Abstract
Aptamers are an emerging class of highly specific targeting ligands. They can be selected in vitro for a large variety of targets, ranging from small molecules to whole cells. Most aptamers selected are nucleic acid-based, allowing chemical synthesis and easy modification. Although their properties make them interesting drug candidates for a broad spectrum of applications and an interesting alternative to antibodies or fusion proteins, they are not yet broadly used. One major drawback of aptamers is their susceptibility to abundant serum nucleases, resulting in their fast degradation in biological fluids. Using modified nucleic acids has become a common strategy to overcome these disadvantages, greatly increasing their half-life under cell culture conditions or even in vivo. Whereas pre-selective modifications of the initial library for aptamer selection are relatively easy to obtain, post-selective modifications of already selected aptamers are still generally very labor-intensive and often compromise the aptamers ability to bind its target molecule.
Here we report the selection, characterization and post-selective modification of a 34 nucleotide (nt) RNA aptamer for a non-dominant, novel target site (domain 3) of the interleukin-6 receptor (IL-6R). We performed structural analyses and investigated the affinity of the aptamer to the membrane-bound and soluble forms (sIL-6R) of the IL-6R. Further, we performed structural analyses of the aptamer in solution using small-angle X-ray scattering and determined its overall shape and oligomeric state. Post-selective exchange of all pyrimidines against their 2′-fluoro analogs increased the aptamers stability significantly without compromising its affinity for the target protein. The resulting modified aptamer could be shortened to its minimal binding motif without loss of affinity.
Keywords: Aptamers, cell biology, cytokine signaling, Interleukin-6 receptor, internalization, molecular modelling, post-selective modification, protein-RNA interactions, SAXS
Abbreviations
- BSA
bovine serum albumin
- CD
circular dichroism
- DMEM
Dulbecco's modified Eagle's medium
- DTT
dithiothreitol
- FBS
fetal bovine serum
- IL-6
interleukin-6
- IL-6R
interleukin-6 receptor
- sIL-6R
soluble interleukin-6 receptor
- gp130
glycoprotein 130
- 2′-F-Py
2′-deoxy-2′-fluoro pyrimidines
- F4
aptamer family 4 consisting of 2 different aptamer variants
- FRA
filter retention assay
- PAGE
polyacrylamide gel electrophoresis
- PBS
phosphate-buffered saline
- RT
room temperature
- SELEX
systematic evolution of ligands by exponential enrichment
Introduction
The pleiotropic cytokine interleukin-6 (IL-6) has multiple beneficial functions in inflammation, hematopoiesis and regeneration. However, deregulation of this cytokine promotes the pathology and progression of a variety of diseases, including rheumatoid arthritis, multiple sclerosis, Crohn's disease and multiple myeloma.1-3
On a cell presenting interleukin-6 receptor (IL-6R), IL-6 signaling occurs by binding of IL-6 to the non-signaling specific IL-6R and subsequent association with a dimer of the cytokine signal transducer glycoprotein 130 (gp130). IL-6R comprises 3 extracellular domains, of which the 2 fibronectin type III-like domains 2 and 3 form the cytokine binding module that directly interacts with IL-6. Both IL-6R and gp130 also exist as soluble forms due to cleavage of the extracellular domains by proteases or alternative splicing.4
In case of the IL-6R, limited proteolysis by the membrane bound A Disintegrin and Metalloproteinase (ADAM) 10 or ADAM17 leads to the release of soluble IL-6R (sIL-6R). IL-6 and sIL-6R form the IL-6/sIL-6R complex, which can activate gp130 on any target cell presenting the ubiquitous gp130, irrespective of the presence of membrane bound IL-6R. This process is called trans-signaling and is inhibited by soluble gp130 (sgp130).5
Whereas gp130 is presented on the plasma membrane of almost all cells, IL-6R production and therefore classic signaling, i.e., signaling through membrane-bound IL-6R, is mainly restricted to hepatocytes, neutrophils, monocytes, and some lymphocytes.6
Classic IL-6 signaling is believed to contribute to tissue regeneration and anti-inflammatory processes, whereas trans-signaling is considered to be mainly pro-inflammatory and plays a significant role in autoimmune and chronic inflammatory diseases as well as inflammation-driven cancers.7
Since the classic IL-6-signaling pathway is involved in many physiological functions, selective inhibition of trans-signaling appears to be the more promising strategy for therapeutic intervention in many indications.
High-potency inhibitors of trans-signaling, apart from derivatives of the natural trans-signaling inhibitor sgp130, e.g., the IgG1-Fc fusion protein sgp130Fc, have not yet been described. The only trans-signaling-specific antibody published so far is vastly inferior to sgp130Fc.8,9 sgp130Fc variants were efficacious in many different animal models of chronic inflammatory diseases10-12 and inflammation-associated cancers.13,14
Despite the very low chances of success due to the special structural situation with 2 practically identical ligands (IL-6/sIL-6R vs. IL-6/IL-6R), the high production costs of large biologics such as sgp130Fc variants drive the search for small-molecule trans-signaling inhibitors.
An alternative approach for the generation of inhibitors and ligands for a broad variety of target molecule classes was introduced in the early 1990s in the form of small nucleic acids termed aptamers. They are generated by an in vitro selection process referred to as systematic evolution of ligands by exponential enrichment (SELEX). SELEX was initially introduced as an approach to scan a pool of up to 1015 individual molecules with randomized nucleotide sequences for those molecules with a specific function, usually binding to a specific target molecule.15,16
Aptamers rapidly made the transition from basic into applied research. Like antibodies, aptamers bind their target molecule with high affinity and specificity. However, unlike their protein counterparts, they are independent of cellular systems for selection and production, show little to non-immunogenicity,17,18 and have lower production costs and batch-to-batch variation. Today, aptamers are available for a huge variety of targets, such as small molecules,19 proteins,20 viruses,21 and even whole cells.22
One major disadvantage of nucleic acids, in particular RNAs, is their susceptibility to nucleases in biological fluids. Pre-selective modification of the starting library and post-selective alterations of the nucleotides as well as the 5′- and 3′-termini were shown to greatly improve the half-life of aptamers in vivo and under cell culture conditions. Macugen™, the first aptamer to be approved by the FDA for the treatment of age-related macular degeneration (AMD), was selected using 2′-deoxy-2′-fluoro-pyrimidines (2′-F-Py) in the enzymatic production of the library during SELEX for increased stability against serum nucleases. Post-selectively, all but 2 purines were replaced by their 2′-deoxy-2′-O-methyl analogs to further increase the ability of the modified aptamer to resist endonuclease degradation. Furthermore, a 3′-3′-linked deoxythymidine was incorporated to shield the aptamer against exonucleases, and a 5′-linked 40-kDa polyethylene glycol moiety was attached for prolonged tissue residence.23-25
Post-selective modifications of an aptamer are generally very time-consuming, can alter its affinity, and often require screening of the effects of single base exchanges in the molecule. We have previously published the selection of a 2′-F-Py aptamer for the IL-6R, termed FAIR-6, with a sequence convergent to an unmodified IL-6R-specific aptamer, AIR-3.26
Further experiments showed that AIR-3, if comprising 2′-F-Py instead of natural ribonucleotides, was also able to bind to its target with comparable affinity as the unmodified form. Shortening of FAIR-6 or 2′-F-Py modified AIR-3, however, led to a complete loss of target binding capability of the aptamers. This phenomenon is rather common with aptamers: most aptamers selected with canonic nucleotides show no or only reduced affinity for their target after post-selective modification of the 2′-moiety, and if their affinity should be retained, shortening of the aptamer will finally compromise or destroy it.27,28
In the present study, we describe the selection, characterization and post-selective modification of an RNA aptamer specific for the IL-6R domain 3 (IL-6R D3) with no apparent sequence similarities to already selected AIR-3 or FAIR-6 and no reduction in affinity, even after minimization. This is, to the best of our knowledge, the first time that the replacement of all 2′-OH pyrimidines by their 2′-F counterparts in addition to shortening of the initially selected aptamer did not impair the affinity of the aptamer for its target molecule.
Results
In vitro selection of RNA aptamers specific for IL-6R domain 3
Previous RNA-SELEX experiments revealed aptamers with a preference for a single binding site on the surface of sIL-6R.26,29 Preliminary results indicate domain 1 of the IL-6R to be the dominant target for nucleic acid ligands in the selection process. This finding is in general agreement with experiences from other SELEX experiments and shows a bias for one target site in the selection of aptamers.30-32
To circumvent these problems and probe new potential sites for aptamer binding on the surface of the sIL-6R, we set up a 2-tier RNA-SELEX. In the first 3 cycles of selection, immobilized sIL-6R was used to ensure enrichment of unmodified RNA aptamers that bind to the native soluble cytokine receptor. We then employed domain 3 of the IL-6R as target for 8 additional selection cycles, aiming at aptamers specific for this site of the receptor. Binding of the enriched nucleic acid library to sIL-6R and IL-6R domain 3 was tested via filter retention assay (FRA) after selection rounds 5, 10 and 11 (data not shown). The enriched pool was cloned after 11 consecutive SELEX rounds. 35 clones were sequenced and their variable core regions aligned using the Jotun-Hein algorithm.33
While seven clones showed unique sequences, the remaining 28 sequences were found at least twice in the alignment. These sequences were grouped into 5 individual families (named F1–F5; Table S1). Representative clones of these 5 families were tested via FRA. Clones representative of 3 families showed binding to sIL-6R with Kd values of 92.7 ± 18.3 nM for F2, 179 ± 117 nM for F3 (Fig. S1) and 34.8 ± 5.8 nM for F4 (Fig. 1A), respectively. F4 was chosen for further characterization due to its superior affinity for the sIL-6R.
Figure 1.
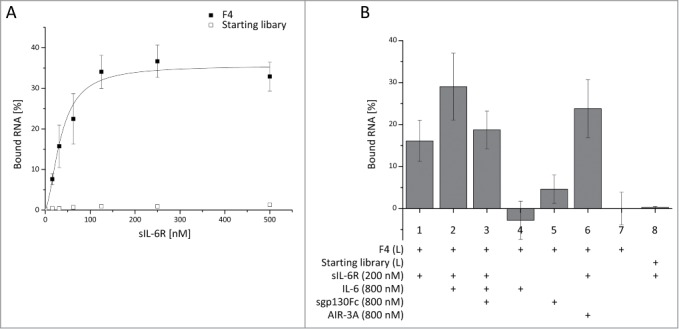
Filter retention assays (FRA) for characterizing the binding properties of the aptamer family 4. (A) A dilution series of sIL-6R was incubated with constant amounts (<1 nM) of a representative clone of 32P-labeled F4 or starting library RNA. F4 binding was analyzed in triplicate; library binding in a single experiment. (B) Binding of 32P-labeled (L) F4 (1–7) to free sIL-6R (1), sIL-6R in context of the IL-6 signaling complex (2, 3), IL-6 (4), sgp130Fc (5) and in concert with the previously published aptamer AIR-3A (6); as a negative control, sIL-6R was incubated with labeled (L) RNA of the starting library (8). All assays were performed in triplicate.
Characterization of the affinity and specificity of F4
To determine the specificity of F4 for sIL-6R alone compared to the IL-6/sIL-6R or IL-6/sIL-6R/sgp130Fc complex, we performed FRA analyses in the presence of the corresponding proteins (Fig. 1B). A concentration of 200 nM sIL-6R was used to ensure saturation of F4 binding (Fig. 1A). 200 nM sIL-6R were pre-incubated with 800 nM sgp130Fc and/or IL-6 prior to aptamer addition to ensure the formation of the IL-6/sIL-6R and IL-6/sIL-6R/sgp130Fc complexes, respectively.
Binding of F4 to sIL-6R was neither influenced by the presence of IL-6 nor by sgp130Fc, indicating a binding epitope of F4 spatially separated from the protein-protein interaction sites in the sIL-6R (Fig. 1B, column 1–3). The stronger signal intensity for the IL-6/sIL-6R complex (Fig. 1B, column 2) may result from increased binding of the protein complex to the nitrocellulose membrane. A saturation of sIL-6R with the previously published aptamer AIR-3A29 had no influence on F4 binding (Fig. 1B, column 6), confirming that F4 and AIR-3A bind to different sites on the sIL-6R. While there was no interaction of F4 with IL-6, slight aptamer binding to sgp130Fc could be observed (Fig. 1B, columns 4 and 5). This off-target effect may result from structural similarities between the fibronectin type III-like domains 4–6 of gp130 and the fibronectin type III-like domain 3 of the sIL-6R.
Identification of the minimal binding motif of F4
In order to identify possible pathways for shortening F4, secondary structure predictions using mFOLD were performed.34 The secondary structure prediction of F4 with the lowest Gibbs free energy was characterized by a stem loop structure with a 14 nt loop between nucleotides 53 and 67 (Fig. 2A). Taking into account the high G content of the F4 core sequence, the G-quadruplex prediction tool QGRS-mapper was used in addition to the putative secondary structure to identify the possible binding motif of the aptamer.35
Figure 2.
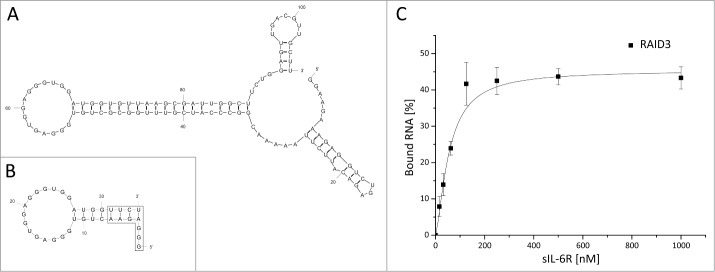
Minimization of the IL-6R-specific aptamer F4 and characterization of the shortened aptamer RAID3. (A) Lowest free energy secondary structure prediction by mFOLD of F4. Nucleotides 1–25 and 85–106 correspond to the constant region of the SELEX library. (B) Lowest free energy secondary structure prediction by mFOLD of the shortened aptamer RAID3. Nucleotides 1–7 were altered for optimization of RNA yield in T7 in vitro transcription; nucleotides 32 and 33 were adapted accordingly for maintenance of the putative terminal stem (boxed nucleotides) of RAID3. (C) FRA evaluation of dilution series of sIL-6R after incubation with a constant amount of <1 nM 32P-labeled RAID3. FRA was performed in triplicate, and mean values were plotted.
Aptamer minimization was based on 2 potentially G-quadruplex forming sections of F4 predicted by the software. The first putative quadruplex comprised the nucleotides 59–71 with either G62 or G64 contributing to one plane and a G-score of 20. The second, alternative quadruplex might comprise the nucleotides 54–67, thereby corresponding to the predicted loop, with the same ambiguity between G62 and G64 and a G-score of 19. The stem leading to the predicted loop was successively shortened. The first 7 and 2 of the last 4 nucleotides of the RNA were altered (boxed nucleotides in Fig. 2B) to achieve better yield in T7 transcription36 and base complementarities in the terminal stem, respectively. The shortened variant of F4 was termed RAID3 (RNA aptamer for interleukin-6 receptor domain 3).
RAID3 showed a Kd of 54.7 ± 5 nM to the sIL-6R in FRA analysis; a dissociation constant similar to that of the full-length aptamer F4 (Kd: 34.8 ± 5.8 nM) (Fig. 2C). However, no binding was observed after removal of the altered 7 5′- and 4 3′-nucleotides of RAID3 (boxed nucleotides in Fig. 2B) or further alteration of the stem in the proximity of the predicted loop to substitute the wobble base pairs U9–G29 and G10–U28 with canonic A–U and G–C base pairs (RAID3 U9C/G10A, Table S2). This indicated the necessity of these nucleotides for correct aptamer folding and/or the availability of G29 and G30 for the formation of the putative G-quadruplex (Fig. 2B).
UV circular dichroism (CD) spectroscopy of RAID3
We performed UV CD spectroscopy to further investigate the putative G-quadruplex predicted by QGRS-mapper in the RAID3 molecule. As previously observed for other G-quadruplex-forming RNAs, RAID3 showed a maximum at 265 nm and a minimum at 240 nm in CD spectra, indicating the formation of a parallel G-quadruplex (Fig. S2A).37,38
As G-quadruplex formation requires the presence of particular monovalent cations, we tested the influence of potassium on the amplitude of the circular dichroism of RAID3. The addition of KCl increased signal intensity at the corresponding wavelengths, supporting the notion of RAID3 adopting a quadruplex folding (Fig. S2A). The negative control AIR 1A (Table S2), a derivative of AIR-3A (Table S2), does not bind to sIL-6R (unpublished data) and fails to form a G-quadruplex under tested conditions. Accordingly, the CD spectrum of AIR 1A shows no alteration after KCl addition (Fig. S2B) and resembles that of RAID3 without potassium present in the buffer. RAID3 U9C/G10A shows a strikingly different behavior (Fig. S2C). In this non-binding variant, 2 G–U wobble base pairs of the putative stem region of RAID3 (Fig. 2B) were replaced by canonic G–C and A–U base pairs. Whereas no strong signals for A-form duplex RNA could be observed in the spectra of RAID3 or AIR-1A, RAID3 U9C/G10A showed a pronounced minimum at 210 nm in concert with a relatively small minimum at 235 nm and a pronounced maximum at 265 nm, indicating the presence of duplex RNA.39
Overall, the amplitudes in the CD spectrum of RAID3 U9C/G10A are much stronger compared to the other RNAs tested. It appears likely that the base exchanges in RAID3 U9C/G10A resulted in the formation of the stem previously predicted for RAID3, and that this structural element was missing in the active aptamer, at least to its full extent. Addition of potassium had no influence on the CD spectrum of RAID3 U9C/G10A, indicating that strengthening of the stem region prevented the formation of the G-quadruplex. This might explain the complete loss of binding of RAID3 U9C/G10A to the sIL-6R.
Structural modeling of RAID3
Based on the predicted secondary structure and putative G-quadruplex comprising nucleotides 18–30 of RAID3, a 3-dimensional structural model of RAID3 was generated using the JUMNA program.40,41 The modeling procedure included structural restraints to stabilize the predicted G-quadruplex structure (QGRS-mapper prediction) and a shortened A-form stem region, comprising only the putative terminal 4 base pairs. For the linker segments, a disordered structure was assumed. The global geometry of the structure is sterically defined by the close vicinity of the G-quadruplex and the stem segment. It served as a working model for fitting to small angle X-ray scattering data.
Small angle X-ray scattering
The SAXS data collected on RAID3 (Fig. 3B) suggested a radius of gyration (Rg) of 2.7 nm and a maximum particle distance (Dmax) of 10 nm. The scattering curve and the characteristic function p(r) (Fig. 3C) suggest an elongated particle shape. The Porod volume computed from the data (26 nm3) indicates a molecular weight (MW) of about 25 ± 3 kDa and points to a dimeric oligomerization state given that the monomer's MW is 11.3 kDa. Since the structural parameters suggested a dimer formation, P2 symmetry was imposed onto the ab initio shape reconstruction of the molecule using DAMMIF.42 It was based on the RAID3 model structure generated by structural modeling (see previous paragraph) employing rigid body refinement. The model obtained has a discrepancy from the experimental data of 0.906 (χ2) (Fig. 3B).
Figure 3.
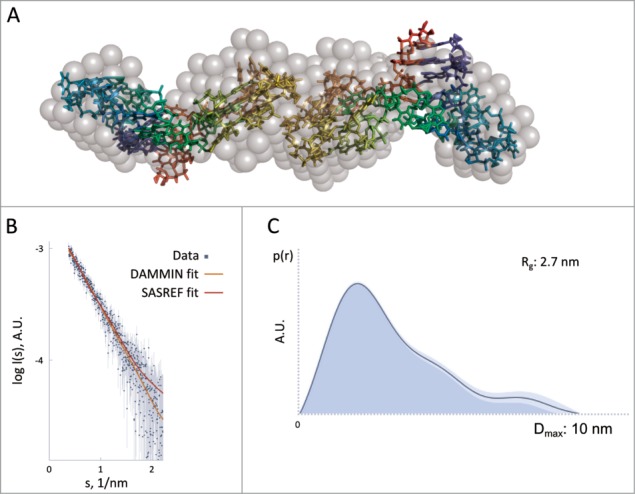
Results of the SAXS experiment. Superimposition of the ab inito and hybrid models obtained using DAMMIN and SASREF, respectively. (A) DAMMIN model is represented as gray semi-transparent spheres, SASREF model is represented as rainbow-colored sticks. The corresponding goodness-of-fits are illustrated in (B) where the SAXS experimental data are represented as blue dots, the fitting of the DAMMIN model (χ2 = 0.906) as an orange curve and the fitting of the SASREF model (χ2 = 1.04) as a red curve. The p(r) distribution is illustrated in figure (C) together with the corresponding structural parameters Rg and Dmax derived from it.
We hypothesized that the interacting nucleotides for the dimer formation are those involved in the quadruplex structure due to the large hydrophobic surface provided by this structural element, and this restrain was imposed as a distance of maximum 8 Å between the 2 quadruplexes (from nt 18 to nt 30). The rigid body modeling reconstruction using SASREF43 was repeated 10 times starting from different initial approximations and yielding comparable results. The obtained dimeric structure fits the experimental data with a χ2 of 1.04 (Fig. 3B) and is well superimposed with the ab initio shape (Fig. 3A).
Microscopic investigation of RAID3 binding to BAF3 cells presenting human IL-6R
To examine if RAID3 was able to bind to membrane-associated IL-6R, 5′-Atto 647N-labeled RAID3 was incubated (30 min at 37°C) with murine BAF3 cells presenting human IL-6R, human gp130, and human tumor necrosis factor α (TNF-α) (BAF3/gp130/IL-6R/TNF) or merely human gp130 (BAF3/gp130). Binding of the aptamer to the cells was analyzed by confocal microscopy (Fig. 4A). A clear fluorescence signal could be detected for BAF3/gp130/IL-6R/TNF cells after incubation with RAID3 Atto 647N. In contrast, BAF3/gp130 cells showed only a weak fluorescence signal after incubation with RAID3 Atto 647N (Fig. 4G). Minor binding of RAID3 Atto 647N to BAF3/gp130 cells suggests some degree of off-target binding of the aptamer to gp130, as observed with sgp130Fc (Fig. 1B).
Figure 4.
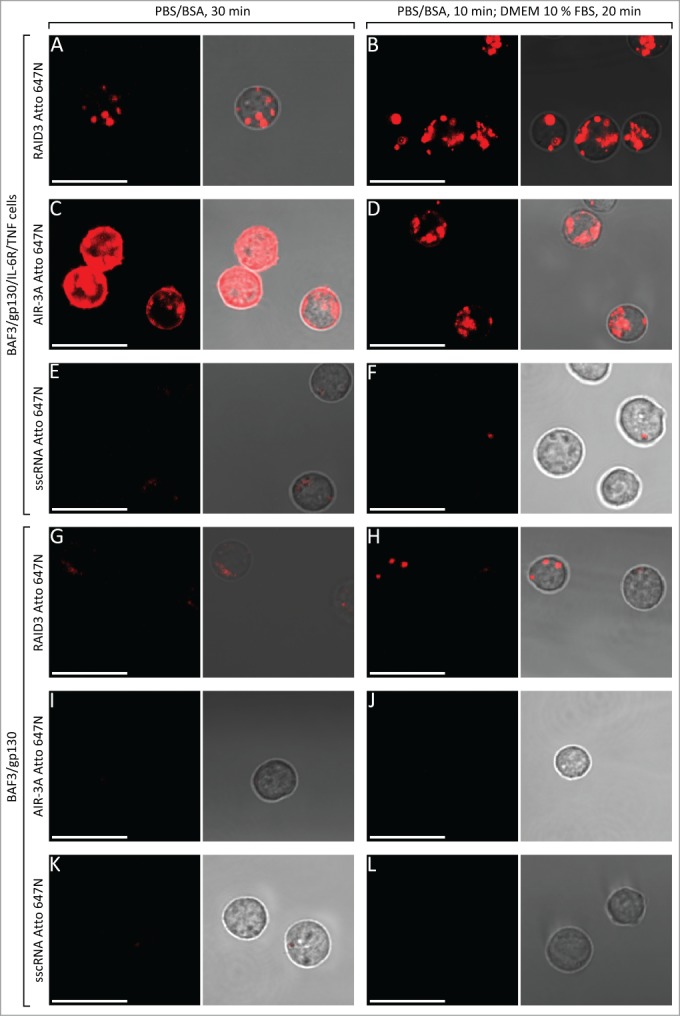
Confocal laser scanning microscopy of BAF3 cells after incubation with 5′-Atto 647N-labeled RNA. BAF3/gp130/IL-6R/TNF (A–F) or BAF3/gp130 cells (G–L) were incubated with 5′-Atto 647N-labeled RAID3 (A, B and G, H), AIR-3A (C, D and I, J) or sscRNA as a control (E, F and K, L). Incubation of the cells with RNA at 37°C was either performed in 1 × PBS/BSA for 30 min (left column) or in 1 × PBS/BSA for 10 min, followed by DMEM supplemented with 10% FBS for 20 min (right column). All fluorescence measurements were performed under constant imaging conditions to ensure comparability of signal intensity between samples. Bars represent 20 µm, each.
BAF3/gp130/IL-6R/TNF cells incubated with AIR-3A Atto 647N as a positive control showed a pronounced fluorescence, confirming the presence of IL-6R on the cells (Fig. 4C). The localization of AIR-3A Atto 647N, however, appeared to be different, when compared to BAF3/gp130/IL-6R/TNF cells incubated with RAID3 Atto 647N (Fig. 4A).
AIR-3A fluorescence seemed to be present both on the cell surface and in the cytoplasm, whereas RAID3 was only observed in apparently compartmentalized regions inside the cells.
To test the hypothesis of an exclusively intracellular localization of RAID3, we treated the cells with Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (DMEM 10% FBS) after 10 min of initial incubation in phosphate-buffered saline (PBS) supplemented with 1 mg/mL bovine serum albumin (BSA) (PBS/BSA) to degrade surface-bound RNA by RNases present in the serum. Although serum factors may influence cellular uptake, unmodified RNAs only have a very short half-life under these conditions and are usually degraded within seconds.26
After 20 min of incubation in DMEM 10% FBS at 37°C, cells were washed and microscopically examined. We detected no qualitative or quantitative difference in fluorescence distribution and overall signal intensity of Atto 647N-labeled RAID3 in BAF3/gp130/IL-6R/TNF cells after serum treatment when compared to PBS/BSA incubation (Fig. 4A, B).
In contrast, addition of DMEM 10% FBS eliminated fluorescence on the surface of BAF3/gp130/IL-6R/TNF cells after incubation with AIR-3A Atto 647N and led to fluorescence patterns resembling that of cells incubated with RAID3 Atto 647N (Fig. 4D). The overall signal intensity of BAF3/gp130/IL-6R/TNF cells incubated with AIR-3A Atto 647N prior to incubation in DMEM 10% FBS appeared to be reduced (Fig. 4C, D), suggesting that surface bound AIR-3A was indeed mainly degraded instead of being taken up to a considerable extent.
No difference could be observed for BAF3/gp130/IL-6R/TNF and BAF3/gp130 cells incubated with single stranded control RNA (sscRNA) Atto 647N solely in PBS/BSA or PBS/BSA and DMEM 10% FBS. This supports the assumption that DMEM 10% FBS treatment did not alter cellular uptake of RNA to a significant degree due to fast degradation of extracellular RNA (Fig. 4E–F and K–L).
Taken together, these results suggest an almost exclusive intracellular localization of RAID3 for the BAF3/gp130/IL-6R/TNF cells due to IL-6R-mediated uptake.
Quantification of RAID3 binding to BAF3 cells presenting human IL-6R
To quantify RAID3 binding, cells were incubated at room temperature (RT) or 37°C with the aptamer and subsequently phenotypically gated (Fig. S3). The median fluorescence of 3 independent measurements of BAF3/gp130/IL-6R/TNF and BAF3/gp130 cells without added RNA and after incubation with Atto 647N-labeled RNA was plotted in a bar chart (Fig. S4). The median auto-fluorescence of the cells was set as baseline for BAF3/gp130/IL-6R/TNF and BAF3/gp130 independently. Only weak fluorescence could be detected when BAF3/gp130/IL-6R/TNF cells were incubated with RAID3 Atto 647N at RT (Fig. S4A). In contrast, incubation with RAID3 at 37°C noticeably increased fluorescence (Fig. S4B). Considering the results of confocal microscopy obtained with the cells after incubation with RAID3 Atto 647N, temperature dependency of fluorescence signal intensity measured by FACS could be explained by an increased rate of aptamer internalization at 37°C. The strong fluorescence signal of cells after incubation with AIR-3A Atto 647N at both RT and 37°C (Fig. S4A, B) most likely resulted from surface-bound RNA and correlates with the localization and signal intensity observed in confocal microscopy (Fig. 3A, C). Due to their susceptibility to serum nucleases, cell experiments with RNA aptamers have to be performed in nuclease free buffer systems. This limits the feasible experimental conditions, especially when working with cells. We therefore wanted to investigate the possibility to increase serum stability of RAID3.
Post-selective modification of RAID3 with 2′-F-Py
To test the susceptibility of RAID3 for post-selective modification, we subjected a 2′-F-Py-modified version of RAID3 (RAID3 2′-F-Py) to FRA analysis (Fig. 5A). RAID3 2′-F-Py showed clear binding to the sIL-6R. No binding of RAID3 U9C/G10A 2′-F-Py to the sIL-6R could be observed in FRA, showing that binding of RAID3 2′-F-Py was most likely not due to unspecific interaction of the modified RNA.
Figure 5.
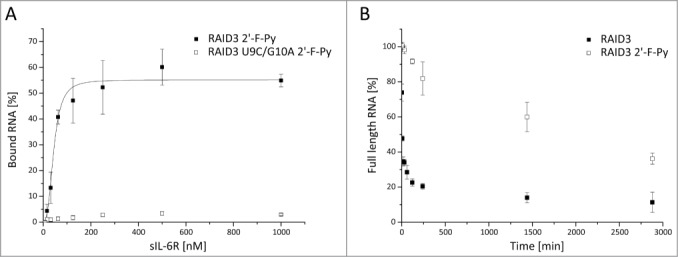
Validation of RAID3 2′-F-Py affinity for the sIL-6R in PBS and its stability under cell culture conditions. (A) Filter retention assay (FRA) evaluation of dilution series of sIL-6R after incubation with a constant concentration of <1 nM 32P-labeled RAID3 2′-F-Py or RAID3 U9C/G10A 2′-F-Py. FRA was performed in triplicate. The 500 and 250 nM sIL-6R spots of one measurement of RAID3 were excluded from utilization due to outlier signals. Mean values of bound RNA were plotted. (B) Stability of RAID3 and RAID3 2′-F-Py in DMEM 10% FBS at 37°C. Aliquots of 32P-labeled RNA were swiftly transferred into RNA loading dye and immediately frozen in liquid N2 in corresponding intervals. Degraded RNA was then electrophoretically separated from full-length intact aptamer by denaturing polyacrylamide gel electrophoresis, and the ratio of full-length to degraded RNA was evaluated with the software Image Lab (BioRad).
The Kd values and the bound fraction of RAID3 2′-F-Py (43.6 ± 2.2 nM) (Fig. 5A) were in the same range as those of the unmodified forms of RAID3 (54.7 ± 5 nM) and F4 (34.8 ± 5.8 nM), respectively (Figs. 1A and 2C). Modifying the 2′-moiety of pyrimidines in RNA greatly improves their stability against abundant serum nucleases like RNase A, which cleaves RNA at the 3′-site of an unpaired U or C. To test the stability of RAID3 2′-F-Py, we incubated 32P-labeled aptamer in DMEM 10% FBS at 37°C over a period of 2 days (Fig. 5B). Even unmodified RAID3 had a relatively long half-life of 1–5 min. 2′-F modification of RAID3, however, led to a pronounced increase in resistance against serum nucleases and a half-life of over 24 h. This is a significant improvement when compared to AIR-3 and its convergent 2′-F-modified analog FAIR-6 with half-lives of less than 2 seconds or 2 hours, respectively.26
Discussion
In this study we present the 2-tier SELEX and significant structural optimization of an RNA aptamer (RAID3) targeting a novel site, domain 3, of the sIL-6R. Although domain 3 is directly involved in the interaction of IL-6R with IL-6 and gp130, providing more than 70% of the interaction surface of the receptor with the cytokine,44 RAID3 binding does not interfere with the formation of the IL-6-dependent signaling complex when tested with the corresponding soluble protein components (IL-6, sIL-6R and sgp130Fc; Fig. 1B). These results indicate that areas favorable for RNA interaction on the surface of the IL-6R are not in close proximity to the interaction sites of the receptor responsible for IL-6 and gp130 binding. Combined with previous results obtained with aptamers like AIR-3 26,29 putatively targeting domain 1 of the IL-6R, our findings suggest that the structure of IL-6R does not favor the selection of aptamers directly interfering with signaling complex formation.
Besides its binding to sIL-6R in cell-free systems, we demonstrate that RAID3 binds to BAF cells presenting IL-6R. Using confocal microscopy, no labeled RAID3 aptamer was detectable on the cell surface, but only in compartmentalized regions inside the cells (Fig. 4A, B). The apparent amount of RAID3 within the cells appeared to be similar to that of another IL-6R binding aptamer, AIR-3A (Fig. 4C, D), and suggests an equal efficiency for internalization. In contrast to RAID3, however, labeled AIR-3A was detected both on the cell surface and in compartments in the cell lumen. The lack of fluorescence at the cell surface may be the result of fast dissociation of RAID3 from the IL-6R with only incorporated aptamer being trapped within the cell after washing. Alternatively, IL-6R domain 3 could be inaccessible to aptamer binding during its residence on the cell surface, with the binding site for the aptamer only becoming available during the internalization process of the receptor.
Reduced binding to cell surface-bound IL-6R might be favorable if RAID3 were to be further modified toward blocking of IL-6 trans-signaling. This application of the aptamer, however, would require a greatly improved resistance of the RNA against abundant serum nucleases. A common method for RNA stabilization is the utilization of 2′-F pyrimidines. Usually, post-selective modification of the pyrimidines disturbs correct aptamer folding and consequently decreases its affinity.27,28 Strikingly, 2′-F-Py modified RAID3 was still able to bind the sIL-6R with an affinity equal to that of the unmodified aptamer. Consequently, this strongly indicates that RAID3 2′-F-Py was still able to undergo correct folding with all pyrimidines replaced by their 2′-F analogs. AIR-3 and FAIR-6, 2 independently selected aptamers sharing a common binding motif, were also able to bind to the sIL-6R either when modified with 2‘-F-Py or comprising only natural nucleotides. Shortening, however, was only possible for the unmodified aptamers, as the 2′-F-Py forms of the shortened aptamers completely lost their ability to bind to the IL-6R. This indicates that the core regions of the modified aptamers were not able to undergo correct folding in the 2′-F-Py modified form of the minimized aptamers and were dependent on the influence of structural elements outside of the core aptamers for target protein binding.
Whereas no changes in affinity could be observed, half-life of RAID3 2′-F-Py in DMEM 10% FBS was significantly improved when compared to the unmodified RAID3.
The putative G-quadruplex folding of RAID3 may explain the resistance of RAID3 against sterical or structural changes induced by 2‘-F-Py. This would imply that the 2′-OH groups of the pyrimidines have no significant impact on the global structure of the aptamer or interaction with the target molecule. Further experiments are necessary to investigate the influence of the surrounding nucleotides on the 3-dimensional structure of the aptamer. This finding could be of great relevance for the post-selective modification and stabilization of other G-quadruplex aptamers selected with canonical RNA nucleotides.
Small angle X-ray scattering experiments suggested a possible dimeric conformation in solution. However, more experimental data are necessary to investigate the relevance of dimer formation for aptamer folding and IL-6R binding.
In the future, we want to focus on further structural characterization of RAID3 and the modified RAID3 2′-F-Py. Using structural modeling, a working 3D model of RAID3 was built which can help to identify nucleotides accessible for interactions and can also help to guide future mutagenesis or chemical probing studies. The results of such studies can, in turn, generate additional restraints to refine the structural model and may eventually allow us to predict how the aptamer interacts with its target protein. In summary, the unparalleled resilience of RAID3 to both minimal binding motif truncation and to post-selective 2′-F-Py modification demonstrates that such modification strategies for G-quadruplex aptamers are not as doomed as the previous literature suggests.
Materials and Methods
Chemicals
If not stated otherwise, chemicals were purchased from Sigma-Aldrich. All buffers were prepared using purified 2 × deionized water (ddH2O) from a Millipore system. For the SELEX, 1 × PBS (137 mM NaCl; 2.7 mM KCl; 6.5 mM Na2HPO4; 1.5 mM KH2PO4) supplemented with 3 mM MgCl2 was used (“SELEX buffer”). Enriched RNA library and single candidate binding tests were also performed in SELEX buffer. As F4 needed no bivalent cations for binding, all further binding tests with the shortened form RAID3 were performed in 1 × PBS.
Oligonucleotides
The oligonucleotides used in this study are listed in Table S2. Unmodified RAID3 and 5′-Atto 647N-labeled RAID3 were purchased from Biomers. 5′-Atto 647N-labeled AIR-3A and single stranded control RNA (sscRNA) were purchased from IBA. DNA templates and primers were purchased from Metabion.
Cell culture
BAF3/gp130/IL-6R/TNF and BAF3/gp130 cells were kindly provided by Dr. Athena Chalaris (Institute of Biochemistry, University of Kiel, Germany). Cell culture was carried out using DMEM (PAA/GE Healthcare, E15-810) with 10% FBS (PAA/GE Healthcare, K41-001), 60 mg/mL penicillin (PAA/GE Healthcare, P11-010) and 100 mg/mL streptomycin (PAA/GE Healthcare, P11-010) at 37°C and in water-saturated atmosphere with 5% CO2. BAF3/gp130/IL-6R/TNF cells were further supplemented with 10 ng/mL human IL-6 (PeproTech, 200-06); BAF3/gp130 cells were supplemented with 10 ng/mL Hyper-IL-6 (a fusion protein of IL-6 and sIL-6R required for growth of these cells).45
Proteins
IL-6 for FRA was purchased from Biochrom (W 1525954002); sIL-6R and sgp130Fc were kindly provided by CONARIS Research Institute AG.
Primer extension and in vitro transcription
We used a primer extension reaction to generate double-stranded template for the in vitro transcription. Denaturing polyacrylamide gel electrophoresis (PAGE) was used to clean template ssDNA prior to further applications. After separation from failed oligonucleotides, full length ssDNA was detected by UV-shadowing and excised out from the gel. The ssDNA was recovered by diffusion elution in 300 mM sodium acetate pH 5.2 at 1,400 rpm and 50°C for 4 hours. Purified ssDNA was ethanol-precipitated and stored in ddH2O.
200 pmol PAGE-purified starting library ssDNA were hybridized with 200 pmol library-specific reverse primer by heating the components in 100 µL 1 × Klenow buffer (Thermo Scientific, EP0051) at 80°C for 5 min and then slowly cooling the reaction down to RT. After hybridization, 1 mM dNTPs and 1.25 U Klenow fragment (Thermo Scientific, EP0051) were added and incubated at 37°C for 2 h, followed by 10 min at 75°C. The fill-in reaction was subsequently precipitated by ethanol and directly used in the T7 in vitro transcription. For the production of RNA, T7 RNA polymerase (0.25 U/µL) was incubated with 200 nM template in 1 × transcription buffer [40 mM Tris HCl pH 8.1, 15 mM MgCL2, 5 mM dithiothreitol (DTT), 2 mM spermidine, 0.01% Triton-X 100, 1.5% PEG 6000] and NTPs (2 mM each) at 37°C for 4 h. The RNA derived from this reaction was PAGE-purified. Template generation for 2′-F-Py RNA production was performed as with the library and other unmodified RNAs. For the transcription of the 2′-F-Py modified RNAs, we employed the T7 polymerase variant Y639F 46 (1.25 U/µL) with the same template, buffer and NTP concentrations as for the conventional transcription. CTP and UTP, however, were replaced by 2′-deoxy-2′-fluorocytidine triphosphate (Metkinen, 104-02) and 2′-deoxy-2′-fluorouridine triphosphate (Metkinen, 104-04).
Biotinylation of sIL-6R and IL-6R domain 3 and immobilization on streptavidin-coated magnetic beads
2.5 nmol sIL-6R or IL-6R domain 3 were mixed with a 3-fold molar excess of EZ-Link Sulfo-NHS-LC-Biotin (Thermo Scientific, PI21335) in a final volume of 100 µL 1 × PBS and incubated at RT for 15 min, followed by 15 min at 4°C. Non-reacted biotin was removed by dialysis against 1 L of 1 × PBS at 4°C overnight using micro-dialysis capsules (QuixSep, QS1-0100) and Spectra/Por dialysis membrane (Spectrum, 132724). The biotinylated protein was immobilized on 4 mg Dynabeads® M-280 Streptavidin (Life Technologies, 11205D) and suspended in 1.25 × PBS with 1.25 µg/µL BSA.
SELEX
RNA generated from 200 pmol library template (∼1 × 1014 molecules) was incubated with ˜100 pmol immobilized sIL-6R in SELEX buffer (containing 1 µg/µL BSA) at 37°C for 30 min. Following incubation, bound RNA was magnetically separated and washed with 200 µL SELEX buffer. Bound RNA was heat-eluted from the beads at 80°C for 3 min in ddH2O and swiftly removed from the immobilized beads by magnetic separation. Eluted RNA was then amplified by RT-PCR. RT-PCR was performed in 1 × DreamTaq buffer (Thermo Scientific, EP0701) and 0.2 × First-Strand buffer (Life Technologies, 18080044) using 1 mM each of forward and reverse primer, 0.3 mM dNTPs and 2 mM DTT. The reaction was then incubated at 65°C for 5 min and snap-cooled on ice. After cooling, 15 U SuperScript™ III Reverse Transcriptase (Life Technologies, 18080044) and 5 U DreamTaq DNA polymerase (Thermo Scientific, EP0701) were added. Reverse transcription was carried out at 54°C for 15 min, directly followed by PCR amplification (30 s at 95°C, 30 s at 60°C and 45 s at 72°C) for an appropriate number of PCR cycles. For the subsequent SELEX cycles, 500 µL of SELEX buffer were used for the washing steps, with one washing step added per selection round. After round 3, immobilized sIL-6R was replaced by immobilized IL-6R domain 3 for 8 additional SELEX cycles.
Filter retention assay (FRA)
FRA was used to investigate the binding kinetics of RNAs to the target proteins. 20 pmol PAGE-purified RNA generated by T7 transcription was 5′-dephosphorylated using FastAP (Thermo Scientific, EF0654) according to the manufacturer's protocol. After heat inactivation of the FastAP, RNA was directly 5′-labeled using [γ-32P]-ATP (370 kBq/µL) (Hartmann Analytics, EP-301) as substrate for T4 polynucleotide kinase (Thermo Scientific, EK0031) following the manufacturer's protocol. Labeled RNA was again PAGE-purified to remove unincorporated [γ-32P]-ATP and then used in constant amounts of <1 nM in protein dilution series for affinity measurements in 1 × SELEX buffer (for enriched library RNA and full-length aptamer candidates) or 1 × PBS (for shortened F4 variants and AIR-3A). After incubation, samples were filtered through a pre-equilibrated nitrocellulose membrane (Carl Roth, HP40.1), driven by negative pressure in a vacuum manifold (Minifold® I Dot-Blot-System; Schleicher & Schuell, no longer available). After binding, the membrane was washed 2 times with either SELEX buffer or PBS (depending on the RNA) and used for exposition and quantification of a phosphor imaging screen (Bio-Rad, 170-7841 and Image Lab 5.1). The software Origin (OriginLab Corporation) was used to determine the Kd (dissociation constant) and maximum binding (Bmax) values. The following equation was used, in which the start value was set as the mean signal of RNA samples bound to the membrane without protein:
All binding analyses were performed in triplicate.
Circular dichroism spectroscopy
RNAs were dissolved in 50 mM Tris pH 7.4, with either no or 10 mM KCl present in the buffer, and CD spectra were recorded using a Jasco J-815 CD spectrometer at 25°C. Each spectrum was accumulated 10 times, and the CD values of a suitable buffer control were subtracted from the corresponding RNA signals for the given wavelength intervals before plotting.
Molecular modeling of RNA quadruplex structure
A modified version of the JUMNA program was used in combination with the Amber 4.1 force field for all calculations.40,41,47 The JUMNA approach utilizes a combination of helicoidal and internal coordinates to describe nucleotide placement and flexibility of a DNA or RNA molecule. The helicoidal description allows easy nucleotide addition and substitution in a given structural motif. Stepwise modeling was performed on the sequence 5′-AGAACUGUGGGAGUGGAGGGUGGAUGGUUCU-3′, excluding the first 3 guanines of RAID3 and following a previously designed protocol.48 Based on the highest-scoring predicted quadruplex segment GGAGGGUGGAUGG (employing the QGRS-mapper program) and similarity to a known RNA quadruplex structure (pdb1My9) a model was generated with the 8 underlined guanine nucleotides forming a quadruplex core.49 Additional nucleotides and the short predicted stem were initially placed close to neighboring nucleotides in the structure. The structure was energy-minimized by keeping only the added nucleotides and neighboring nucleotides mobile and locking helical coordinates of the central core formed by guanine nucleotides. Finally, the entire model including a potassium ion placed in the central pore of the quadruplex was energy-minimized resulting in a structure with stable quadruplex and stem duplex but disordered linker segments.
Small angle X-ray scattering
X-ray synchrotron radiation scattering data from a solution of RAID3 in PBS with 137 mM NaCl, 2.7 mM KCl, 6.5 mM Na2HPO4 and 1.5 mM KH2PO4 were collected on the B21 camera on the storage ring DIAMOND (Oxfordshire, UK) using a Pilatus 2M detector. The range of momentum transfer 0.025 < s < 4.010 nm was covered (s = 4π sin θ/λ, where 2θ is the scattering angle). One single but highly diluted solution with a concentration of 0.20 mg/mL was measured because of the tendency of the sample to form unspecific aggregates at higher concentrations. The data were normalized to the intensity of the transmitted beam and radially averaged, the scattering of the solvent-blank was subtracted and the curve was scaled for RNA concentration using the ATSAS software suite.50
The p(r) distribution function was calculated using GNOM,51 the ab initio model was obtained using DAMMIN42 and the rigid body models using SASREF.43 The collected data and models have been deposited in the SASBDB with the accession code SASDA88.52
Confocal Laser Scanning Microscopy (CLSM)
BAF3/gp130/IL-6R/TNF and BAF3/gp130 cells were washed 3 × with 1 × PBS/BSA (1 µg/µL), and 100,000 cells per sample were then incubated for either 30 min in 100 µL PBS/BSA (1 µg/µL) or for 10 min in 100 µL PBS/BSA (1 µg/µL) plus 20 min in 100 µL DMEM supplemented with 10% FBS at 37°C with 500 nM 5′-Atto 647N-labeled RAID3, AIR-3A or sscRNA, respectively. After incubation with the RNAs, the cells were washed with PBS/BSA (1 µg/µL) and imaged with an LSM 510 ConfoCor2 system (Carl Zeiss). The basic adjustments used were as follows: HeNe laser (633 nm), 5–15% laser power, 260 µm pinhole diameter, beam splitters: HFT514/633 nm and NFT 545 nm, LP 650 nm filter.
Flow cytometry
BAF3/gp130/IL-6R/TNF and BAF3/gp130 cells were washed 3 × with 1 × PBS/BSA (1 µg/µL), and 100,000 cells per sample were then incubated for either 30 min in 200 µL PBS/BSA (1 µg/µL) at RT or at 37°C, either without RNA or with 200 nM 5′-Atto 647N-labeled RAID3, AIR-3A or sscRNA, respectively. After washing with PBS/BSA (1 µg/µL), fluorescence intensities were determined by analysis with a FACSAria III (BD Bioscience) using the software FACSDiva Version 6.1.3.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Anna Rath for carefully proofreading this manuscript.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Ataie-Kachoie P, Pourgholami MH, Morris DL. Inhibition of the IL-6 signaling pathway: a strategy to combat chronic inflammatory diseases and cancer. Cytokine Growth Factor Rev 2013; 24:163-73; PMID:23107589; http://dx.doi.org/ 10.1016/j.cytogfr.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 2.Kishimoto T. Factors affecting B-cell growth and differentiation. Ann Rev Immunol 1985; 3:133-57; PMID:3933529; http://dx.doi.org/ 10.1146/annurev.iy.03.040185.001025 [DOI] [PubMed] [Google Scholar]
- 3.Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol 2014; 6:a016295; PMID:25190079; http://dx.doi.org/ 10.1101/cshperspect.a016295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garbers C, Janner N, Chalaris A, Moss ML, Floss DM, Meyer D, Koch-Nolte F, Rose-John S, Scheller J. Species specificity of ADAM10 and ADAM17 proteins in interleukin-6 (IL-6) trans-signaling and novel role of ADAM10 in inducible IL-6 receptor shedding. J Biol Chem 2011; 286:14804-11; PMID:21454673; http://dx.doi.org/ 10.1074/jbc.M111.229393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rose-John S, Heinrich PC. Soluble receptors for cytokines and growth factors: generation and biological function. Biochem J 1994; 300 (Pt 2):281-90; PMID:8002928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheller J, Garbers C, Rose-John S. Interleukin-6: from basic biology to selective blockade of pro-inflammatory activities. Semin Immunol 2014; 26:2-12; PMID:24325804; http://dx.doi.org/ 10.1016/j.smim.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 7.Baran P, Nitz R, Grotzinger J, Scheller J, Garbers C. Minimal interleukin 6 (IL-6) receptor stalk composition for IL-6 receptor shedding and IL-6 classic signaling. J Biol Chem 2013; 288:14756-68; PMID:23564454; http://dx.doi.org/ 10.1074/jbc.M113.466169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbers C, Thaiss W, Jones GW, Waetzig GH, Lorenzen I, Guilhot F, Lissilaa R, Ferlin WG, Grötzinger J, Jones SA, et al.. Inhibition of classic signaling is a novel function of soluble glycoprotein 130 (sgp130), which is controlled by the ratio of interleukin 6 and soluble interleukin 6 receptor. J Biol Chem 2011; 286:42959-70; PMID:21990364; http://dx.doi.org/ 10.1074/jbc.M111.295758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lissilaa R, Buatois V, Magistrelli G, Williams AS, Jones GW, Herren S, Shang L, Malinge P, Guilhot F, Chatel L, et al.. Although IL-6 trans-signaling is sufficient to drive local immune responses, classical IL-6 signaling is obligate for the induction of T cell-mediated autoimmunity. J Immunol 2010; 185:5512-21; http://dx.doi.org/ 10.4049/jimmunol.1002015 [DOI] [PubMed] [Google Scholar]
- 10.Atreya R, Mudter J, Finotto S, Mullberg J, Jostock T, Wirtz S, Schütz M, Bartsch B, Holtmann M, Becker C, et al.. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in crohn disease and experimental colitis in vivo. Nat Med 2000; 6:583-8; PMID:10802717; http://dx.doi.org/ 10.1038/75068 [DOI] [PubMed] [Google Scholar]
- 11.Nowell MA, Richards PJ, Horiuchi S, Yamamoto N, Rose-John S, Topley N, Williams AS, Jones SA. Soluble IL-6 receptor governs IL-6 activity in experimental arthritis: blockade of arthritis severity by soluble glycoprotein 130. J Immunol 2003; 171:3202-9; http://dx.doi.org/ 10.4049/jimmunol.171.6.3202 [DOI] [PubMed] [Google Scholar]
- 12.Tsantikos E, Maxwell MJ, Putoczki T, Ernst M, Rose-John S, Tarlinton DM, Hibbs ML. Interleukin-6 trans-signaling exacerbates inflammation and renal pathology in lupus-prone mice. Arthritis Rheum 2013; 65:2691-702; PMID:23818297 [DOI] [PubMed] [Google Scholar]
- 13.Becker C, Fantini MC, Schramm C, Lehr HA, Wirtz S, Nikolaev A, Burg J, Strand S, Kiesslich R, Huber S, et al.. TGF-beta suppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity 2004; 21:491-501; PMID:15485627; http://dx.doi.org/ 10.1016/j.immuni.2004.07.020 [DOI] [PubMed] [Google Scholar]
- 14.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, et al.. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 2009; 15:103-13; PMID:19185845; http://dx.doi.org/ 10.1016/j.ccr.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature 1990; 346:818-22; PMID:1697402; http://dx.doi.org/ 10.1038/346818a0 [DOI] [PubMed] [Google Scholar]
- 16.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990; 249:505-10; PMID:2200121; http://dx.doi.org/ 10.1126/science.2200121 [DOI] [PubMed] [Google Scholar]
- 17.Foy JW, Rittenhouse K, Modi M, Patel M. Local tolerance and systemic safety of pegaptanib sodium in the dog and rabbit. J Ocul Pharmacol Ther 2007; 23:452-66; PMID:17900226; http://dx.doi.org/ 10.1089/jop.2006.0149 [DOI] [PubMed] [Google Scholar]
- 18.Zhu G, Ye M, Donovan MJ, Song E, Zhao Z, Tan W. Nucleic acid aptamers: an emerging frontier in cancer therapy. Chem Commun 2012; 48:10472-80; PMID:22951893; http://dx.doi.org/ 10.1039/c2cc35042d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holeman LA, Robinson SL, Szostak JW, Wilson C. Isolation and characterization of fluorophore-binding RNA aptamers. Fold Des 1998; 3:423-31; PMID:9889155; http://dx.doi.org/ 10.1016/S1359-0278(98)00059-5 [DOI] [PubMed] [Google Scholar]
- 20.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and Characterization of Nuclease-stabilized RNA Molecules That Bind Human Prostate Cancer Cells via the Prostate-specific Membrane Antigen. Cancer Res 2002; 62:4029-33; PMID:12124337 [PubMed] [Google Scholar]
- 21.Binning JM, Leung DW, Amarasinghe GK. Aptamers in virology: recent advances and challenges. Front Microbiol 2012; 3:29; PMID:22347221; http://dx.doi.org/ 10.3389/fmicb.2012.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal Chem 2007; 79:4900-7; PMID:17530817; http://dx.doi.org/ 10.1021/ac070189y [DOI] [PubMed] [Google Scholar]
- 23.Ng EW, Shima DT, Calias P, Cunningham ET Jr., Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Dis 2006; 5:123-32; PMID:16518379; http://dx.doi.org/ 10.1038/nrd1955 [DOI] [PubMed] [Google Scholar]
- 24.Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjić N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem 1998; 273:20556-67; PMID:9685413; http://dx.doi.org/ 10.1074/jbc.273.32.20556 [DOI] [PubMed] [Google Scholar]
- 25.Wilson C, Keefe AD. Building oligonucleotide therapeutics using non-natural chemistries. Curr Opin Chem Biol 2006; 10:607-14; PMID:17049298; http://dx.doi.org/ 10.1016/j.cbpa.2006.10.001 [DOI] [PubMed] [Google Scholar]
- 26.Meyer C, Berg K, Eydeler-Haeder K, Lorenzen I, Grotzinger J, Rose-John S, Hahn U. Stabilized Interleukin-6 receptor binding RNA aptamers. RNA Biol 2014; 11:57-65; PMID:24440854; http://dx.doi.org/ 10.4161/rna.27447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adler A, Forster N, Homann M, Goringer HU. Post-SELEX chemical optimization of a trypanosome-specific RNA aptamer. Comb Chem High Throughput Screen 2008; 11:16-23; PMID:18220540; http://dx.doi.org/ 10.2174/138620708783398331 [DOI] [PubMed] [Google Scholar]
- 28.Shigdar S, Macdonald J, O'Connor M, Wang T, Xiang D, Al Shamaileh H, Qiao L, Wei M, Zhou SF, Zhu Y, et al.. Aptamers as theranostic agents: modifications, serum stability and functionalisation. Sensors 2013; 13:13624-37; PMID:24152925; http://dx.doi.org/ 10.3390/s131013624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer C, Eydeler K, Magbanua E, Zivkovic T, Piganeau N, Lorenzen I, Grötzinger J, Mayer G, Rose-John S, Hahn U. Interleukin-6 receptor specific RNA aptamers for cargo delivery into target cells. RNA Biol 2012; 9:67-80; PMID:22258147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong Q, Wang J, Ahmad KM, Csordas AT, Zhou J, Nie J, Stewart R, Thomson JA, Rossi JJ, Soh HT. Selection strategy to generate aptamer pairs that bind to distinct sites on protein targets. Anal Chem 2012; 84:5365-71; PMID:22624874; http://dx.doi.org/ 10.1021/ac300873p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, Janjic N. Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents. Mol Ther Nucleic Acids 2014; 3:e201; PMID:25291143; http://dx.doi.org/ 10.1038/mtna.2014.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi H, Fan X, Sevilimedu A, Lis JT. RNA aptamers directed to discrete functional sites on a single protein structural domain. Proc Natl Acad Sci U S A 2007; 104:3742-6; PMID:17360423; http://dx.doi.org/ 10.1073/pnas.0607805104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hein J. Unified approach to alignment and phylogenies. Methods in enzymology 1990; 183:626-45; PMID:2314296; http://dx.doi.org/ 10.1016/0076-6879(90)83041-7 [DOI] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; http://dx.doi.org/ 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikin O, D'Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res 2006; 34:W676-82; PMID:16845096; http://dx.doi.org/ 10.1093/nar/gkl253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanov SA, Welz R, Gottikh MB, Muller S. ; RNA synthesis by T7 RNA polymerase supported primer extension. Molekuliarnaia biologiia 2004; 38:798-803; PMID:15554183 [PubMed] [Google Scholar]
- 37.Balagurumoorthy P, Brahmachari SK, Mohanty D, Bansal M, Sasisekharan V. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res 1992; 20:4061-7; PMID:1508691; http://dx.doi.org/ 10.1093/nar/20.15.4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang AY, Balasubramanian S. The kinetics and folding pathways of intramolecular G-quadruplex nucleic acids. J Am Chem Soc 2012; 134:19297-308; PMID:23113843; http://dx.doi.org/ 10.1021/ja309851t [DOI] [PubMed] [Google Scholar]
- 39.Mitsuoka Y, Kodama T, Ohnishi R, Hari Y, Imanishi T, Obika S. A bridged nucleic acid, 2′,4′-BNA COC: synthesis of fully modified oligonucleotides bearing thymine, 5-methylcytosine, adenine and guanine 2′,4′-BNA COC monomers and RNA-selective nucleic-acid recognition. Nucleic Acids Res 2009; 37:1225-38; PMID:19136459; http://dx.doi.org/ 10.1093/nar/gkn1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavery R, Zakrzewska K, Sklenar H. JUMNA: Junction Minimisation of Nucleic Acids. Computer Physics Communications 1995; 91:135-58; http://dx.doi.org/ 10.1016/0010-4655(95)00046-I [DOI] [Google Scholar]
- 41.Zacharias M. Conformational analysis of DNA-Trinucleotide-hairpin-loop structures using a continuum solvent model. Biophys J 2001; 80:2350-63; PMID:11325735; http://dx.doi.org/ 10.1016/S0006-3495(01)76205-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 1999; 76:2879-86; PMID:10354416; http://dx.doi.org/ 10.1016/S0006-3495(99)77443-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petoukhov MV, Svergun DI. Joint use of small-angle X-ray and neutron scattering to study biological macromolecules in solution. Eur Biophys J 2006; 35:567-76; PMID:16636827; http://dx.doi.org/ 10.1007/s00249-006-0063-9 [DOI] [PubMed] [Google Scholar]
- 44.Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science 2003; 300:2101-4; PMID:12829785; http://dx.doi.org/ 10.1126/science.1083901 [DOI] [PubMed] [Google Scholar]
- 45.Fischer M, Goldschmitt J, Peschel C, Brakenhoff JP, Kallen KJ, Wollmer A, Grötzinger J, Rose-John S. I. A bioactive designer cytokine for human hematopoietic progenitor cell expansion. Nat Biotechnol 1997; 15:142-5; PMID:9035138; http://dx.doi.org/ 10.1038/nbt0297-142 [DOI] [PubMed] [Google Scholar]
- 46.Sousa R, Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J 1995; 14:4609-21; PMID:7556104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cornell WD, Cieplak P, Bayly CI, Gould IR, Kenneth M. Merz J, Ferguson DM, et al.. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chemical Society 1995; 117:5179-97; http://dx.doi.org/ 10.1021/ja00124a002 [DOI] [Google Scholar]
- 48.Homann M, Goringer HU. Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res 1999; 27:2006-14; PMID:10198434; http://dx.doi.org/ 10.1093/nar/27.9.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu H, Matsugami A, Katahira M, Uesugi S. A dimeric RNA quadruplex architecture comprised of two G:G(:A):G:G(:A) hexads, G:G:G:G tetrads and UUUU loops. J Mol Biol 2002; 322:955-70; PMID:12367521; http://dx.doi.org/ 10.1016/S0022-2836(02)00876-8 [DOI] [PubMed] [Google Scholar]
- 50.Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HD, Konarev PV, Svergun DI. New developments in the program package for small-angle scattering data analysis. J App Crystallogr 2012; 45:342-50; PMID:25484842; http://dx.doi.org/ 10.1107/S0021889812007662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J App Crystallogr 1992; 25:495-503; http://dx.doi.org/ 10.1107/S0021889892001663 [DOI] [Google Scholar]
- 52.Valentini E, Kikhney AG, Previtali G, Jeffries CM, Svergun DI. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res 2015; 43:D357-63; PMID:25352555; http://dx.doi.org/ 10.1093/nar/gku1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


