decapping enhancers stimulate the activity of Dcp2, like Edc1-3, Dhh1, Lsm1-7.50 In the mammalian genome, 20 2 genes of the Nudix family of hydrolases can be found.74 Similarly to Dcp2, they contain a conserved consensus motif GX5EX7REUXEEXGU, where X represents any residue and U a hydrophobic one. Among them, cytoplasmic Nudt16 (X29), first identified in Xenopus laevis, also possesses decapping activity and RNA binding properties.75,76 Decapped mRNA is a substrate for subsequent 5′>3′ degradation by Xrn1 exonuclease and this process is also associated and regulated by the decapping efficiency, since Xrn1 directly interacts with Hedls (Edc4) (part of the decapping complex).56
Exosome-mediated 3′ to 5′ mRNA degradation
In the cytoplasmic deadenylation-dependent pathway, alternatively to 5′>3′ decay, mRNA can be degraded in the 3′>5′ direction by the cytoplasmic exosome; the remaining cap is hydrolyzed by a scavenger decapping enzyme, DcpS, which shows a specificity toward short RNA fragments (Fig. 1). The exosome is a multi-subunit complex involved in the degradation, processing and quality control of many groups of RNA in eukaryotic cells. The cytoplasmic enzyme consists of 9 core subunits and the enzymatically active ribonuclease.77 Exosome composition is extremely well conserved among different organisms. Six subunits, Rrp41, Rrp42, Rrp43, Rrp45, Rrp46 and Mtr3, share 20-30% sequence similarity with E. coli polynucleotide phosphorylase (PNPase) and RNase PH domains. Moreover, they form a hexameric core ring.78 Rrp4, Rrp40 and Csl4 possess RNA binding properties through their, also well conserved, S1 and KH motifs and are present across the eukaryotic domain. The tenth essential subunit of the cytoplasmic eukaryotic exosome, Rrp44 (Dis3), a homolog of bacterial RNase II,79,80,81 is an active 3′- 5′ nuclease which contains N-terminal PIN domain responsible for endonucleolytic activity and the exosome ring attachment.82,83,84 In higher eukaryotes, this subunit is present in more than one form. Three or 4 cofactors called superkillers or Ski proteins associate and regulate the activity of the exosome: Ski2, Ski3 and Ski8 which form a heterotetramer in the stoichiometry of 1:1:285 and yeast Ski7 protein, which can associate both with the Ski complex and the exosome.86 After the degradation of the mRNA body by the exosome complex, the remaining m7GpppN cap is hydrolyzed by the m7G-specific pyrophosphatase, DcpS. This enzyme, unlike Dcp2, carries a histidine triad motif (HXHXH) and forms an asymmetric homodimer.87,88
Saccharomyces cerevisiae
In yeast, mRNA degradation from the 3′ end is a minor pathway. However, most of the proteins involved in this process are well conserved and have been extensively studied in this organism. The S. cerevisiae cytoplasmic exosome consists of a 9-subunit, enzymatically inactive, ring, formed by Rrp41, Rrp42, Rrp43, Rrp45, Rrp46 and Mtr3 and cap made of Rrp4, Rrp40 and Csl4. Endo- and exoribonucleolytic activity is provided by the Rrp44 (Dis3) protein, which possesses S1, PIN, RNB and 2 cold-shock (CSD1 and CSD2) domains.79,82-84 Broad biochemical and structural studies of yeast exosome and exosome-associated Ski complex allowed for the in-depth understanding of the mechanism of RNA substrates′ degradation.85,89-91 The exosome is functionally and physically associated with the Ski complex. In yeast, the Ski complex is formed by Ski2, the active helicase, Ski3 and 2 Ski8 proteins that modulate the RNA-binding and ATPase properties of Ski2.85 Moreover, the Ski7 protein bridges the interaction between the Ski complex and the exosome.86 This protein has an interesting evolutionary history since most fungi, excluding yeasts, have a single Ski7/Hbs1 gene which is alternatively spliced into Ski7 and Hbs1-like proteins.
The yeast genome encodes 2 homologs of mammalian DcpS scavenger decapping enzyme, Dcs1 and Dcs2.92 They are both members of the HIT family of pyrophosphatases; however, only the Dcs1 protein has been shown to hydrolyze the cap structure.92
Schizosaccharomyces pombe
Very recently, the 10-subunit cytoplasmic exosome was purified from Schizosaccharomyces pombe, confirming its existence in this organism and probable role in cytoplasmic mRNA degradation.93 The Ski complex as well as Ski7 was also identified,94 pointing to the connection of the exosome with its partner. Interestingly, an independent duplication of Ski7/Hsb1 gene occurred in S. pombe.94
Drosophila melanogaster
To date, the entire exosome complex in flies, as the main enzyme responsible for the 3′>5′ mRNA degradation, has not been studied, even though the Drosophila melanogaster genome encodes 9 main cytoplasmic exosome subunits: Mtr3, Ski6/Rrp41, Rrp42, Rrp45, Rrp46, Rrp40, Rrp4, Csl4, Dis3. The Rrp43 protein could not be purified as a part of the core exosome.95,96 This fact does not exclude, however, the possibility that it is incorporated into the complex in vivo. In vitro studies of fly Dis3 showed its endonucleolytic activity and main localization in the nucleus.97 Moreover, immunolocalization experiments pointed out that in D. melanogaster, different sub-complexes might be formed, apart from the core exosome, with distinct functionalities.95,98 Yeast homologs of the Ski complex were also identified in fruit fly. While in yeast they are the regulators of 3′>5′mRNA degradation by the exosome, in Drosophila, they play an important role in degradation of the 5′ends in miRNA-mediated mRNA degradation.99 The ortholog of Ski7 has not been identified.
Plants
In the Arabidopsis thaliana genome, the first exosome subunits identified were Rrp4 and Rrp41. Interestingly, unlike in yeast and humans, Rrp4 was shown to be catalytically active 3′>5′ phosphorolytic ribonuclease from the RNase PH family.100 Further experiments allowed for the recognition of the whole 9-subunit core: KH or S1 containing domain Rrp4, Rrp40 and Csl4, and with the RNase PH domain, Rrp41, Rrp42, Rrp43, Rrp45B, Rrp46 and Mtr3.101 Genes encoding Rrp40 and Rrp45 are duplicated. However, only one of each gene isoform was purified with the complex.101 Two Dis3 (Rrp44) paralogs were identified as Rrp44A and Rrp44B/SOV. Nonetheless, the cytoplasmic catalytic subunit remains to be determined, as Rrp44A was proposed to be nuclear and Rrp44B/SOV does not interact with the core exosome due to the absence of a PIN domain.102 The A. thaliana genome encodes 3 genes of the Rrp6 family, 2 of which are nuclear (RRP6L1 and RRP6L2) and one which is strictly cytoplasmic (RRP6L3); thus, the latter is a plausible candidate for a cytoplasmic catalytic subunit with hydrolytic activity associated with the core exosome.103 The Ski complex but not Ski7 has been characterized in plants.
Mammals
The human exosome, as the main enzyme responsible for 3′>5′ transcripts degradation, has been widely studied. The crystal structure reveals the conserved composition: Rrp41, Rrp45, Rrp46, Rrp43, Mtr3 and Rrp42 form a PH domain ring, Csl4, Rrp4 and Rrp40 form a cap.78 Enzymatic activity is performed, like in yeast, by the Dis3 protein. Nevertheless, there is an additional exosome-associated Dis3 homolog in humans, called Dis3L. While Dis3 is mainly localized in the nucleus, Dis3L is cytoplasmic.77,104 Dis3 and Dis3L possess conserved S1, CSD1, CSD2, PIN and RNB domains and associate with the exosome core.104 The residual cap structure, a product of mRNA degradation by the exosome, is hydrolyzed by a scavenger decapping enzyme, DcpS.88 DcpS forms a functional homodimer and decaps substrates with no more than 10 nucleotides.87 Human homologs of the superkiller family – Ski2 (SKIV2L, Ski2W), Ski3 (TTC37) and Ski8 (WDR61), form a complex.105 The Ski8 homolog is also a part of the PAF complex.105
Uridylation-dependent mRNA degradation
CytoplasmiC-terminal uridyl transferases, although absent in budding yeast, are present in a variety of organisms.106 Nowadays, new roles for 3′ end uridylation in the cytoplasmic mRNA degradation pathways are becoming more evident, yet still remain abstruse.
In S. pombe cells, the Cid1 protein is responsible for terminal mono-, di- and oligouridylation of polyadenylated transcripts which promotes Lsm-dependent decapping and deadenylation-independent mRNA decay.64 Interestingly, a new ribonuclease from the RNase II family, a homolog of human Dis3L2, was identified in fission yeast.93 As in humans, Dis3L2 does not have a conserved PIN domain and does not associate with the exosome. Both human and fission yeast Dis3L2 proteins function independently of the exosome complex and show a preference for 3′ uridylated RNAs.93,107,108 What is more, human Dis3L2 is responsible for the degradation of mammalian pre-let-7 precursor miRNA oligouridylated by Lin28.109 On the other hand, monouridylation of pre-let-7 favors maturation of this miRNA.110,111 The addition of uridyl residues was also shown to promote the degradation of miRNA and siRNA in Caenorhabditis elegans, zebrafish, Chlamydomonas reinhardtii and Arabidopsis.112-115 Human Dis3L2 is the closest homolog of A. thaliana's cytoplasmic protein AtRRP44B/SOV (Suppressor of Varicose), which indicates that AtRRP44B/SOV might also be involved in this degradation process102 (Fig. 2).
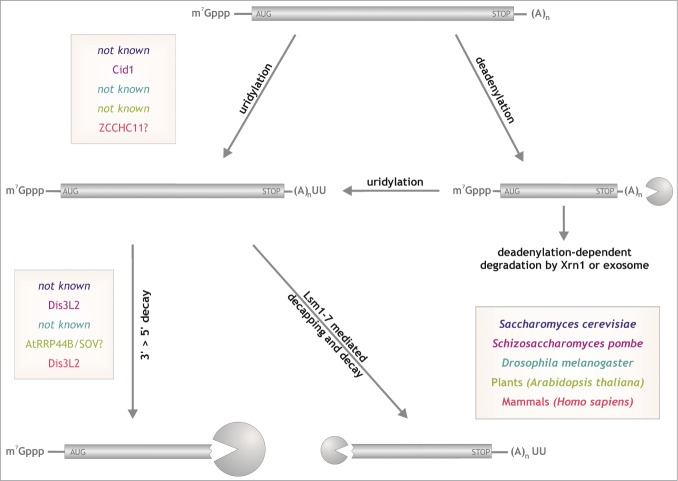
Figure 2.
Uridylation-dependent degradation. Properly synthesized mRNAs may be subjected to uridylation even after initial shortening of the poly(A) tail.64 Uridylation can lead to Dis3L2-mediated 3’>5’ degradation or Lsm1-7-mediated decapping and decay. Color coding and question marks as in Figure 1.
Moreover, metazoan replication-dependent histone mRNA degradation again involves 3′ end uridylation. As levels of those transcripts undergo significant changes during the cell cycle, and they have to be rapidly degraded after S-phase when DNA replication is completed, new effective decay mechanisms have emerged during evolution.116 Histone transcripts form a conserved stem-loop at their 3′ end rather than a poly(A) tail, as in lower eukaryotes, and this structure is indispensable for their replication-dependent degradation. The RNA hairpin forms a complex with SLBP protein (hairpin-binding factor, HBF in S. cerevisiae) and a conserved exoribonuclease Eri1 (human 3′hExo).117,118 Histone mRNA decay is triggered when SLBP recruits NMD-related UPF1 protein119 and TUTase(s) (most likely ZCCHC11120) which adds an oligo(U) tail on the 3′ end of the transcript.121 The oligo(U) tract serves as a binding site for the Lsm1-7 heptamer, which enhances 3′hExo activity and exposes a single-stranded transcript to subsequent degradation by the exosome.117 Decapping factors Lsm1 and Dcp2 were also shown to play a role in histone mRNA decay.117,122
Thus far, conservation of known TUTases and emerging evidence of their impact on RNA stability suggest that uridylation might be another common posttranscriptional gene expression regulating mechanism which remains to be determined.
Aberrant mRNA Degradation
There are several cytoplasmic RNA quality-control mechanisms that prevent the formation of aberrantly synthesized and potentially toxic proteins. According to the error present in the mRNA, nonsense-mediated decay (NMD), no-go decay (NGD) or non-stop decay (NSG) can be triggered.123,124 Moreover, mutated 18S rRNA transcripts are degraded by the non-functional 18S rRNA decay pathway.
Nonsense-mediated decay (NMD)
NMD eliminates transcripts containing premature termination codon (a PTC can be inserted into the molecule as a result of a transcription error, genetic mutation or during splicing125), normal mRNA molecules with long 3′ UTRs, transcripts with an upstream ORF (uORF) in the 5′ UTR and mRNAs with introns present in their 3′ UTRs.126-128 A model of how the PTC-containing mRNAs are recognized and targeted to degradation is based on the synergy between the kinetic difference of ribosome release from normal and premature stop codon and the interaction between the terminating ribosome and downstream cis-acting signals that vary across species (e.g. Hrp1 helicase in yeast and exon-junction complex EJC in mammals)129,130 (Fig. 3).
Figure 3.
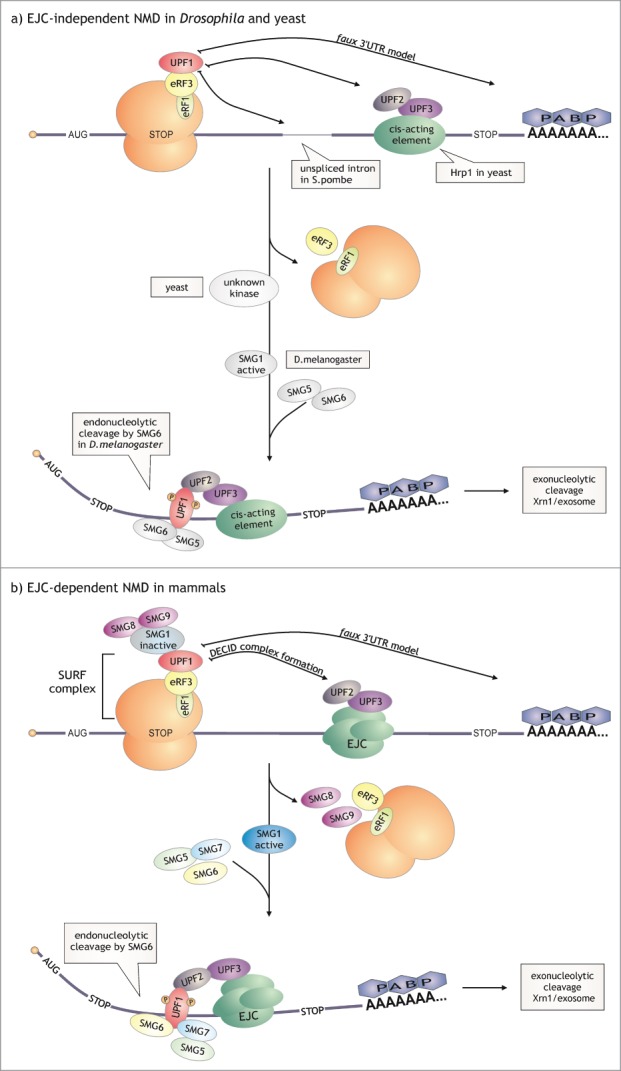
Nonsense-mediated decay. NMD in mammals often depends on the insertion of the exon-junction complex downstream of the PTC, which is not true in the case of lower eukaryotes. So far, the only model that appears to be conserved among species is the faux 3’ UTR model. a) EJC-independent NMD. In S. cerevisiae, S. pombe and D. melanogaster NMD is independent of EJC. Hrp1 downstream of a PTC (S. cerevisiae, S. pombe?), unspliced introns (S. pombe, D. melanogaster?) and extended 3’ UTRs (S. cerevisiae, S. pombe?, D. melanogaster) are the elements that can trigger NMD in these organisms. In D. melanogaster SMG1 activates UPF1 by phosphorylation. In S. cerevisiae UPF1 undergoes phosphorylation-dephosphorylation cycles,193,194 which may indicate that, similar to higher eukaryotes, this protein is activated by a yet unknown kinase. Activation of NMD in S. cerevisiae (and probably in S. pombe) results in rapid decapping and accelerated deadenylation followed by an exonucleolytic degradation. In fruit fly cells, SMG6 endonuclease cleaves the aberrant transcripts before exonucleolytic decay. b) EJC-dependent NMD. The mammalian NMD model assumes the formation of SURF and subsequent DECID complex (ribosome:SURF:EJC). Active SMG1 phopshorylates UPF1, which causes interaction between the helicase and SMG5, SMG6 and SMG7. After the endonucleolytic cleavage by SMG6, the resulting fragments are degraded by the exosome and Xrn1.
Despite the fact that there are huge differences between species in the conservation of NMD factors (i.e. SMG1 and SMG5-9), 3 core proteins are present in each examined case. They were initially identified in S. cerevisiae as UPF1-3 (up frameshift) and in C. elegans as SMG2-4 (suppressor with morphological effect on genitalia).131-135
The key player, UPF1, is an ATP-dependent RNA helicase136 with N-terminal Cys- and His-rich zinc-finger domain, 2 recombinase A (RecA)-like domains common to superfamily I (SFI) helicases and 2 regulatory domains (1B and 1C).137 UPF1 binds to UPF2, an acidic protein with 3 conserved domains that are homologous to eIF4G.138 UPF2 in turn interacts with UPF3, a protein with an unspecific RNP-type RNA-binding domain.139 UPF1, along with UPF2 and UPF3, promote accelerated mRNA degradation by endonucleolytic cleavage, 5′>3′ decay or 3′>5′ decay. Furthermore, UPF1 and presumably other UPF proteins play an important role in post-termination ribosome release from PTC and in its recycling.140,141
Saccharomyces cerevisiae
UPF1 helicase plays a central, nonetheless not completely understood, role in yeast NMD. This protein is most likely recruited to the prematurely terminating ribosome in an inactive form, as UPF1 interaction with release factors eRF1 and eRF3 inhibits UPF1 ATPase activity.142,143 Direct interaction between UPF1 and UPF2144 takes place after recognition of the PTC by the UPF2-UPF3 complex and causes activation of the NMD pathway. The N-terminal domain of UPF1 interacts with decapping factors: Edc3, Pat1 and Dcp2 (indirectly, through Edc3) which stimulates rapid, deadenylation-independent decapping followed by Xrn1 degradation.145 Alternatively, UPF1 stimulates accelerated deadenylation and 3′>5′ decay orchestrated by the Ski complex and performed by the exosome.146 The exact mechanism of deadenylation stimulation has not been described, but it may relate to UPF1 helicase activity and potential mRNP structure destabilization.
NMD in S. cerevisiae is triggered by at least 2 cis-acting elements. Firstly, the surveillance complex interacts with downstream sequence elements (DSE) associated with Hrp1 helicase, which subsequently interacts with phosphorylated UPF2.147 An absence of this interaction in Hrp1 mutants stabilizes nonsense-containing mRNAs.147 Secondly, deletion of the coding region between PTC and 3′ UTR causes stabilization of the NMD substrates.148 What is more, transcripts with extended 3′ UTRs are NMD substrates in S. cerevisiae, D. melanogaster, human and plant cells (faux 3′ UTR model),127,149,150 which suggests the role of poly(A) binding protein (or other 3′ UTR-associated proteins) and proper mRNP structure in the recognition of a PTC.
Schizosaccharomyces pombe
Fission yeast genome contains introns in about 43% of genes.151 UPF1 and UPF2 homologs were identified in S. pombe138,152 and the UPF3 sequence was predicted in the genome.153 Despite this, S. pombe was not widely studied in the context of NMD. Recent discoveries indicate, however, that fission yeast NMD could possibly lay somewhere in between mammalian (dependent on exon-junction complex) and budding yeast pathways: introns enhance NMD in S. pombe but most probably in an EJC-independent manner and it is not relevant whether PTC is inserted downstream or upstream of an intron.154
Drosophila melanogaster
Similar to fission yeast, the fly exon-junction complex (EJC) components are dispensable for NMD.155 Drosophila genome encodes all 3 UPF proteins and homologs of mammalian SMG1 (UPF1 phosphorylating kinase), SMG5 and SMG6 (endonuclease with PIN domain156), but not SMG7.155 It is believed that fruit fly cells initiate NMD only by SMG6-directed endonucleolytic cleavage and subsequent exonucleolytic decay.157
Plants
In plant cells, NMD substrates are recognized either by their long 3′ UTRs or by the presence of EJC-like complex downstream of the PTC127,158. UPF1, UPF2, UPF3 and SMG7 are conserved and essential for both types of NMD159 and UPF1 undergoes phosphorylation.160 SMG7 mediates PTC-containing transcript to degradation by Xrn4-independent pathway.161 No SMG1, SMG5 and SMG6 orthologs were found in A. thaliana.
Mammals
Mammalian genomes encode all UPF proteins – UPF1, UPF2 and 2 UPF3 paralogs, UPF3A and UPF3B (or UPF3 and UPF3X, respectively). UPF3B is believed to be slightly more effective in NMD.162 Additional factors include SMG1 and SMG5-9.
Even though NMD can act independently of the splicing event,163 the best studied mammalian model describes the role of the exon-junction complex (EJC) in the recognition of PTC. After the event of splicing, the EJC is deposited on the mRNA molecule ∼20-24 nucleotides upstream of the exon-exon junction.164 Introduction of the PTC upstream of the EJC triggers the NMD degradation pathway through UPF3, which is deposited on the EJC during splicing.
It is believed that stalling of the ribosome on the PTC causes the formation of the SURF complex between NMD and release factors (SMG1-UPF1-eRF1-eRF3).165 SMG1 kinase activity is inhibited by SMG8/SMG9 until the ribosome-SURF locates an EJC bound to the UPF2-UPF3 complex.166 Phosphorylation of UPF1 by SMG1 causes the interaction between the helicase and SMG5, SMG6 and SMG7. SMG6 has an endonucleolytic activity, cleaves a PTC-containing transcript and creates 5′ and 3′ mRNA fragments which are subsequently degraded by the exosome and Xrn1, respectively.156,167 SMG5 and SMG7 form a heterodimer that associates with UPF1 and provokes mRNA degradation.168
Dom34-Hbs1: at the intersection of NGD, NSD and NRD
There are several different RNA defects causing translational stalling of the ribosome.169 According to the error present in the RNA molecule, no-go (NGD), non-stop (NSG) or non-functional 18S rRNA decay (NRD) is triggered. Recently, it has been proposed that the evolutionarily conserved Dom34-Hbs1 complex plays a role in all 3 of those pathways by allowing the dissociation of ribosomal subunits170,171 (Fig. 4). However, all of the correlations between NGD, NSD and 18S NRD are not yet clear.
Figure 4.

A unified model of mRNA degradation pathways triggered by ribosome stalling. a) Ribosome stalling leads to recruitment of Dom34-Hbs1 complex and yeast Ski7 protein during NGD, NSD and 18S NRD pathways. b) In the case of NGD and NSD, the recruitment of Dom34-Hbs1 induces mRNA endonucleolytic cleavage by an unknown endonuclease (mainly upstream of the stalled ribosome).183 GTP hydrolysis results in Hbs1 dissociation and causes conformational changes in Dom34.178 Rli1 binds to Dom34. c) ATP hydrolysis enables subunit dissociation.186 In the case of NGD and NSD, fragments of endonucleotically cleaved mRNA are degraded by the exosome and Xrn1; nascent peptide is eliminated from the cytoplasm by the proteasome. Color coding and question marks as in Figure 1.
No-go decay (NGD) takes place when the elongation complex (EC) is blocked during translation by e.g., mRNA secondary structures or more than 6 consecutive positively charged amino acid residues present in a newly synthesized polypeptide. In those cases, subsequent to transcript degradation, Asc1 (RACK1 in mammals) mediates translation arrest and thereby leads to degradation of the aberrant protein by the E3 ubiquitin ligases Ltn1 and NOT4 (which is outside the scope of this review).172-174 Transcripts lacking an in-frame stop codon, triggering non-stop decay (NSD), can be generated by cryptic polyadenylation signals within the ORF or premature 3′ polyadenylation.175 The ribosome therefore proceeds along the poly(A) tail and is finally stalled/slowed down, probably by electrostatic interaction between positively charged poly-lysine residues and the negatively charged ribosomal tunnel.176,177 Finally, defects in ribosomal 18S rRNA may be caused by mutations, chemical damage or faulty biogenesis and lead to translational stalls and subsequent non-functional 18S rRNA decay (NRD).171
Saccharomyces cerevisiae
During NDG and NSD, the Dom34-Hbs1 complex recognizes a stalled ribosome, induces subunit dissociation178-180 and stimulates mRNA endonucleolytic cleavage by unknown endonuclease(s).181,182 It is dispensable but facilitates no-go and non-stop mRNA degradation by the exosome and is required for the complete degradation of NGD and NSD intermediates.170,183
Neither PAN2-PAN3, nor the CCR4-NOT deadenylase activity is involved in those aberrant transcripts′ degradation – the exosome is stimulated by Ski7 and is dependent on Ski2-Ski3-Ski8 heterotrimer.170,184 No-go and non-stop reporter transcripts and their intermediates are also more abundant in xrn1Δ mutant.183-185
The yeast ATPase Rli1 along with Dom34-Hbs1 complex significantly accelerates dissociation of stalled ribosome subunits,186 whereas Dom34 andRli1 are involved in the final round of ribosomal cytoplasmic maturation.187
The same factors, i.e., Dom34-Hbs1, Ski7, exosome and Xrn1, are involved in 18S NRD. However, the mechanistic differences became evident when a Dom34 interaction disrupting mutation was introduced to Hbs1 – it strongly impaired NGD but had almost no effect on 18S NRD.179 On the other hand, Dom34 mutants were defective in both NGD and 18S-NRD.183
Schizosaccharomyces pombe
In 2010, the crystal structure of fission yeast Dom34-Hbs1 complex was solved.188 Dom34 is a paralog of eRF1189 with 2 similar (M and C) and one significantly different N-terminal domain.188 Hbs1 is a conserved member of GTPase family and a paralog of eRF3.189,190 It contains a GTPase domain, and 2 domains (II and III) by which it interacts with Dom34.188 The crystal structure of fission yeast Dom34-Hbs1 and overall shape of eRF1-eRF3-GTP are alike.188
As all of the Ski complex subunits and Ski7 are present in S. pombe cells, it is reasonable to assume that NGD and NSD pathways are similar to the one in S. cerevisiae.
Drosophila melanogaster
Homologs of both Dom34 (Pelota) and Hbs1 are encoded in the fruit fly genome. Nevertheless, there are no studies concerning their involvement in RNA decay pathways in D. melanogaster. It was shown that Pelota is required for male meiosis and controls the self-renewal of germline stem cells in Drosophila.191
Plants
The A. thaliana genome encodes Pelota, homolog of Dom34. Nothing is known, however, about its involvement in RNA decay.
Mammals
Pelota is a mammalian homolog of Dom34, which forms a complex with Hbs1. Unlike yeast Dom34-Hbs1 complex, Pelota-Hbs1 has the ability to dissociate stalled ribosomes only in the presence of ABCE1 (homolog of yeast Rli1), a member of an ATP-binding cassette (ABC) transporters′ superfamily. This dissociation depends on Pelota and ABCE1 and Hbs1 has only stimulatory effect. Ribosomes which are stalled on transcripts containing more than 9 nt downstream of the P-site are not disassembled. This suggests that – contrary to that seen in the S. cerevisiae complex – Pelota-Hbs1-ABCE1 may be involved only in mammalian non-stop decay.192 Degradation by exosome pathways is plausible, as Hbs1L co-purifies with human Dis3L.104
The ability of dissociating vacant 80S ribosomes192 and the fact that Dom34-Rli1 (ABCE1 yeast homolog) is involved in the final round of ribosomal cytoplasmic maturation187 imply that this complex may be also involved in other cellular processes.
Concluding remarks
Pathways and enzymes of cytoplasmic mRNA degradation have been extensively studied in many different organisms. However, many questions still remain unanswered. Most information has been provided by studies in S. cerevisiae and humans, but there are still many gaps concerning other species. For instance, no-go, non-stop and 18S NRD decay in S. pombe, D. melanogaster and plants have not been studied and only limited information is available. Uridylation-dependent mRNA degradation is a newly discovered and thus poorly understood pathway involved in posttranscriptional gene expression regulation. Another interesting aspect is the tissue specific expression of various components of mRNA degradation pathway in metazoa, as still limited information is available on this matter.
What is more, genomes of various organisms encode many different enzymes assigned to perform the same processes, as in the case of deadenylases. Identification of their specific mRNA substrates, the exact composition of complexes or establishing possible cooperative action between them would be crucial to understand the process of, for example, the removal of the poly(A) tail, as a trigger for mRNA degradation.
Moreover, structural information is often indispensable in revealing the exact function and the mechanism of action of a single protein or a whole protein complex. Structural changes across the various species provide insight into the evolution or conservation of the protein architecture. For instance, the structure of ring-shaped exonucleases from bacteria, archaea and eukaryotes has broadened our understanding of mRNA substrate degradation mechanisms. Thus, structural studies should be performed regarding other single proteins or macromolecular complexes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by the ERC Starting grant (to AD) and a scholarship form Foundation for Polish Science International PhD programme (to MU).
References
- 1. Pavlopoulou A, Vlachakis D, Balatsos NAA, Kossida S. A comprehensive phylogenetic analysis of deadenylases. Evol Bioinforma Online 2013; 9:491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 2007; 8:113-26; PMID:17245413; http://dx.doi.org/ 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- 3. Chen C-YA, Shyu A-B. Protein segregase meddles in remodeling of mRNA-protein complexes. Genes Dev 2013; 27:980-4; PMID:23651853; http://dx.doi.org/ 10.1101/gad.219469.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 2005; 33:7138-50; PMID:16391004; http://dx.doi.org/ 10.1093/nar/gki1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen C-YA, Shyu A-B. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol 2003; 23:4805-13; PMID:12832468; http://dx.doi.org/ 10.1128/MCB.23.14.4805-4813.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A 2006; 103:4034-9; PMID:16495412; http://dx.doi.org/ 10.1073/pnas.0510928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA N Y N 2009; 15:21-32; http://dx.doi.org/ 10.1261/rna.1399509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shyu AB, Greenberg ME, Belasco JG. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev 1989; 3:60-72; PMID:2496006; http://dx.doi.org/ 10.1101/gad.3.1.60 [DOI] [PubMed] [Google Scholar]
- 9. Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol 2008; 9:337-44; PMID:18334997; http://dx.doi.org/ 10.1038/nrm2370 [DOI] [PubMed] [Google Scholar]
- 10. Thore S, Mauxion F, Séraphin B, Suck D. X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep 2003; 4:1150-5; PMID:14618157; http://dx.doi.org/ 10.1038/sj.embor.7400020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim Biophys Acta 2013; 1829:561-70; PMID:23337855; http://dx.doi.org/ 10.1016/j.bbagrm.2013.01.003 [DOI] [PubMed] [Google Scholar]
- 12. Collart MA. Global control of gene expression in yeast by the Ccr4-Not complex. Gene 2003; 313:1-16; PMID:12957374; http://dx.doi.org/ 10.1016/S0378-1119(03)00672-3 [DOI] [PubMed] [Google Scholar]
- 13. Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J Mol Biol 2001; 314:683-94; PMID:11733989; http://dx.doi.org/ 10.1006/jmbi.2001.5162 [DOI] [PubMed] [Google Scholar]
- 14. Chen J, Chiang Y-C, Denis CL. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J 2002; 21:1414-26; PMID:11889047; http://dx.doi.org/ 10.1093/emboj/21.6.1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clark LB, Viswanathan P, Quigley G, Chiang Y-C, McMahon JS, Yao G, Chen J, Nelsbach A, Denis CL. Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J Biol Chem 2004; 279:13616-23; PMID:14734555; http://dx.doi.org/ 10.1074/jbc.M313202200 [DOI] [PubMed] [Google Scholar]
- 16. Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J 2002; 21:1427-36; PMID:11889048; http://dx.doi.org/ 10.1093/emboj/21.6.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 2004; 11:121-7; PMID:14749774; http://dx.doi.org/ 10.1038/nsmb724 [DOI] [PubMed] [Google Scholar]
- 18. Bhaskar V, Roudko V, Basquin J, Sharma K, Urlaub H, Séraphin B, Conti E. Structure and RNA-binding properties of the Not1-Not2-Not5 module of the yeast Ccr4-Not complex. Nat Struct Mol Biol 2013; 20:1281-8; PMID:24121231; http://dx.doi.org/ 10.1038/nsmb.2686 [DOI] [PubMed] [Google Scholar]
- 19. Panasenko OO, Collart MA. Not4 E3 ligase contributes to proteasome assembly and functional integrity in part through Ecm29. Mol Cell Biol 2011; 31:1610-23; PMID:21321079; http://dx.doi.org/ 10.1128/MCB.01210-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boeck R, Tarun S. Jr, Rieger M, Deardorff JA, Müller-Auer S, Sachs AB. The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J Biol Chem 1996; 271:432-8; PMID:8550599; http://dx.doi.org/ 10.1074/jbc.271.1.432 [DOI] [PubMed] [Google Scholar]
- 21. Brown CE, Tarun SZ, Jr, Boeck R, Sachs AB. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol 1996; 16:5744-53; PMID:8816488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 2001; 104:377-86; PMID:11239395; http://dx.doi.org/ 10.1016/S0092-8674(01)00225-2 [DOI] [PubMed] [Google Scholar]
- 23. Parker R. RNA degradation in Saccharomyces cerevisae. Genetics 2012; 191:671-702; PMID:22785621; http://dx.doi.org/ 10.1534/genetics.111.137265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim D-U, Hayles J, Kim D, Wood V, Park H-O, Won M, Yoo H-S, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 2010; 28:617-23; PMID:20473289; http://dx.doi.org/ 10.1038/nbt.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hayles J, Wood V, Jeffery L, Hoe K-L, Kim D-U, Park H-O, Salas-Pino S, Heichinger C, Nurse P. A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol 2013; 3:130053; PMID:23697806; http://dx.doi.org/ 10.1098/rsob.130053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andersen KR, Jonstrup AT, Van LB, Brodersen DE. The activity and selectivity of fission yeast Pop2p are affected by a high affinity for Zn2+ and Mn2+ in the active site. RNA N Y N 2009; 15:850-61; http://dx.doi.org/ 10.1261/rna.1489409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marasovic M, Zocco M, Halic M. Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol Cell 2013; 52:173-83; PMID:24095277; http://dx.doi.org/ 10.1016/j.molcel.2013.08.046 [DOI] [PubMed] [Google Scholar]
- 28. Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J 2004; 23:2862-71; PMID:15215893; http://dx.doi.org/ 10.1038/sj.emboj.7600273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E. Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA N Y N 2010; 16:1356-70; http://dx.doi.org/ 10.1261/rna.2145110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huntzinger E, Kuzuoglu-Öztürk D, Braun JE, Eulalio A, Wohlbold L, Izaurralde E. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res 2013; 41:978-94; PMID:23172285; http://dx.doi.org/ 10.1093/nar/gks1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chiba Y, Green PJ. mRNA Degradation Machinery in Plants. J Plant Biol 2009; 52:114-24; http://dx.doi.org/ 10.1007/s12374-009-9021-2 [DOI] [Google Scholar]
- 32. Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol 2010; 152:866-75; PMID:19955262; http://dx.doi.org/ 10.1104/pp.109.149005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walley JW, Kelley DR, Savchenko T, Dehesh K. Investigating the function of CAF1 deadenylases during plant stress responses. Plant Signal Behav 2010; 5:802-5; PMID:20421740; http://dx.doi.org/ 10.4161/psb.5.7.11578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abbasi N, Park Y-I, Choi S-B. RNA deadenylation and decay in plants. J Plant Biol 2013; 56:198-207; http://dx.doi.org/ 10.1007/s12374-013-0201-8 [DOI] [Google Scholar]
- 35. Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA N Y N 2004; 10:1200-14; http://dx.doi.org/ 10.1261/rna.7540204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Doidge R, Mittal S, Aslam A, Winkler GS. Deadenylation of cytoplasmic mRNA by the mammalian Ccr4-Not complex. Biochem Soc Trans 2012; 40:896-901; PMID:22817755; http://dx.doi.org/ 10.1042/BST20120074 [DOI] [PubMed] [Google Scholar]
- 37. Lau N-C, Kolkman A, van Schaik FMA, Mulder KW, Pijnappel WWMP, Heck AJR, Timmers HTM. Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J 2009; 422:443-53; PMID:19558367; http://dx.doi.org/ 10.1042/BJ20090500 [DOI] [PubMed] [Google Scholar]
- 38. Mittal S, Aslam A, Doidge R, Medica R, Winkler GS. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death and senescence. Mol Biol Cell 2011; 22:748-58; PMID:21233283; http://dx.doi.org/ 10.1091/mbc.E10-11-0898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Uchida N, Hoshino S-I, Katada T. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J Biol Chem 2004; 279:1383-91; PMID:14583602; http://dx.doi.org/ 10.1074/jbc.M309125200 [DOI] [PubMed] [Google Scholar]
- 40. Yamashita A, Chang T-C, Yamashita Y, Zhu W, Zhong Z, Chen C-YA, Shyu A-B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 2005; 12:1054-63; PMID:16284618; http://dx.doi.org/ 10.1038/nsmb1016 [DOI] [PubMed] [Google Scholar]
- 41. Chen C-YA, Shyu A-B. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA 2011; 2:167-83; PMID:21957004; http://dx.doi.org/ 10.1002/wrna.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gao M, Fritz DT, Ford LP, Wilusz J. Interaction between a Poly(A)-Specific Ribonuclease and the 5? Cap Influences mRNA Deadenylation Rates In Vitro. Mol Cell 2000; 5:479-88; PMID:10882133; http://dx.doi.org/ 10.1016/S1097-2765(00)80442-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip Rev RNA 2010; 1:253-65; PMID:21935889; http://dx.doi.org/ 10.1002/wrna.15 [DOI] [PubMed] [Google Scholar]
- 44. Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol 2004; 39:197-216; PMID:15596551; http://dx.doi.org/ 10.1080/10409230490513991 [DOI] [PubMed] [Google Scholar]
- 45. Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA N Y N 2003; 9:231-8; http://dx.doi.org/ 10.1261/rna.2151403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J 1999; 18:5411-22; PMID:10508173; http://dx.doi.org/ 10.1093/emboj/18.19.5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gavin A-C, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon A-M, Cruciat C-M, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 2002; 415:141-7; PMID:11805826; http://dx.doi.org/ 10.1038/415141a [DOI] [PubMed] [Google Scholar]
- 48. She M, Decker CJ, Svergun DI, Round A, Chen N, Muhlrad D, Parker R, Song H. Structural basis of dcp2 recognition and activation by dcp1. Mol Cell 2008; 29:337-49; PMID:18280239; http://dx.doi.org/ 10.1016/j.molcel.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ling SHM, Qamra R, Song H. Structural and functional insights into eukaryotic mRNA decapping. Wiley Interdiscip Rev RNA 2011; 2:193-208; PMID:21957006; http://dx.doi.org/ 10.1002/wrna.44 [DOI] [PubMed] [Google Scholar]
- 50. Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem 2004; 73:861-90; PMID:15189161; http://dx.doi.org/ 10.1146/annurev.biochem.73.011303.074032 [DOI] [PubMed] [Google Scholar]
- 51. Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell 2008; 32:605-15; PMID:19061636; http://dx.doi.org/ 10.1016/j.molcel.2008.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tharun S. Lsm1-7-Pat1 complex: A link between 3′ and 5′-ends in mRNA decay? RNA Biol 2009; 6:228-32; PMID:19279404; http://dx.doi.org/ 10.4161/rna.6.3.8282 [DOI] [PubMed] [Google Scholar]
- 53. Sharif H, Conti E. Architecture of the Lsm1-7-Pat1 complex: a conserved assembly in eukaryotic mRNA turnover. Cell Rep 2013; 5:283-91; PMID:24139796; http://dx.doi.org/ 10.1016/j.celrep.2013.10.004 [DOI] [PubMed] [Google Scholar]
- 54. Chowdhury A, Mukhopadhyay J, Tharun S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 2007; 13:998-1016; PMID:17513695; http://dx.doi.org/ 10.1261/rna.502507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell 2010; 39:773-83; PMID:20832728; http://dx.doi.org/ 10.1016/j.molcel.2010.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Braun JE, Truffault V, Boland A, Huntzinger E, Chang C-T, Haas G, Weichenrieder O, Coles M, Izaurralde E. A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5′ exonucleolytic degradation. Nat Struct Mol Biol 2012; 19:1324-31; PMID:23142987; http://dx.doi.org/ 10.1038/nsmb.2413 [DOI] [PubMed] [Google Scholar]
- 57. Sun M, Schwalb B, Pirkl N, Maier KC, Schenk A, Failmezger H, Tresch A, Cramer P. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell 2013; 52:52-62; PMID:24119399; http://dx.doi.org/ 10.1016/j.molcel.2013.09.010 [DOI] [PubMed] [Google Scholar]
- 58. Harigaya Y, Jones BN, Muhlrad D, Gross JD, Parker R. Identification and analysis of the interaction between Edc3 and Dcp2 in Saccharomyces cerevisiae. Mol Cell Biol 2010; 30:1446-56; PMID:20086104; http://dx.doi.org/ 10.1128/MCB.01305-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wu D, Muhlrad D, Bowler MW, Jiang S, Liu Z, Parker R, Song H. Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation. Cell Res 2014; 24:233-46; PMID:24247251; http://dx.doi.org/ 10.1038/cr.2013.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sakuno T, Araki Y, Ohya Y, Kofuji S, Takahashi S, Hoshino S, Katada T. Decapping reaction of mRNA requires Dcp1 in fission yeast: its characterization in different species from yeast to human. J Biochem (Tokyo) 2004; 136:805-12; http://dx.doi.org/ 10.1093/jb/mvh190 [DOI] [PubMed] [Google Scholar]
- 61. She M, Decker CJ, Chen N, Tumati S, Parker R, Song H. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat Struct Mol Biol 2006; 13:63-70; PMID:16341225; http://dx.doi.org/ 10.1038/nsmb1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fromm SA, Truffault V, Kamenz J, Braun JE, Hoffmann NA, Izaurralde E, Sprangers R. The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex. EMBO J 2012; 31:279-90; PMID:22085934; http://dx.doi.org/ 10.1038/emboj.2011.408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang C-Y, Chen W-L, Wang S-W. Pdc1 functions in the assembly of P bodies in Schizosaccharomyces pombe. Mol Cell Biol 2013; 33:1244-53; PMID:23319050; http://dx.doi.org/ 10.1128/MCB.01583-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 2009; 16:616-23; PMID:19430462; http://dx.doi.org/ 10.1038/nsmb.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Szankasi P, Smith GR. Requirement of S. pombe exonuclease II, a homologue of S. cerevisiae Sep1, for normal mitotic growth and viability. Curr Genet 1996; 30:284-93; PMID:8781170; http://dx.doi.org/ 10.1007/s002940050134 [DOI] [PubMed] [Google Scholar]
- 66. Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V. Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol 2008; 28:6695-708; PMID:18765641; http://dx.doi.org/ 10.1128/MCB.00759-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol 2010; 189:289-302; PMID:20404111; http://dx.doi.org/ 10.1083/jcb.200910141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Xu J, Yang J-Y, Niu Q-W, Chua N-H. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell 2006; 18:3386-98; PMID:17158604; http://dx.doi.org/ 10.1105/tpc.106.047605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xu J, Chua N-H. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell 2009; 21:3270-9; PMID:19855049; http://dx.doi.org/ 10.1105/tpc.109.070078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci U S A 2000; 97:13985-90; PMID:11106401; http://dx.doi.org/ 10.1073/pnas.97.25.13985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rymarquis LA, Souret FF, Green PJ. Evidence that XRN4, an Arabidopsis homolog of exoribonuclease XRN1, preferentially impacts transcripts with certain sequences or in particular functional categories. RNA N Y N 2011; 17:501-11; http://dx.doi.org/ 10.1261/rna.2467911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Piccirillo C, Khanna R, Kiledjian M. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA N Y N 2003; 9:1138-47; http://dx.doi.org/ 10.1261/rna.5690503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fenger-Grøn M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 2005; 20:905-15; PMID:16364915; http://dx.doi.org/ 10.1016/j.molcel.2005.10.031 [DOI] [PubMed] [Google Scholar]
- 74. Mildvan AS, Xia Z, Azurmendi HF, Saraswat V, Legler PM, Massiah MA, Gabelli SB, Bianchet MA, Kang L-W, Amzel LM. Structures and mechanisms of Nudix hydrolases. Arch Biochem Biophys 2005; 433:129-43; PMID:15581572; http://dx.doi.org/ 10.1016/j.abb.2004.08.017 [DOI] [PubMed] [Google Scholar]
- 75. Taylor MJ, Peculis BA. Evolutionary conservation supports ancient origin for Nudt16, a nuclear-localized, RNA-binding, RNA-decapping enzyme. Nucleic Acids Res 2008; 36:6021-34; PMID:18820299; http://dx.doi.org/ 10.1093/nar/gkn605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Song M-G, Li Y, Kiledjian M. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell 2010; 40:423-32; PMID:21070968; http://dx.doi.org/ 10.1016/j.molcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chlebowski A, Lubas M, Jensen TH, Dziembowski A. RNA decay machines: the exosome. Biochim Biophys Acta 2013; 1829:552-60; PMID:23352926; http://dx.doi.org/ 10.1016/j.bbagrm.2013.01.006 [DOI] [PubMed] [Google Scholar]
- 78. Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 2006; 127:1223-37; PMID:17174896; http://dx.doi.org/ 10.1016/j.cell.2006.10.037 [DOI] [PubMed] [Google Scholar]
- 79. Dziembowski A, Lorentzen E, Conti E, Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 2007; 14:15-22; PMID:17173052; http://dx.doi.org/ 10.1038/nsmb1184 [DOI] [PubMed] [Google Scholar]
- 80. Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell 2007; 27:324-31; PMID:17643380; http://dx.doi.org/ 10.1016/j.molcel.2007.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Frazão C, McVey CE, Amblar M, Barbas A, Vonrhein C, Arraiano CM, Carrondo MA. Unravelling the dynamics of RNA degradation by ribonuclease II and its RNA-bound complex. Nature 2006; 443:110-4; http://dx.doi.org/ 10.1038/nature05080 [DOI] [PubMed] [Google Scholar]
- 82. Schaeffer D, Meaux S, Clark A, van Hoof A. Determining in vivo activity of the yeast cytoplasmic exosome. Methods Enzymol 2008; 448:227-39; PMID:19111179; http://dx.doi.org/ 10.1016/S0076-6879(08)02612-8 [DOI] [PubMed] [Google Scholar]
- 83. Lebreton A, Tomecki R, Dziembowski A, Séraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 2008; 456:993-6; PMID:19060886; http://dx.doi.org/ 10.1038/nature07480 [DOI] [PubMed] [Google Scholar]
- 84. Schaeffer D, Tsanova B, Barbas A, Reis FP, Dastidar EG, Sanchez-Rotunno M, Arraiano CM, van Hoof A. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol 2009; 16:56-62; PMID:19060898; http://dx.doi.org/ 10.1038/nsmb.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Halbach F, Reichelt P, Rode M, Conti E. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell 2013; 154:814-26; PMID:23953113; http://dx.doi.org/ 10.1016/j.cell.2013.07.017 [DOI] [PubMed] [Google Scholar]
- 86. Araki Y, Takahashi S, Kobayashi T, Kajiho H, Hoshino S, Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J 2001; 20:4684-93; PMID:11532933; http://dx.doi.org/ 10.1093/emboj/20.17.4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J 2002; 21:4699-708; PMID:12198172; http://dx.doi.org/ 10.1093/emboj/cdf448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chen N, Walsh MA, Liu Y, Parker R, Song H. Crystal structures of human DcpS in ligand-free and m7GDP-bound forms suggest a dynamic mechanism for scavenger mRNA decapping. J Mol Biol 2005; 347:707-18; PMID:15769464; http://dx.doi.org/ 10.1016/j.jmb.2005.01.062 [DOI] [PubMed] [Google Scholar]
- 89. Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell 2009; 139:547-59; PMID:19879841; http://dx.doi.org/ 10.1016/j.cell.2009.08.042 [DOI] [PubMed] [Google Scholar]
- 90. Malet H, Topf M, Clare DK, Ebert J, Bonneau F, Basquin J, Drazkowska K, Tomecki R, Dziembowski A, Conti E, et al. RNA channelling by the eukaryotic exosome. EMBO Rep 2010; 11:936-42; PMID:21072061; http://dx.doi.org/ 10.1038/embor.2010.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Drazkowska K, Tomecki R, Stodus K, Kowalska K, Czarnocki-Cieciura M, Dziembowski A. The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res 2013; 41:3845-58; PMID:23404585; http://dx.doi.org/ 10.1093/nar/gkt060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Liu S-W, Jiao X, Liu H, Gu M, Lima CD, Kiledjian M. Functional analysis of mRNA scavenger decapping enzymes. RNA N Y N 2004; 10:1412-22; http://dx.doi.org/ 10.1261/rna.7660804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 2013; 32:1842-54; PMID:23503588; http://dx.doi.org/ 10.1038/emboj.2013.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Marshall AN, Montealegre MC, Jiménez-López C, Lorenz MC, van Hoof A. Alternative splicing and subfunctionalization generates functional diversity in fungal proteomes. PLoS Genet 2013; 9:e1003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kiss DL, Andrulis ED. The exozyme model: a continuum of functionally distinct complexes. RNA N Y N 2011; 17:1-13; http://dx.doi.org/ 10.1261/rna.2364811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 2002; 420:837-41; PMID:12490954; http://dx.doi.org/ 10.1038/nature01181 [DOI] [PubMed] [Google Scholar]
- 97. Mamolen M, Smith A, Andrulis ED. Drosophila melanogaster Dis3 N-terminal domains are required for ribonuclease activities, nuclear localization and exosome interactions. Nucleic Acids Res 2010; 38:5507-17; PMID:20421210; http://dx.doi.org/ 10.1093/nar/gkq295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Graham AC, Kiss DL, Andrulis ED. Differential distribution of exosome subunits at the nuclear lamina and in cytoplasmic foci. Mol Biol Cell 2006; 17:1399-409; PMID:16407406; http://dx.doi.org/ 10.1091/mbc.E05-08-0805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 2005; 11:459-69; PMID:15703439; http://dx.doi.org/ 10.1261/rna.7231505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Chekanova JA, Shaw RJ, Wills MA, Belostotsky DA. Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J Biol Chem 2000; 275:33158-66; PMID:10930416; http://dx.doi.org/ 10.1074/jbc.M005493200 [DOI] [PubMed] [Google Scholar]
- 101. Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 2007; 131:1340-53; PMID:18160042; http://dx.doi.org/ 10.1016/j.cell.2007.10.056 [DOI] [PubMed] [Google Scholar]
- 102. Kumakura N, Otsuki H, Tsuzuki M, Takeda A, Watanabe Y. Arabidopsis AtRRP44A Is the Functional Homolog of Rrp44/Dis3, an Exosome Component, Is Essential for Viability and Is Required for RNA Processing and Degradation. PLoS ONE 2013; 8:e79219; PMID: 24244451; http://dx.doi.org/ 10.1371/journal.pone.0079219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Lange H, Holec S, Cognat V, Pieuchot L, Ret ML, Canaday J, Gagliardi D. Degradation of a Polyadenylated rRNA Maturation By-Product Involves One of the Three RRP6-Like Proteins in Arabidopsis thaliana. Mol Cell Biol 2008; 28:3038-44; PMID:18285452; http://dx.doi.org/ 10.1128/MCB.02064-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J 2010; 29:2342-57; PMID:20531386; http://dx.doi.org/ 10.1038/emboj.2010.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zhu B, Mandal SS, Pham A-D, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev 2005; 19:1668-73; PMID:16024656; http://dx.doi.org/ 10.1101/gad.1292105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Scott DD, Norbury CJ. RNA decay via 3′ uridylation. Biochim Biophys Acta 2013; 1829:654-65; PMID:23385389; http://dx.doi.org/ 10.1016/j.bbagrm.2013.01.009 [DOI] [PubMed] [Google Scholar]
- 107. Chang H-M, Triboulet R, Thornton JE, Gregory RI. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 2013; 497:244-8; PMID:23594738; http://dx.doi.org/ 10.1038/nature12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lubas M, Damgaard CK, Tomecki R, Cysewski D, Jensen TH, Dziembowski A. Exonuclease hDIS3L2 specifies an exosome-independent 3′-5′ degradation pathway of human cytoplasmic mRNA. EMBO J 2013; 32:1855-68; PMID:23756462; http://dx.doi.org/ 10.1038/emboj.2013.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA N Y N 2013; 19:1632-8; http://dx.doi.org/ 10.1261/rna.040055.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell 2008; 32:276-84; PMID:18951094; http://dx.doi.org/ 10.1016/j.molcel.2008.09.014 [DOI] [PubMed] [Google Scholar]
- 111. Heo I, Ha M, Lim J, Yoon M-J, Park J-E, Kwon SC, Chang H, Kim VN. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell 2012; 151:521-32; PMID:23063654; http://dx.doi.org/ 10.1016/j.cell.2012.09.022 [DOI] [PubMed] [Google Scholar]
- 112. Van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell 2009; 139:135-48; PMID:19804759; http://dx.doi.org/ 10.1016/j.cell.2009.09.012 [DOI] [PubMed] [Google Scholar]
- 113. Ibrahim F, Rymarquis LA, Kim E-J, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A 2010; 107:3906-11; PMID:20142471; http://dx.doi.org/ 10.1073/pnas.0912632107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJT, Roovers EF, Ladurner P, Berezikov E, Ketting RF. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J 2010; 29:3688-700; PMID:20859253; http://dx.doi.org/ 10.1038/emboj.2010.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol CB 2012; 22:689-94; http://dx.doi.org/ 10.1016/j.cub.2012.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Yang X, Purdy M, Marzluff WF, Dominski Z. Characterization of 3′hExo, a 3′ exonuclease specifically interacting with the 3′ end of histone mRNA. J Biol Chem 2006; 281:30447-54; PMID:16912046; http://dx.doi.org/ 10.1074/jbc.M602947200 [DOI] [PubMed] [Google Scholar]
- 117. Hoefig KP, Rath N, Heinz GA, Wolf C, Dameris J, Schepers A, Kremmer E, Ansel KM, Heissmeyer V. Eri1 degrades the stem-loop of oligouridylated histone mRNAs to induce replication-dependent decay. Nat Struct Mol Biol 2013; 20:73-81; PMID:23202588; http://dx.doi.org/ 10.1038/nsmb.2450 [DOI] [PubMed] [Google Scholar]
- 118. Tan D, Marzluff WF, Dominski Z, Tong L. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3′hExo ternary complex. Science 2013; 339:318-21; PMID:23329046; http://dx.doi.org/ 10.1126/science.1228705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol 2005; 12:794-800; PMID:16086026; http://dx.doi.org/ 10.1038/nsmb972 [DOI] [PubMed] [Google Scholar]
- 120. Schmidt M-J, West S, Norbury CJ. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA N Y N 2011; 17:39-44; http://dx.doi.org/ 10.1261/rna.2252511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev 2008; 22:50-65; PMID:PMID:18172165; http://dx.doi.org/ 10.1101/gad.1622708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Su W, Slepenkov SV, Slevin MK, Lyons SM, Ziemniak M, Kowalska J, Darzynkiewicz E, Jemielity J, Marzluff WF, Rhoads RE. mRNAs containing the histone 3′ stem-loop are degraded primarily by decapping mediated by oligouridylation of the 3′ end. RNA N Y N 2013; 19:1-16; http://dx.doi.org/ 10.1261/rna.034470.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol 2012; 19:594-601; PMID:22664987; http://dx.doi.org/ 10.1038/nsmb.2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet 2012; 13:246-59; PMID:22392217; http://dx.doi.org/ 10.1038/nrg3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Popp MW-L, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 2013; 47:139-65; PMID:24274751; http://dx.doi.org/ 10.1146/annurev-genet-111212-133424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Baker KE, Parker R. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr Opin Cell Biol 2004; 16:293-9; PMID:15145354; http://dx.doi.org/ 10.1016/j.ceb.2004.03.003 [DOI] [PubMed] [Google Scholar]
- 127. Kertész S, Kerényi Z, Mérai Z, Bartos I, Pálfy T, Barta E, Silhavy D. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res 2006; 34:6147-57; http://dx.doi.org/ 10.1093/nar/gkl737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Nyikó T, Sonkoly B, Mérai Z, Benkovics AH, Silhavy D. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol Biol 2009; 71:367-78; http://dx.doi.org/ 10.1007/s11103-009-9528-4 [DOI] [PubMed] [Google Scholar]
- 129. Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr Opin Cell Biol 2005; 17:316-25; PMID:15901503; http://dx.doi.org/ 10.1016/j.ceb.2005.04.005 [DOI] [PubMed] [Google Scholar]
- 130. Maquat LE, Tarn W-Y, Isken O. The pioneer round of translation: features and functions. Cell 2010; 142:368-74; PMID:20691898; http://dx.doi.org/ 10.1016/j.cell.2010.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev 1995; 9:423-36; PMID:7883167; http://dx.doi.org/ 10.1101/gad.9.4.423 [DOI] [PubMed] [Google Scholar]
- 132. Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 1991; 5:2303-14; PMID:1748286; http://dx.doi.org/ 10.1101/gad.5.12a.2303 [DOI] [PubMed] [Google Scholar]
- 133. Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol 1992; 12:2165-77; PMID:1569946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev 1993; 7:1885-97; PMID:8104846; http://dx.doi.org/ 10.1101/gad.7.10.1885 [DOI] [PubMed] [Google Scholar]
- 135. Cali BM, Kuchma SL, Latham J, Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans. Genetics 1999; 151:605-16; PMID:9927455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA N Y N 2000; 6:1226-35; http://dx.doi.org/ 10.1017/S1355838200000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J 2007; 26:253-64; PMID:17159905; http://dx.doi.org/ 10.1038/sj.emboj.7601464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol 2000; 20:8944-57; PMID:11073994; http://dx.doi.org/ 10.1128/MCB.20.23.8944-8957.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol 2004; 11:330-7; PMID:15004547; http://dx.doi.org/ 10.1038/nsmb741 [DOI] [PubMed] [Google Scholar]
- 140. Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense- mediated mRNA decay. Cell 2010; 143:938-50; PMID:21145460; http://dx.doi.org/ 10.1016/j.cell.2010.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ghosh S, Ganesan R, Amrani N, Jacobson A. Translational competence of ribosomes released from a premature termination codon is modulated by NMD factors. RNA N Y N 2010; 16:1832-47; http://dx.doi.org/ 10.1261/rna.1987710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev 1998; 12:1665-77; PMID:9620853; http://dx.doi.org/ 10.1101/gad.12.11.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Wang W, Czaplinski K, Rao Y, Peltz SW. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J 2001; 20:880-90; PMID:11179232; http://dx.doi.org/ 10.1093/emboj/20.4.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev 1995; 9:437-54; PMID:7883168; http://dx.doi.org/ 10.1101/gad.9.4.437 [DOI] [PubMed] [Google Scholar]
- 145. Swisher KD, Parker R. Interactions between Upf1 and the decapping factors Edc3 and Pat1 in Saccharomyces cerevisiae. PloS One 2011; 6:e26547; PMID:22065998; http://dx.doi.org/ 10.1371/journal.pone.0026547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′->5′ degradation. Mol Cell 2003; 11:1405-13; PMID:12769863; http://dx.doi.org/ 10.1016/S1097-2765(03)00190-4 [DOI] [PubMed] [Google Scholar]
- 147. González CI, Ruiz-Echevarría MJ, Vasudevan S, Henry MF, Peltz SW. The yeast hnRNP-like protein Hrp1/Nab4 marks a transcript for nonsense-mediated mRNA decay. Mol Cell 2000; 5:489-99; http://dx.doi.org/ 10.1016/S1097-2765(00)80443-8 [DOI] [PubMed] [Google Scholar]
- 148. Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 2004; 432:112-8; PMID:15525991; http://dx.doi.org/ 10.1038/nature03060 [DOI] [PubMed] [Google Scholar]
- 149. Kebaara BW, Atkin AL. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae. Nucleic Acids Res 2009; 37:2771-8; PMID:19270062; http://dx.doi.org/ 10.1093/nar/gkp146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J 2007; 26:1591-601; PMID:17318186; http://dx.doi.org/ 10.1038/sj.emboj.7601588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Wood V, Gwilliam R, Rajandream M-A, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature 2002; 415:871-80; PMID:11859360; http://dx.doi.org/ 10.1038/nature724 [DOI] [PubMed] [Google Scholar]
- 152. Rodríguez-Gabriel MA, Watt S, Bähler J, Russell P. Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol Cell Biol 2006; 26:6347-56; http://dx.doi.org/ 10.1128/MCB.00286-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Wood V, Harris MA, McDowall MD, Rutherford K, Vaughan BW, Staines DM, Aslett M, Lock A, Bähler J, Kersey PJ, et al. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res 2012; 40:D695-699; PMID:22039153; http://dx.doi.org/ 10.1093/nar/gkr853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Wen J, Brogna S. Splicing-dependent NMD does not require the EJC in Schizosaccharomyces pombe. EMBO J 2010; 29:1537-51; PMID:20360683; http://dx.doi.org/ 10.1038/emboj.2010.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J 2003; 22:3960-70; PMID:12881430; http://dx.doi.org/ 10.1093/emboj/cdg371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA N Y N 2008; 14:2609-17; http://dx.doi.org/ 10.1261/rna.1386208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Frizzell KA, Rynearson SG, Metzstein MM. Drosophila mutants show NMD pathway activity is reduced, but not eliminated, in the absence of Smg6. RNA N Y N 2012; 18:1475-86; http://dx.doi.org/ 10.1261/rna.032821.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 2012; 40:2454-69; PMID:22127866; http://dx.doi.org/ 10.1093/nar/gkr932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Riehs-Kearnan N, Gloggnitzer J, Dekrout B, Jonak C, Riha K. Aberrant growth and lethality of Arabidopsis deficient in nonsense-mediated RNA decay factors is caused by autoimmune-like response. Nucleic Acids Res 2012; 40:5615-24; PMID:22379136; http://dx.doi.org/ 10.1093/nar/gks195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kerényi F, Wawer I, Sikorski PJ, Kufel J, Silhavy D. Phosphorylation of the N- and C-terminal UPF1 domains plays a critical role in plant nonsense-mediated mRNA decay. Plant J Cell Mol Biol 2013; 76:836-48; http://dx.doi.org/ 10.1111/tpj.12346 [DOI] [PubMed] [Google Scholar]
- 161. Mérai Z, Benkovics AH, Nyikó T, Debreczeny M, Hiripi L, Kerényi Z, Kondorosi E, Silhavy D. The late steps of plant nonsense-mediated mRNA decay. Plant J Cell Mol Biol 2012; 73:50-62; PMID: 22974464; http://dx.doi.org/ 10.1111/tpj.12015 [DOI] [PubMed] [Google Scholar]
- 162. Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA N Y N 2006; 12:1015-22; http://dx.doi.org/ 10.1261/rna.12506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Metze S, Herzog VA, Ruepp M-D, Mühlemann O. Comparison of EJC-enhanced and EJC-independent NMD in human cells reveals two partially redundant degradation pathways. RNA N Y N 2013; 19:1432-48; http://dx.doi.org/ 10.1261/rna.038893.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 2000; 19:6860-9; PMID:11118221; http://dx.doi.org/ 10.1093/emboj/19.24.6860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 2006; 20:355-67; PMID:16452507; http://dx.doi.org/ 10.1101/gad.1389006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev 2009; 23:1091-105; PMID:19417104; http://dx.doi.org/ 10.1101/gad.1767209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 2009; 16:49-55; PMID:19060897; http://dx.doi.org/ 10.1038/nsmb.1530 [DOI] [PubMed] [Google Scholar]
- 168. Jonas S, Weichenrieder O, Izaurralde E. An unusual arrangement of two 14-3-3-like domains in the SMG5-SMG7 heterodimer is required for efficient nonsense-mediated mRNA decay. Genes Dev 2013; 27:211-25; PMID:23348841; http://dx.doi.org/ 10.1101/gad.206672.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Graille M, Séraphin B. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat Rev Mol Cell Biol 2012; 13:727-35; PMID:23072885; http://dx.doi.org/ 10.1038/nrm3457 [DOI] [PubMed] [Google Scholar]
- 170. Van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 2002; 295:2262-4; PMID:11910110; http://dx.doi.org/ 10.1126/science.1067272 [DOI] [PubMed] [Google Scholar]
- 171. Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell 2009; 34:440-50; PMID:19481524; http://dx.doi.org/ 10.1016/j.molcel.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Dimitrova LN, Kuroha K, Tatematsu T, Inada T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem 2009; 284:10343-52; PMID:19204001; http://dx.doi.org/ 10.1074/jbc.M808840200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K, Inada T. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep 2010; 11:956-61; PMID:21072063; http://dx.doi.org/ 10.1038/embor.2010.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Bengtson MH, Joazeiro CAP. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 2010; 467:470-3; PMID:20835226; http://dx.doi.org/ 10.1038/nature09371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 2010; 143:1018-29; PMID:21145465; http://dx.doi.org/ 10.1016/j.cell.2010.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Lu J, Kobertz WR, Deutsch C. Mapping the electrostatic potential within the ribosomal exit tunnel. J Mol Biol 2007; 371:1378-91; PMID:17631312; http://dx.doi.org/ 10.1016/j.jmb.2007.06.038 [DOI] [PubMed] [Google Scholar]
- 177. Lu J, Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol 2008; 384:73-86; PMID:18822297; http://dx.doi.org/ 10.1016/j.jmb.2008.08.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 2010; 330:369-72; PMID:20947765; http://dx.doi.org/ 10.1126/science.1192430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Van den Elzen AMG, Henri J, Lazar N, Gas ME, Durand D, Lacroute F, Nicaise M, van Tilbeurgh H, Séraphin B, Graille M. Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nat Struct Mol Biol 2010; 17:1446-52; PMID:21102444; http://dx.doi.org/ 10.1038/nsmb.1963 [DOI] [PubMed] [Google Scholar]
- 180. Kobayashi K, Kikuno I, Kuroha K, Saito K, Ito K, Ishitani R, Inada T, Nureki O. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1α complex. Proc Natl Acad Sci U S A 2010; 107:17575-9; PMID:20876129; http://dx.doi.org/ 10.1073/pnas.1009598107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 2006; 440:561-4; PMID:16554824; http://dx.doi.org/ 10.1038/nature04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Schaeffer D, van Hoof A. Different nuclease requirements for exosome-mediated degradation of normal and nonstop mRNAs. Proc Natl Acad Sci U S A 2011; 108:2366-71; PMID:21262801; http://dx.doi.org/ 10.1073/pnas.1013180108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell 2012; 46:518-29; PMID:22503425; http://dx.doi.org/ 10.1016/j.molcel.2012.03.013 [DOI] [PubMed] [Google Scholar]
- 184. Frischmeyer PA, Hoof A van, O′Donnell K, Guerrerio AL, Parker R, Dietz HC. An mRNA Surveillance Mechanism That Eliminates Transcripts Lacking Termination Codons. Science 2002; 295:2258-61; PMID:11910109; http://dx.doi.org/ 10.1126/science.1067338 [DOI] [PubMed] [Google Scholar]
- 185. Inada T, Aiba H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J 2005; 24:1584-95; PMID:15933721; http://dx.doi.org/ 10.1038/sj.emboj.7600636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci U S A 2011; 108:E1392-1398; PMID:22143755; http://dx.doi.org/ 10.1073/pnas.1113956108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell 2012; 150:111-21; PMID:22770215; http://dx.doi.org/ 10.1016/j.cell.2012.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, Parker R, Song H. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol 2010; 17:1233-40; PMID:20890290; http://dx.doi.org/ 10.1038/nsmb.1922 [DOI] [PubMed] [Google Scholar]
- 189. Atkinson GC, Baldauf SL, Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol 2008; 8:290; PMID:18947425; http://dx.doi.org/ 10.1186/1471-2148-8-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Inagaki Y, Ford Doolittle W. Evolution of the eukaryotic translation termination system: origins of release factors. Mol Biol Evol 2000; 17:882-9; PMID:10833194; http://dx.doi.org/ 10.1093/oxfordjournals.molbev.a026368 [DOI] [PubMed] [Google Scholar]
- 191. Xi R, Doan C, Liu D, Xie T. Pelota controls self-renewal of germline stem cells by repressing a Bam-independent differentiation pathway. Dev Camb Engl 2005; 132:5365-74. [DOI] [PubMed] [Google Scholar]
- 192. Pisareva VP, Skabkin MA, Hellen CUT, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 2011; 30:1804-17; PMID:21448132; http://dx.doi.org/ 10.1038/emboj.2011.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Wang W, Cajigas IJ, Peltz SW, Wilkinson MF, González CI. Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol Cell Biol 2006; 26:3390-400; PMID:16611983; http://dx.doi.org/ 10.1128/MCB.26.9.3390-3400.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Lasalde C, Rivera AV, León AJ, González-Feliciano JA, Estrella LA, Rodríguez-Cruz EN, Correa ME, Cajigas IJ, Bracho DP, Vega IE, et al. Identification and functional analysis of novel phosphorylation sites in the RNA surveillance protein Upf1. Nucleic Acids Res 2014; 42:1916-29; PMID:24198248; http://dx.doi.org/ 10.1093/nar/gkt1049 [DOI] [PMC free article] [PubMed] [Google Scholar]