Abstract
Dysbiosis of the intestinal microbial community is considered a risk factor for development of chronic intestinal inflammation as well as other diseases such as diabetes, obesity and even cancer. Study of the innate and adaptive immune pathways controlling bacterial colonization has however proven difficult in rodents, considering the extensive cross-talk between bacteria and innate and adaptive immunity. Here, we used the zebrafish to study innate and adaptive immune processes controlling the microbial community. Zebrafish lack a functional adaptive immune system in the first weeks of life, enabling study of the innate immune system in the absence of adaptive immunity. We show that in wild type zebrafish, the initial lack of adaptive immunity associates with overgrowth of Vibrio species (a group encompassing fish and human pathogens), which is overcome upon adaptive immune development. In Rag1-deficient zebrafish (lacking adaptive immunity) Vibrio abundance remains high, suggesting that adaptive immune processes indeed control Vibrio species. Using cell transfer experiments, we confirm that adoptive transfer of T lymphocytes, but not B lymphocytes into Rag1-deficient recipients suppresses outgrowth of Vibrio. In addition, ex vivo exposure of intestinal T lymphocytes to Rag1-deficient microbiota results in increased interferon-gamma expression by these T lymphocytes, compared to exposure to wild type microbiota. In conclusion, we show that T lymphocytes control microbial composition by effectively suppressing the outgrowth of Vibrio species in the zebrafish intestine.
Keywords: lymphocytes, microbiota, mucosal immunity, Vibrio, zebrafish
Abbreviations
- wpf
weeks post fertilization
Introduction
Intestinal dysbiosis is associated with several diseases.1-5 Dysbiosis may refer to disturbances in the microbial community composition, but can also refer to disturbed interaction between the microbiota and the host. Disturbance of the microbial community due to for example antibiotic use can lead to outgrowth of so-called pathobionts. A pathobiont, according to Mazmanian and Round, is ‘a symbiont that is able to promote pathology only when specific genetic or environmental conditions are altered in the host’.6-8 Experimental animal models show that modification of the microbial composition can have a profound influence on the health status of the host. For example, colonization of mice with segmented filamentous bacteria (SFB) induces Th17 cells in the lamina propria and results in resistance towards intestinal inflammation caused by Citrobacter rodentium.9 Furthermore, spontaneous colitis in IL10-deficient mice can be prevented by narrow and broad spectrum antibiotics or through inoculation in early life with a mix of Clostridium species.10,11 We previously reported that the microbial composition affects development and severity of zebrafish enterocolitis by regulating the composition and number of infiltrating cells.12 In addition to physical constrains such as pH, acidity and availability of nutrients, the host immune system has an effect on the microbial colonization of the intestines as well. This indicates that the host immune system not only adapts to microbial composition but indeed actively creates the terms of microbial survival and thus composition in the intestine.13,14
Despite the great relevance to elucidate mechanisms by which microbes are able to colonize their hosts, dissection of the innate and adaptive immune signaling pathways controlling colonization has proven difficult due to the extensive cross-talk. High throughput 16S rRNA sequence analysis of faeces from several mouse models deficient for innate signaling, revealed that these pathways in part control intestinal microbial composition. Nod2-deficient, MyD88-deficient and Mmp7-deficient mice all display microbial communities that deviate from wild type littermates.15-17 As for the role of adaptive immunity on bacterial colonization, much attention was directed towards the role of IgA-secreting B cells.18-21 IgA can prevent colonization of certain bacteria and can allow colonization of others.22 Certain subsets of T lymphocytes can influence microbial composition. For example, it was shown that CD1d-deficient mice lacking NKT-cells display differences in intestinal microbial composition resulting from altered Paneth cell function.23 These data suggest a role for specific subsets of T lymphocytes in the establishment of intestinal microbiota. Furthermore, certain bacteria are able to induce regulatory T lymphocytes24,25 and large numbers of intra-epithelial lymphocytes reside within the epithelial cell layer that separates the underlying mucosa from the intestinal lumen, suggesting an important role in maintaining intestinal homeostasis with the microbiota.26
To clarify the role of lymphocytes on microbial community composition and intestinal milieu we made use of zebrafish. Zebrafish develop ex-utero and are unique in that during the first weeks of life only innate immunity is functional. From two to three weeks of age the adaptive immune system develops.27-29 This is in contrast to mice and humans, where adaptive immune cells are present in the intestines during the first weeks of bacterial colonization.30 Thus, the zebrafish model allowed us to specifically investigate the early bacterial colonization period in the absence of fully functional adaptive immunity and the impact of adaptive immune development on colonization thereafter. We show that zebrafish T lymphocytes can actively shape the intestinal microbial composition by effectively suppressing the outgrowth of Vibrio; a group of bacteria that is known to encompass human and fish-pathogens.
Results
Adult Rag1-deficient zebrafish harbor a different intestinal microbial community compared to wild type siblings
In order to establish whether the presence of adaptive immune cells dictates a difference in microbial community we performed sequence analysis of bacterial 16S rRNA of intestinal content from adult Rag1-deficient zebrafish and wild type siblings at 14 wpf (weeks post fertilization). Rag1-deficient zebrafish harbour a distinct and significantly different microbial composition compared to wild type and heterozygous siblings (Figure 1A and B). The microbial profiles of wild type and heterozygote siblings are different yet cluster together, while the microbial profile of Rag1-deficient siblings is clearly distinct from this cluster (Figure 1A). Figure 1B shows a phylogenetic tree of the different microbiota including a bar containing information about the species retrieved from the 16S rRNA sequences. Interestingly, most of the species retrieved in both Rag1-sufficient and Rag-deficient zebrafish belonged to the Fusobacteria. Further specification of these Fusobacteria revealed that all operational taxonomic units (OTUs) belonging to these Fusobacteria are identified as Cetobacterium somerae, a commonly found bacterium in freshwater fish (table S1). Figure 1B again shows that while the wild type and heterozygote intestinal microbial communities cluster together phylogenetically, Rag1-deficient intestinal microbiota form a separate cluster. Although minor variations exist in the relative abundance of different bacteria between wild type and heterozygous zebrafish, the difference with Rag1-deficient siblings is prominent. Especially, the 16S rRNA sequences retrieved from the Rag1-deficient intestinal contents revealed higher relative abundance of species belonging to the order Vibrio compared to wild type siblings (brown bar, Figure 1B) Our sampling depth was very high relative to alpha diversity as measured by Shannon diversity index (figure S1). Shannon's diversity index is a commonly used diversity index that takes into account both abundance and evenness of species present in the community. A high value of Shannon's diversity index is representative of a diverse and equally distributed community and lower values represent less diverse community. A value of 0 would represent a community with just one species. Based on a Random Forest Analysis the presence of Vibrio could clearly predict the genotype of our fish. Random Forest Analysis depicts to what extent each Operational Taxonomic Unit (OTU; bacterial species) can predict the genotype (wild type, heterozygote or Rag1-deficient). 31 In our analysis, we considered an OTU to be highly predictive if its importance score was at least 0.001.32 (Figure S2).
Figure 1.

Microbiota composition is dependent on genotype. (A) Classified sample diagram of Redundancy analysis (RDA). Analysis was performed using Canoco5. Taxonomic composition at the genus level was used as response data, genotype as explanatory variable. The variation explained by the ordination axis is significantly higher than random (p<0.002, permutation test). Red symbols represent genotype, open symbols are the individual samples. The colored lines are envelopes connecting samples of the same genotype, (B) Hierarchical clustering of Rag1-deficient, heterozygous and wild type siblings (n = 4/genotype) and intestinal composition as analyzed by 16S rRNA sequence analysis. Robustness of the clustering was estimated using jack-knifing analysis (20 replicates; indicated as a fraction). (C) Real Time quantitative PCR of Cetobacterium somerae and (D) Vibrio log copy number in the zebrafish intestines over time (1, 5 and 14 weeks post fertilization, one experiment). Black bars: wild type, white bars: Rag1-deficient zebrafish. Data are represented as mean +/− standard error of the mean. Statistics: Data tested for normal distribution by Kolmogorov-Smirnov test (not normally distributed). Mann-Whitney test, *:p = 0.04, **: p = 0.0016. Zebrafish are derived from the same clutch of eggs and raised in the same aquarium tank.
Vibrio abundance decreases over time in wild type zebrafish but not in Rag1-deficient zebrafish
Since two major groups of bacteria -Fusobacteriales (specifically Cetobacterium somerae) and Vibrio- constitute >90% of all intestinal bacterial species in Rag1-deficient zebrafish and their siblings, we designed specific Real Time qPCR primers to determine the abundance of these groups of over time in the intestines. We confirmed the 16S rRNA sequence data by Real Time quantitative PCR in adults displaying increased abundance of Vibrio in the Rag1-deficient zebrafish compared to wild type siblings (Figure 1C and D). We analyzed the microbiota at 1 wpf (innate), 5 wpf (adaptive immune development) and 14 wpf (innate and adaptive immunity) in Rag1-deficient and wild type littermates. This analysis showed that the difference in intestinal species composition between Rag1-deficient zebrafish and wild type siblings is apparent from 5 wpf onwards (Figure 1C, D). While Vibionales species are abundant in both Rag1-deficient and wild type zebrafish at 1 wpf (both only have innate immunity), their abundance decreases from 1010 to 106 in the wild type intestinal community from 5 wpf onwards. In contrast, in Rag1-deficient zebrafish the relative abundance of Vibrio remains equal over time (Figure 1D). Conversely, the presence of Cetobacterium somerae is slightly (though not significantly) reduced in Rag1-deficient zebrafish compared to wild type siblings from 5 wpf onwards (Figure 1C).
Adoptive transfer of T lymphocytes, but not B lymphocytes reduces the relative abundance of Vibrio
To evaluate the role of lymphocytes in shaping the microbial community, we sorted T and Non-T (B and NK-like cells) lymphocytes from conventionally raised T lymphocyte reporter (Lck-GFP) transgenic zebrafish, based on lymphocyte scatter and GFP fluorescence using flow cytometry. Immediately after sorting, we adoptively transferred sorted T cells or non-T cells into Rag1-deficient recipients. At one week post cell transfer, we analyzed log copy numbers of Cetobacterium somerae and Vibrio by Real time quantitative PCR and we assessed whether we could detect T lymphocytes in the intestines of the recipients by flow cytometry. Indeed, GFP positive events were detected in the intestines of most (but not all) zebrafish (Figure 2A, solid and grey line). Evaluation of the intestinal microbes by qPCR one week after transfer revealed that when T lymphocytes were transferred into Rag1-deficient zebrafish, and a positive intestinal GFP signal was detected, the relative abundance of Vibrio decreased to wild type levels, while Cetobacterium somerae levels tended to be higher compared to PBS injected zebrafish (p = 0.06) (Figure 1D, 2B, C). Adoptive transfer of Non-T lymphocytes containing B cells and NK-like cells did not modulate Vibrio abundance (striped line, Figure 2A and 2C). Notably, in one of the experiments, two Rag1-deficient zebrafish did not show GFP positive events in the intestine after adoptive transfer of wild type T lymphocytes (Figure 2A, grey line) suggesting that transfer failed in these fish. Importantly, in these fish the Vibrio species were retained at the elevated levels seen in the non-transferred Rag1-deficient fish, and were thus not reduced to wild type levels (Supplemental Figure 4). This again supports the role of T lymphocytes in the repression of Vibrio observed in the wild type fish. Overall, a small negative correlation was observed between the GFP-positive events (T lymphocytes) found in the intestines at 1 week post injection (wpi) and the intestinal abundance of Vibrio (Spearman rank, two-tailed p = 0.04, Figure 2D), whereas adoptive transfer of lymphocytes in wild type zebrafish did not alter the microbial composition (data not shown).
Figure 2.
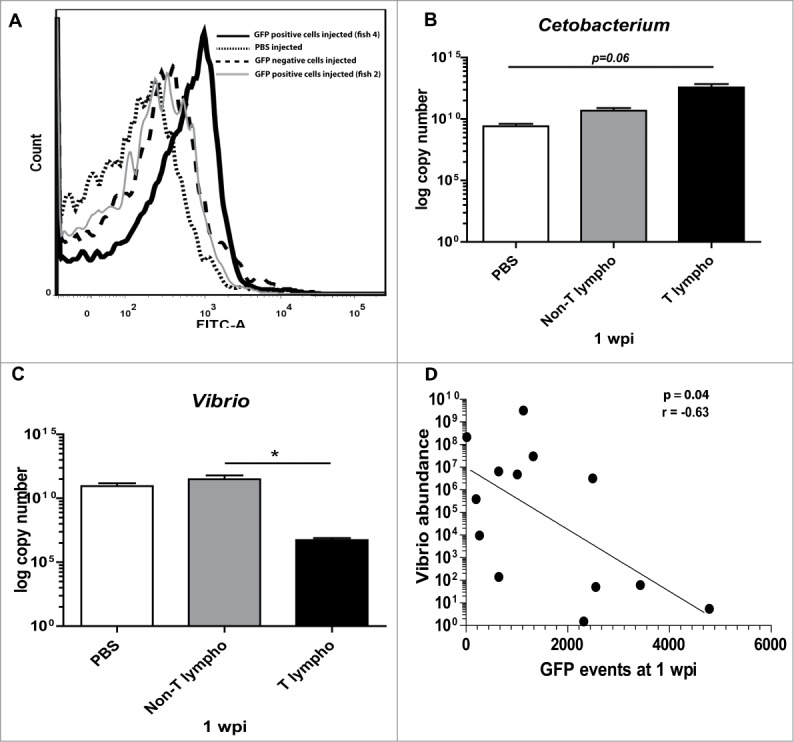
Adoptive transfer of T but not Non-T lymphocytes reduces the abundance of Vibrio 1 week after transfer. (A) Representative flow cytometry plot for GFP signal in the intestines one week after adoptive transfer of PBS, sorted non-T lymphocytes (GFP-negative cells), sorted T lymphocytes (GFP-positive cells) in Rag1-deficient zebrafish. Real Time PCR for Cetobacterium somerae (B) and Vibrio (C) abundance in zebrafish that received PBS, T lymphocytes or Non-T lymphocytes in the intestines 1 week after transfer. (D) Correlation of GFP-positive events with Vibrio abundance (Spearman Rank, p = 0.04).
Sorted T lymphocytes show increased in vitro responses towards Rag-deficient but not wild type intestinal bacteria
Does the modulated microbiota in Rag-deficient zebrafish have an immunological consequence to T cell activation and/or effector function? To address this question, we sorted T lymphocytes to assess a possible T cell-mediated response upon direct exposure to intestinal bacteria from wild type and Rag1-deficient zebrafish. After seeding in 24 wells plates, T lymphocytes were exposed to either wild type or Rag-deficient intestinal microbiota for 6 hours. After 6 hours cytokine responses were assessed. Rag1-deficient microbiota was able to induce ifnγ expression in sorted T lymphocytes to a greater extent than wild type microbiota (Figure 3A). We did not detect significant differences in il10 and tnfα expression between wild type and Rag1-deficient microbiota exposed T lymphocytes (Figure 3C and D). Expression of il17 was not detected in any of the groups (data not shown). From these ex vivo experiments it can be concluded that microbes derived from rag1 mutant zebrafish (containing high Vibrio abundance) induce a stronger T lymphocyte response than microbes derived from wild type siblings. Interestingly, counting the cells after 6 hours revealed that stimulation with Rag1-deficient microbiota had increased the number of cells 3 fold compared to t0, while no difference was seen in cell numbers after 6 hours stimulation with wild type microbial content or medium (Figure S3).
Figure 3.
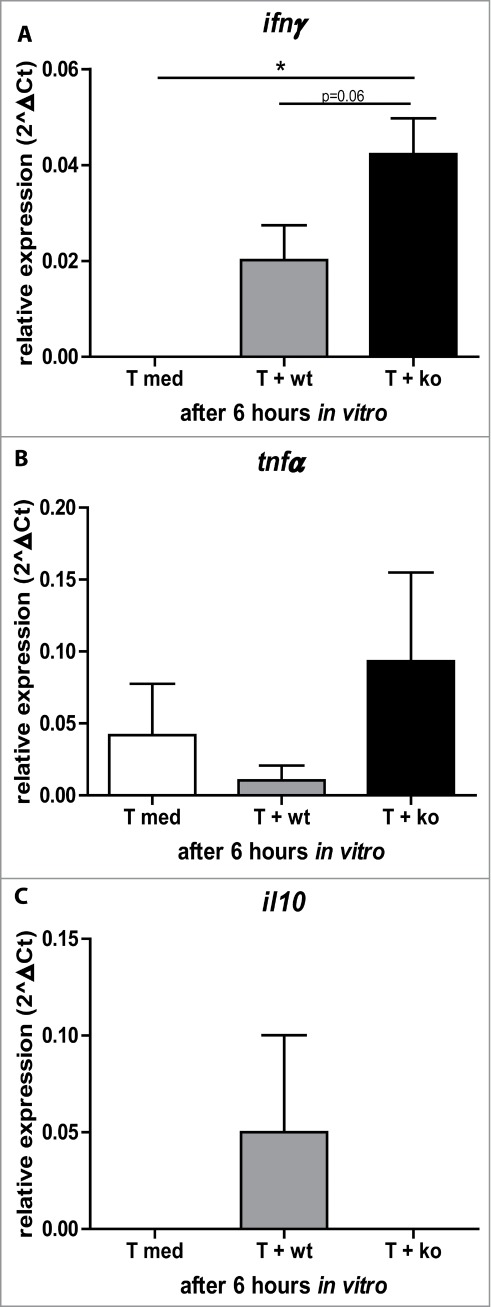
T lymphocyte responses towards Rag1-deficient and wild type microbiota. Relative expression of ifnγ (A), tnfα (B) and il10 (C) in T lymphocytes left in medium or exposed to either wild type or rag1-deficient intestinal microbiota for 6 hours.
Intestinal Immune activation in Rag1-deficient zebrafish at adult age
We finally considered that the altered intestinal microbial community and lack of lymphocytes in Rag1-deficient zebrafish may coincide with an inflamed intestinal milieu. To this end, we determined intestinal cytokine expression by Real Time quantitative PCR. Several cytokines were differently expressed in Rag1-deficient zebrafish and wild type siblings (Figure 4). Expression of il1β and cxcl8-l2 was increased in Rag1-deficient zebrafish at 14 wpf indicating immune activation at these time points (Figure 4A and B). Expression of interleukin(il)17f2, il10 and ifnγ was significantly lower in Rag1-deficient zebrafish intestines at 14 wpf, which relates to the absence of T cells that normally produce these cytokines (Figure 4C, E, and F). We did not observe a difference in expression of tnf-a between Rag1-deficient zebrafish and wild type siblings at 14 wpf (Figure 4D). From 14 weeks onwards Rag1-deficient zebrafish develop dropsy (oedema of the tissues caused by bacterial infection) or become severely anorexic (Figure S5). Thus, absence of lymphocytes results in an altered microbial community from 5 weeks post fertilization onwards and an inflamed intestinal milieu later in life.
Figure 4.

Relative expression levels of intestinal cytokines in wild type and Rag1-deficient zebrafish at 14 wpf. Relative expression of il1β (A), cxcl8-l2 (B), il10 (C), tnfα (D), ifnγ (E) and il17f2 (F) in the intestines of zebrafish of 14 wpf as measured by Real Time PCR.
Discussion
In this study we report that T lymphocytes have a profound effect on intestinal microbial composition. T lymphocytes actively repress outgrowth of Vibrio species in the zebrafish intestines. Since in zebrafish adaptive immunity is absent in the first weeks of life and gradually develops from 2 to 3 weeks post fertilization27, we could specifically address the effect of adaptive immune development on intestinal bacterial colonisation patterns.
In our study we observed high abundance of Vibrio species at larval (innate) stages of zebrafish development, in agreement with observations made by Rawls et al.33 In their study Rawls et al. compared microbial communities of zebrafish at different time points. From 6 to 30 days post fertilization the number of sequences belonging to the Vibrio group go down from 207 to 12 in conventionally raised fish, however at adult age 113 sequences are found. This high number of Vibrio species found at adult age is due to an increase in the number of Vibrio navarrensis sequences. In contrast, we do not observe an increase at 14 wpf in Vibrio species in our wild type zebrafish in fact the abundance stays low (table S1). In addition, in their study, Rawls and colleagues do not find Cetobacterium somerae species until adult age, while we find these bacteria throughout life in our fish, wild type and Rag1-deficient. In contrast, Rawls et al report more Aeromonas, Lactococcus and Pseudomonas species. In a separate study, Roeselers and colleagues also reported the presence of Vibrio in adult wild type zebrafish. 34 Diversity in microbial composition between different studies can be attributed to the different aquatic environment and diets used, since diet and tank water are important factors that determine microbial composition. For this reason we took great care to minimize and control for these differences in our experiments. Comparisons between wild type and mutant zebrafish are between siblings which have been raised in the same tank.
The data presented in our manuscript expand on previous studies in mice showing T cell involvement in intestinal homeostasis. For example, mice lacking a specific subset of T-lymphocytes, i.e. NKT lymphocytes display a different microbial composition. These CD1d-deficient mice have more epithelial attaching segmented filamentous bacteria (SFB) and increased bacterial colonization rate upon mono-colonization with specific bacteria.35 Furthermore, it has been reported that severe combined immuno-deficient (SCID) mice that lack T and B cells show increased and prolonged colonization upon mono-association with SFB compared to heterozygous littermates.22 In addition, Dimitriu et al. also reported differences in bacterial community composition in faeces between WT mice and mice lacking adaptive immunity (CD45, RAG and 45RAG double knock-out) as analyzed by Principal coordinates analysis (PCoA) plots of bacterial community composition (T-RFLP fingerprints).36 Interestingly, some bacterial species (and possible pathobionts) were only detected in the CD45 knockout mice, such as Alcaligenaceae and Helicobacteraceae.36 In contrast, thymectomy of tadpoles (Xenopus laevis), a model in between zebrafish and higher vertebrates, did not result in differences in microbial composition.37 Although, Rag-deficiency and thymectomy cannot directly be compared, further investigation into this difference might yield clues concerning the mechanism by which T cells regulate microbial composition independently of B cells in zebrafish. In corroboration with our data, Kawamoto and colleagues showed that not only Rag1–/– mice, but also mice lacking only B cells (Ighm–/–) or only T cells (Cd3e–/–) had reduced bacterial diversity and bacterial communities, indicating that T lymphocytes do have a B lymphocyte independent effect on the microbiota in mice.38 The order Vibrio contains species that are known fish pathogens. For example, Vibrio anguillarum as well as Vibrio salmonicida can cause vibriosis, a deadly haemorrhagic septicaemic disease. This disease can affect marine as well as fresh water fish.39, 40 Furthermore, species such as Vibrio parahaemolyticus, Vibrio vulnificus and Vibrio cholerae can induce disease in humans.41-44 In spite of the high abundance of Vibrio species throughout life, our Rag1-deficient zebrafish do not appear to have any symptoms of disease until after 14 wpf. After 14 wpf, however, fish displayed increased pro-inflammatory cytokine responses (cxcl8-l2 and il1β) in the intestines, which might reflect the continuous burden of infections due to a lack of adaptive immunity. Furthermore, it has been shown that Foxp3+ regulatory T cells contribute to diversification of gut microbiota and regulation of subsequent inflammatory responses in mice. Although transcription factor Foxp3 has been reported in zebrafish, it is very difficult to identify this distinct Treg subset to date.45
In addition, most fish become anorexic and die, when not euthanized, before 16 wpf. In a few cases, we have observed dropsy (1% of Rag1-deficient fish) at >14 wpf. Dropsy is an accumulation of fluids in the body cavity due to internal organ failure. Dropsy can be caused by bacterial and viral infections as well as husbandry imbalances (salt concentration).46 In light of this, it is apparent that Rag1-deficient zebrafish are more susceptible to disease just like their murine counterparts. Rag1-deficient zebrafish harbor increased abundance of Vibrio species from early age and yet do not show pathology until adult age. Nevertheless, life-long dysbiosis together with their immunocompromised state might render these fish more susceptible to additional triggers such as bacterial and viral infections as well as other environmental factors.
The outgrowth of Vibrio in Rag1-deficient zebrafish suggests a specific role for lymphocytes in modulation of the microbial community. Indeed, adoptive transfer of T lymphocytes was able to reduce the relative abundance of Vibrio. In the absence of adaptive immunity, in Rag1-deficient zebrafish or larval zebrafish, Vibrio abundance is high. A recent report on establishment of mouse intestinal microbiota also suggests that during the early stages of development typical pathobionts, in this case Helicobacter and Sphingomonas, are able to colonize despite activation of innate immunity.47,48 Similar to our zebrafish data, development of adaptive immunity (from day 4-7) in these conventionalized mice coincided with decreased abundance of these pathobionts. We hypothesize that these (host-specific) pathobionts (such as Helicobacter, Sphingomonas and Vibrio spp.) activate T lymphocytes, which subsequently results in repression of these species. Indeed, we observed that sorted T lymphocytes specifically respond by proliferation and expression of ifnγ when exposed to Rag1-deficient microbial content (with increased abundance of Vibrio), while this response is seen less upon exposure to wild type intestinal content. Interestingly, recently it was shown that CD4+ T lymphocyte derived IFNγ can induce MHC class II on intestinal epithelial cells which is able to protect against colitis.49 Furthermore, Do and colleagues showed that IFN-γ signalling in non-hematopoietic cells may control inflammation.50 Additionally, we have recently shown that transfer of T lymphocytes can induce epithelial cxcl8-l1 in zebrafish, suggesting that T lymphocytes can also activate epithelial cell immune responses.51 Future investigation into the different intestinal T lymphocyte subsets in zebrafish will aim to dissect whether these T lymphocytes are directly activated by bacterial ligands or whether memory T lymphocytes are responsible for further immune activation and suppression of Vibrio species.
In conclusion, by using the zebrafish as our model system we were able to specifically study the effect of adaptive immune development on intestinal bacterial community establishment. As the number of available transgenic (immune cell) reporter zebrafish keeps rising, and protocols are developed to easily modify these fish both genetically and environmentally, we anticipate that the use of this model system will greatly enhance or understanding of bacterial-host interaction. The findings presented here show that early overgrowth of Vibrio species induces adaptive pro-inflammatory cascades, which initiates full maturation of the zebrafish mucosal immune system and might be necessary to establish mucosal homeostasis between the zebrafish host and the microbiota.
Materials and Methods
Animals
All zebrafish strains were maintained under standard husbandry conditions at the Hubrecht Institute Utrecht (14 hours light, 10 hours dark regime). The larval fish are fed with Tetrahymena pyriformis (CCAP) (from 5-14 dpf), Novotom with dried Artemia (1-3 wpf) and a combination of Artemia (230.000 npg, Ocean nutrition) and Tetramin Flakes (Tetra, Aqua Schwartz) twice daily (3-20 wpf). The rag1-/- line (rag1t26683) was generated at the Hubrecht institute52 and for all analyses siblings of rag1+/- in-crossed fish were used and kept in the same tank to control for environmental and maternal effects on microbial composition. The lck:GFP zebrafish were purchased from ZIRC.28 All animal experiments were approved by the Animal Experimentation Committee (DEC) of University Medical Centre Utrecht. Animals were monitored by skilled personnel at the Hubrecht Institute. All experiments described were carried out following guidelines of the Animal Experimentation Committee and Dutch Law on Animal Experimentation.
DNA isolation intestinal microbes
Intestinal contents are washed from intestinal tissue, collected in 50 μl of sterile water and immediately stored at −20ºC until further processing. DNA was isolated with the Invitek DNA Stool kit (Isogen, cat nr 1038110200) according to the manufacturer's instructions.
16S rRNA sequencing intestinal content
DNA samples were adjusted to a concentration of 20ng/ul and the V1 and V2 region of the 16S ribosomal RNA gene was amplified by PCR. The degenerated forward primers (27F-DegS) contained the titanium sequencing adaptor A at the 5’ end, followed by a variable eight nucleotide barcode sequence (Table 1).53-55 Reverse primers consisted of an equimolar mix of two primer sequences56 based on previously published probes EUB 338 I, II and III57, both containing the titanium sequencing adaptor B at the 5’ end (Table 1). Adaptor sequences were kindly provided by GATC-Biotech (Konstanz, Germany). PCRs were performed in a total volume of 100ul containing 2U of Phusion Hot start II High-Fidelity DNA polymerase (Thermo Scientific cat nr. F-537S), 1x High Fidelity PCR buffer, 2 μl PCR Grade Nucleotide Mix (Roche, cat nr 11581295001), 500nM forward primer, 500nM reverse primer and 40ng DNA. Negative control PCRs were performed without addition of template. An internal control for the reliability of the pyrosequencing technique was performed by including one sample twice, using different forward primers (primer 27F-DegS-1 and 27F-DegS-13 for sample Rag +/+ No.1). The PCR programme employed initiated with a 30 seconds denaturing step at 98°C, followed by 30 cycles of denaturation for 10 seconds at 98°C, annealing for 20 seconds at 56°C and elongation for 20 seconds at 72°C, followed by a final elongation step for 10 minutes at 72°C (GS0001 Thermocycler Westburg). The amplicon size was verified by agarose gel electrophoresis and purified using a High Pure PCR Cleanup Micro Kit (Roche, cat nr 04983955001) according to the manufacturers’ instructions. An equimolar mix of all 13 samples with a total DNA concentration of 13ug was prepared. Purified PCR products were mixed in equimolar amounts followed by agarose gel electrophoresis, band-excision, and purification using the High Pure PCR purification kit (Roche, cat nr 04983955001). The purified pooled amplicons were pyrosequenced using 454 titanium sequencing.
Table 1.
Barcoded sequencing primers
| Primer name | 5’ to 3’ sequence (titanium adaptor (– barcode) – primer sequence) |
|---|---|
| 27F-DegS-1 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCAACC – GTTYGATYMTGGCTCAG |
| 27F-DegS-2 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCAAGG – GTTYGATYMTGGCTCAG |
| 27F-DegS-3 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCATCG – GTTYGATYMTGGCTCAG |
| 27F-DegS-4 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCATGC – GTTYGATYMTGGCTCAG |
| 27F-DegS-5 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCGCAT – GTTYGATYMTGGCTCAG |
| 27F-DegS-6 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCGCTA – GTTYGATYMTGGCTCAG |
| 27F-DegS-7 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCGGAA – GTTYGATYMTGGCTCAG |
| 27F-DegS-8 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCGGTT – GTTYGATYMTGGCTCAG |
| 27F-DegS-9 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCTACG – GTTYGATYMTGGCTCAG |
| 27F-DegS-10 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCTAGC – GTTYGATYMTGGCTCAG |
| 27F-DegS-11 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCTTCC – GTTYGATYMTGGCTCAG |
| 27F-DegS-12 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACCTTGG – GTTYGATYMTGGCTCAG |
| 27F-DegS-13 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACGAACG – GTTYGATYMTGGCTCAG |
| 27F-DegS-14 | CCATCTCATCCCTGCGTGTCTCCGACTCAG – AACGAAGC – GTTYGATYMTGGCTCAG |
| titAdapB338R-I | CCTATCCCCTGTGTGCCTTGGCAGTCTCAG – GCWGCCTCCCGTAGGAGT |
| titAdapB338R-II | CCTATCCCCTGTGTGCCTTGGCAGTCTCAG – GCWGCCACCCGTAGGTGT |
16S rRNA sequence data analysis
Pyrosequencing data analysis was carried out using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline58 using settings as recommended in the QIIME 1.2 tutorial with the following exceptions: reads were filtered for chimeric sequences using Chimera Slayer59; and OTU clustering was performed with an identity threshold of 97%, using parameters as recommended in the QIIME newsletter of December 17th 2010 (http://qiime.wordpress.com/2010/12/17/new-default-parameters-for-uclust-otu-pickers/). The Ribosomal Database Project (RDP) classifier version 2.232,59,60 was used for taxonomic classifications. Hierarchical clustering of samples was performed using UPGMA with weighted UniFrac as a distance measure. Robustness of the clustering was estimated using jackknifing analysis (20 replicates) as implemented in Qiime 1.2. Redundancy analysis (RDA) was performed using Canoco5, using default settings of the analysis type “Constrained-supplementary”. For the permutation tests, 500 randomizations were done. Details on the underlying mathematics can be found in ter Braak et al.{ter Braak CJF, 2012 #179}
Quantitative Real Time PCR Fusobacteriales and Vibrio
Primers for Cetobacterium somerae and Vibrio sp. were designed using the oligo 6.22 program and were checked for specificity against the rdp-database (http://rdp.cme.msu.edu/misc/rel10info.jsp). Table 2 depicts the primer sequences used. Pure clinical isolates of Fusobacteria sp. and Vibrio sp. (kindly provided by the department of Microbiology, UMCU) were used to test primer efficiency and specificity. Real Time PCR was performed using SyBr Green amplification (BioRad). The PCR program used: 95°C 3 min., 40x [95°C 30 sec., 60°C 60 sec., 72°C 90 sec.], followed by a melting curve 95°C 30 sec., 65°C 5 sec., increase to 95°C in 0.5°C steps. Log copy numbers were calculated from standard curves of the different primer pairs on pure cultures and corrected per 100 ng input DNA.
Table 2.
Real Time quantitative PCR primer sets
| Primer name | Sequence 5’- 3’ |
|---|---|
| Cetobacterium somerae FW | 5’ GCTTGCCGGAACTTAGT 3’ |
| Cetobacterium somerae Rev | 5’ TCATCGCAGGCAGTATC 3’ |
| Vibrio FW | 5’ AGACGCTGGAGTGCCT 3’ |
| Vibrio Rev | 5’ GTGCTGGCAAACAAGG 3’ |
| Cxcl8-l2 FW | 5’ TGTTTTCCTGGCATTTCTGACC 3’ |
| Cxcl8-l2 Rev | 5’ TTTACAGTGTGGGCTTGGAGGG 3’ |
| il17f2 FW | 5’ AACCGGTTGTGTGATACTG 3’ |
| il17f2 Rev | 5’ CTGGGCTTCAAAGATGAC 3’ |
| il-10 FW | 5’ AGGGCTTTCCTTTAAGACTG 3’ |
| il-10 Rev | 5’ ATATCCCGCTTGAGTTCC 3’ |
| tnfα FW | 5’ CAGGGCAATCAACAAGA 3’ |
| tnfα FW | 5’ CCTGGTCCTGGTCATCT 3’ |
| ifnγ FW | 5’ TTGGGTTGGAAAATCTGT 3’ |
| ifnγ Rev | 5’ TCTTGGAAAATGTCTTCATAGA 3’ |
| il-1β FW | 5’ TGCGGGCAATATGAAGTCA 3’ |
| il-1β Rev | 5’ TTCGCCATGAGCATGTCC 3’ |
| β-actin FW | 5’ ACCGCTGCCTCTTCTT 3’ |
| β-actin Rev | 5’ GCAATGCCAGGGTACA 3’ |
Adoptive transfer of T and non-T lymphocytes
T cell reporter zebrafish (Lck:GFP) were used to isolate GFP-positive T lymphocytes and GFP-negative non-T lymphocytes. Pooled intestinal tissues were strained over a 40 μm filter (Greiner Cat. No. 542 040) and the cell suspension was collected in supplemented L-15 medium as previously described.51 Cells were washed twice with medium and filtered over 40 μm filters. Subsequently, T and non-T lymphocytes were FACS sorted on basis of FSC and SSC scatter and GFP-positivity or negativity, respectively (FACS Aria, BD). Next, T or non-T lymphocytes were adoptively transferred into 5 wpf old Rag1-deficient zebrafish and wild type siblings. A total of 15.000 cells or PBS was injected i.p. in a volume of 10 μl under anaesthesia with MS222 (Tricaine, Sigma Aldrich, Cat no. E10521).
In vitro stimulation of T lymphocytes
T cell reporter zebrafish (Lck:GFP) were used to isolate GFP-positive T lymphocytes. Per well 100.000 T lymphocytes were seeded. T lymphocytes were left in medium alone or exposed to 100 μl intestinal content of wild type or Rag1-deficient zebrafish. The intestinal content was freshly prepared and consisted of pooled intestinal content (n = 5) resuspended in 500 μl PBS. This mixture was centrifuged to pellet dietary particles and the supernatant was used as microbial mix. T lymphocytes were stimulated with this mixture for 6 hours after which the cells were counted, pelleted and collected in TriPure (Roche, Cat No. 11667157001) for RNA isolation.
RNA isolation and cDNA synthesis from intestinal tissue
Total intestinal tissue was collected in TriPure (Roche, Cat No. 11667157001) and RNA was extracted by phenol/chloroform extraction. cDNA was synthesized from RNA by iScript (BioRad, Cat No. 170-8890) reverse transcriptase according to the manufacturer's instructions.
Quantitative Real Time PCR intestinal cytokine expression
Primers were designed using the oligo 6.22 program and blasted against the EMBL database. All primers were designed and selected to have an annealing temperature of 64-66°C, with a difference between the annealing temperature of the forward and reverse primer <0.5ºC and primer r2 of >0.95. Accession numbers for the different genes analysed were: cxcl8-l1 (ZDB-GENE-081104-317), cxcl8-l2 (ZDB-GENE-101026-3), il17f2 (BX294375), il-10 (AY887900), tnfα (ENSDARG00000009511), ifnγ (NM001020793), IL-1β (NM212844), β-actin (NM131031). Primer sequences and references are depicted in Table 2. Real Time quantitative PCR was performed using SyBr Green amplification (BioRad, Cat. No.172-5270). The PCR program used: 95°C 3 min., 40x [95°C 10 sec., 60°C 10 sec., 72°C 30 sec.], followed by a melting curve 95°C 30 sec., 65°C 5 sec., increase to 95°C in 0.5°C steps. Relative expression was assessed by calculating relative expression compared to beta-actin; 2٨-(Cqtarget-Cqbeta-actin).
Statistical analysis
Data were tested for normal distribution by Kolmogorov-Smirnov test. Mann-Whitney tests were performed on data sets not normally distributed. Unpaired t-test was used on normally distributed data sets to calculate statistical significance. For correlation statistics of intestinal Vibrio and GFP count, Spearman Rank was used. The statistical tests used and p values obtained are mentioned in figure legends.
Acknowledgements
The authors would like to thank Prof. Schulte-Merker at the Hubrecht Institute who kindly provided the necessary infrastructure for the experiments. Furthermore, we would like to thank Dr. Maria Forlenza of Wageningen University for kindly providing us with SPF carp serum. We would like to thank Dr. Renshaw for the mpx:eGFP transgenic line. Additionally, we would like to thank the animal caretakers at the Hubrecht Institute for excellent care of the zebrafish. Lieke Reubsaet kindly provided us with bacterial isolates. Finally, we would like to thank Dr. P. van Baarlen and Prof. B. Prakken and the lab-members of the Center for Molecular and Cellular Intervention, Laboratory for Translational Immunology (CMCI-LTI) Utrecht for useful discussions and critical review of the manuscript.
Funding
This work was generously funded by the Wilhelmina Children's Hospital Research Fund.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Bultman S.J., Emerging roles of the microbiome in cancer. Carcinogenesis, 2014. 35(2): p. 249-55; PMID:24302613; http://dx.doi.org/ 10.1093/carcin/bgt392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sartor R.B., Microbial-host interactions in inflammatory bowel diseases and experimental colitis. Nestle Nutr Workshop Ser Pediatr Program, 2009. 64: p. 121-32; discussion 132-7, 251-7; PMID:19710519; http://dx.doi.org/ 10.1159/000235787 [DOI] [PubMed] [Google Scholar]
- 3. Turnbaugh P.J., Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. , A core gut microbiome in obese and lean twins. Nature, 2009. 457(7228): p. 480-4; PMID:19043404; http://dx.doi.org/ 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brugman S., Klatter FA, Visser JT, Wildeboer-Veloo AC, Harmsen HJ, Rozing J, Bos NA. Antibiotic treatment partially protects against type 1 diabetes in the Bio-Breeding diabetes-prone rat. Is the gut flora involved in the development of type 1 diabetes? Diabetologia, 2006. 49(9): p. 2105-8; PMID:16816951; http://dx.doi.org/ 10.1007/s00125-006-0334-0 [DOI] [PubMed] [Google Scholar]
- 5. Yeoh N, Burton JP, Suppiah P, Reid G, Stebbings S. The role of the microbiome in rheumatic diseases. Curr Rheumatol Rep, 2013. 15(3): p. 314; PMID:23378145; http://dx.doi.org/ 10.1007/s11926-012-0314-y [DOI] [PubMed] [Google Scholar]
- 6. Mazmanian S.K., Capsular polysaccharides of symbiotic bacteria modulate immune responses during experimental colitis. J Pediatr Gastroenterol Nutr, 2008. 46 Suppl 1: p. E11-2; PMID:18354314 [DOI] [PubMed] [Google Scholar]
- 7. Mazmanian S.K., Round J.L., and Kasper D.L., A microbial symbiosis factor prevents intestinal inflammatory disease. Nature, 2008. 453(7195): p. 620-5; PMID:18509436; http://dx.doi.org/ 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- 8. Round J.L. and Mazmanian S.K., The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol, 2009. 9(5): p. 313-23; PMID:19343057; http://dx.doi.org/ 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. , Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell, 2009. 139(3): p. 485-98; PMID:19836068; http://dx.doi.org/ 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hoentjen F., Harmsen HJ, Braat H, Torrice CD, Mann BA, Sartor RB, Dieleman LA. Antibiotics with a selective aerobic or anaerobic spectrum have different therapeutic activities in various regions of the colon in interleukin 10 gene deficient mice. Gut, 2003. 52(12): p. 1721-7; PMID:14633949; http://dx.doi.org/ 10.1136/gut.52.12.1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. , Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature, 2013. 500(7461): p. 232-6; PMID:23842501; http://dx.doi.org/ 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- 12. Brugman S, Liu KY, Lindenbergh-Kortleve D, Samsom JN, Furuta GT, Renshaw SA, Willemsen R, Nieuwenhuis EE. Oxazolone-induced enterocolitis in zebrafish depends on the composition of the intestinal microbiota. Gastroenterology, 2009. 137(5): p. 1757-67 e1; PMID:19698716; http://dx.doi.org/ 10.1053/j.gastro.2009.07.069 [DOI] [PubMed] [Google Scholar]
- 13. Rawls J.F., Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell, 2006. 127(2): p. 423-33; PMID:17055441; http://dx.doi.org/ 10.1016/j.cell.2006.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brugman S. and Nieuwenhuis E.E., Mucosal control of the intestinal microbial community. J Mol Med (Berl), 2010. 88(9): p. 881-8; PMID:20523962; http://dx.doi.org/ 10.1007/s00109-010-0639-9 [DOI] [PubMed] [Google Scholar]
- 15. Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS, Nod2 is required for the regulation of commensal microbiota in the intestine. Proc Natl Acad Sci U S A, 2009. 106(37): p. 15813-8; PMID:19805227; http://dx.doi.org/ 10.1073/pnas.0907722106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Larsson E., Tremaroli V, Lee YS, Koren O, Nookaew I, Fricker A, Nielsen J, Ley RE, Bäckhed F, Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut, 2012. 61(8): p. 1124-31; PMID:22115825; http://dx.doi.org/ 10.1136/gutjnl-2011-301104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salzman N.H, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al. , Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol, 2010. 11(1): p. 76-83; PMID:19855381; http://dx.doi.org/ 10.1038/ni.1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bos N.A., Jiang H.Q., and Cebra J.J., T cell control of the gut IgA response against commensal bacteria. Gut, 2001. 48(6): p. 762-4; PMID:11358892; http://dx.doi.org/ 10.1136/gut.48.6.762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kushnir N., Bos NA, Zuercher AW, Coffin SE, Moser CA, Offit PA, Cebra JJ. B2 but not B1 cells can contribute to CD4+ T-cell-mediated clearance of rotavirus in SCID mice. J Virol, 2001. 75(12): p. 5482-90; PMID:11356955; http://dx.doi.org/ 10.1128/JVI.75.12.5482-5490.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shroff K.E., Meslin K., and Cebra J.J., Commensal enteric bacteria engender a self-limiting humoral mucosal immune response while permanently colonizing the gut. Infect Immun, 1995. 63(10): p. 3904-13; PMID:7558298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shroff K.E. and Cebra J.J., Development of mucosal humoral immune responses in germ-free (GF) mice. Adv Exp Med Biol, 1995. 371A: p. 441-6; PMID:8525962http://dx.doi.org/ 10.1007/978-1-4615-1941-6_92 [DOI] [PubMed] [Google Scholar]
- 22. Jiang H.Q., Thurnheer MC, Zuercher AW, Boiko NV, Bos NA, Cebra JJ. Interactions of commensal gut microbes with subsets of B- and T-cells in the murine host. Vaccine, 2004. 22(7): p. 805-11; PMID:15040931; http://dx.doi.org/ 10.1016/j.vaccine.2003.11.022 [DOI] [PubMed] [Google Scholar]
- 23. Nieuwenhuis E.E., Matsumoto T, Lindenbergh D, Willemsen R, Kaser A, Simons-Oosterhuis Y, Brugman S, Yamaguchi K, Ishikawa H, Aiba Y, et al. , Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J Clin Invest, 2009. 119(5): p. 1241-50; PMID:19349688; http://dx.doi.org/ 10.1172/JCI36509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duchmann R, Kaiser I, Hermann E, Mayet W, Ewe K, Meyer zum Büschenfelde KH. Tolerance exists towards resident intestinal flora but is broken in active inflammatory bowel disease (IBD). Clin Exp Immunol, 1995. 102(3): p. 448-55; PMID:8536356; http://dx.doi.org/ 10.1111/j.1365-2249.1995.tb03836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nutsch K.M. and Hsieh C.S., T cell tolerance and immunity to commensal bacteria. Curr Opin Immunol, 2012. 24(4): p. 385-91; PMID:22613090; http://dx.doi.org/ 10.1016/j.coi.2012.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kayama H. and Takeda K., Regulation of intestinal homeostasis by innate and adaptive immunity. Int Immunol, 2012. 24(11): p. 673-80; PMID:22962437; http://dx.doi.org/ 10.1093/intimm/dxs094 [DOI] [PubMed] [Google Scholar]
- 27. Lam S.H., Chua HL, Gong Z, Lam TJ, Sin YM. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev Comp Immunol, 2004. 28(1): p. 9-28; PMID:12962979; http://dx.doi.org/ 10.1016/S0145-305X(03)00103-4 [DOI] [PubMed] [Google Scholar]
- 28. Langenau D.M, Ferrando AA, Traver D, Kutok JL, Hezel JP, Kanki JP, Zon LI, Look AT, Trede NS. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci U S A, 2004. 101(19): p. 7369-74; PMID:15123839; http://dx.doi.org/ 10.1073/pnas.0402248101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Page D.M, Wittamer V, Bertrand JY, Lewis KL, Pratt DN, Delgado N, Schale SE, McGue C, Jacobsen BH, Doty A, et al. , An evolutionarily conserved program of B-cell development and activation in zebrafish. Blood, 2013. 122(8): p. e1-11; PMID:23861249; http://dx.doi.org/ 10.1182/blood-2012-12-471029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. El Aidy S., van Baarlen P, Derrien M, Lindenbergh-Kortleve DJ, Hooiveld G, Levenez F, Doré J, Dekker J, Samsom JN, Nieuwenhuis EE, et al. , Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol, 2012. 5(5): p. 567-79; PMID:22617837; http://dx.doi.org/ 10.1038/mi.2012.32 [DOI] [PubMed] [Google Scholar]
- 31. R Core Team (2012). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from: http://www.R-project.org/ [Google Scholar]
- 32. Touw W.G, Bayjanov JR, Overmars L,Backus L, Boekhorst J, Wels M, van Hijum SA. Data mining in the Life Sciences with Random Forest: a walk in the park or lost in the jungle? Brief Bioinform, 2013. 14(3): p. 315-26; PMID:22786785; http://dx.doi.org/ 10.1093/bib/bbs034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rawls J.F., Samuel B.S., and Gordon J.I., Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A, 2004. 101(13): p. 4596-601; PMID:15070763; http://dx.doi.org/ 10.1073/pnas.0400706101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. Evidence for a core gut microbiota in the zebrafish. ISME J, 2011. 5(10): p. 1595-608; PMID:21472014; http://dx.doi.org/ 10.1038/ismej.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jiang H.Q., Bos N.A., and Cebra J.J., Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect Immun, 2001. 69(6): p. 3611-7; PMID:11349021; http://dx.doi.org/ 10.1128/IAI.69.6.3611-3617.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dimitriu P.A, Boyce G, Samarakoon A, Hartmann M, Johnson P, Mohn WW. Temporal stability of the mouse gut microbiota in relation to innate and adaptive immunity. Environ Microbiol Rep, 2013. 5(2): p. 200-10; PMID:23584963; http://dx.doi.org/ 10.1111/j.1758-2229.2012.00393.x [DOI] [PubMed] [Google Scholar]
- 37. Mashoof S, Goodroe A, Du CC, Eubanks JO, Jacobs N, Steiner JM, Tizard I, Suchodolski JS, Criscitiello MF. Ancient T-independence of mucosal IgXA: gut microbiota unaffected by larval thymectomy in Xenopus laevis. Mucosal Immunol, 2013. 6(2): p. 358-68; PMID:22929561; http://dx.doi.org/ 10.1038/mi.2012.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, Tsutsui Y, Qin H, Honda K, Okada T, et al. , Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity, 2014. 41(1): p. 152-65; PMID:25017466; http://dx.doi.org/ 10.1016/j.immuni.2014.05.016 [DOI] [PubMed] [Google Scholar]
- 39. Frans I, Michiels CW, Bossier P, Willems KA, Lievens B, Rediers H. Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J Fish Dis, 2011. 34(9): p. 643-61; PMID:21838709; http://dx.doi.org/ 10.1111/j.1365-2761.2011.01279.x [DOI] [PubMed] [Google Scholar]
- 40. Bjelland A.M, Fauske AK, Nguyen A, Orlien IE, Ostgaard IM, Sørum H. Expression of Vibrio salmonicida virulence genes and immune response parameters in experimentally challenged Atlantic salmon (Salmo salar L.). Front Microbiol, 2013. 4: p. 401; PMID:24391635; http://dx.doi.org/ 10.3389/fmicb.2013.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bej A.K, Patterson DP, Brasher CW, Vickery MC, Jones DD, Kaysner CA. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J Microbiol Methods, 1999. 36(3): p. 215-25; PMID:10379807; http://dx.doi.org/ 10.1016/S0167-7012(99)00037-8 [DOI] [PubMed] [Google Scholar]
- 42. Sigman M. and Luchette F.A., Cholera: something old, something new. Surg Infect (Larchmt), 2012. 13(4): p. 216-22; PMID:22913779; http://dx.doi.org/ 10.1089/sur.2012.127 [DOI] [PubMed] [Google Scholar]
- 43. Nair G.B, Ramamurthy T, Bhattacharya SK, Dutta B, Takeda Y, Sack DA. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin Microbiol Rev, 2007. 20(1): p. 39-48; PMID:17223622; http://dx.doi.org/ 10.1128/CMR.00025-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jones M.K. and Oliver J.D., Vibrio vulnificus: disease and pathogenesis. Infect Immun, 2009. 77(5): p. 1723-33; http://dx.doi.org/ 10.1128/IAI.01046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quintana F.J, Iglesias AH, Farez MF, Caccamo M, Burns EJ, Kassam N, Oukka M, Weiner HL. Adaptive autoimmunity and Foxp3-based immunoregulation in zebrafish. PLoS One, 2010. 5(3): p. e9478; PMID:20221429; http://dx.doi.org/ 10.1371/journal.pone.0009478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matthews J.L., Common diseases of laboratory zebrafish. Methods Cell Biol, 2004. 77: p. 617-43; PMID:15602935; http://dx.doi.org/ 10.1016/S0091-679X(04)77033-8 [DOI] [PubMed] [Google Scholar]
- 47. El Aidy S, Derrien M, Aardema R, Hooiveld G, Richards SE, Dane A, Dekker J, Vreeken R, Levenez F, Doré J, et al. , Transient inflammatory-like state and microbial dysbiosis are pivotal in establishment of mucosal homeostasis during colonisation of germ-free mice. Benef Microbes, 2014. 5(1): p. 67-77; PMID:24322881; http://dx.doi.org/ 10.3920/BM2013.0018 [DOI] [PubMed] [Google Scholar]
- 48. El Aidy S, Merrifield CA, Derrien M, van Baarlen P, Hooiveld G, Levenez F, Doré J, Dekker J, Holmes E, Claus SP, et al. , The gut microbiota elicits a profound metabolic reorientation in the mouse jejunal mucosa during conventionalisation. Gut, 2013. 62(9): p. 1306-14; PMID:22722618; http://dx.doi.org/ 10.1136/gutjnl-2011-301955 [DOI] [PubMed] [Google Scholar]
- 49. Thelemann C, Eren RO, Coutaz M, Brasseit J, Bouzourene H, Rosa M, Duval A, Lavanchy C, Mack V, Mueller C, et al. , Interferon-gamma induces expression of MHC class II on intestinal epithelial cells and protects mice from colitis. PLoS One, 2014. 9(1): p. e86844; PMID:24489792; http://dx.doi.org/ 10.1371/journal.pone.0086844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Do J.S, Asosingh K, Baldwin WM, 3rd, Min B. Cutting edge: IFN-gammaR signaling in non-T cell targets regulates T cell-mediated intestinal inflammation through multiple mechanisms. J Immunol, 2014. 192(6): p. 2537-41; PMID:24523506; http://dx.doi.org/ 10.4049/jimmunol.1303101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Brugman S, Witte M, Scholman RC, Klein MR, Boes M, Nieuwenhuis EE. T lymphocyte-dependent and -independent regulation of Cxcl8 expression in zebrafish intestines. J Immunol, 2014. 192(1): p. 484-91; PMID:24277695; http://dx.doi.org/ 10.4049/jimmunol.1301865 [DOI] [PubMed] [Google Scholar]
- 52. Wienholds E, Schulte-Merker S, Walderich B, Plasterk RH. Target-selected inactivation of the zebrafish rag1 gene. Science, 2002. 297(5578): p. 99-102; PMID:12098699; http://dx.doi.org/ 10.1126/science.1071762 [DOI] [PubMed] [Google Scholar]
- 53. van den Bogert B, de Vos WM, Zoetendal EG, Kleerebezem M. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol, 2011. 77(6): p. 2071-80; PMID:21257804; http://dx.doi.org/ 10.1128/AEM.02477-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hamady M, Widmann J, Copley SD, Knight R. MotifCluster: an interactive online tool for clustering and visualizing sequences using shared motifs. Genome Biol, 2008. 9(8): p. R128; PMID:18706079; http://dx.doi.org/ 10.1186/gb-2008-9-8-r128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol, 1999. 22(3): p. 434-44; PMID:10553296; http://dx.doi.org/ 10.1016/S0723-2020(99)80053-8 [DOI] [PubMed] [Google Scholar]
- 56. Guss A.M, Roeselers G, Newton IL, Young CR, Klepac-Ceraj V, Lory S, Cavanaugh CM. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J, 2011. 5(1): p. 20-9; PMID:20631810; http://dx.doi.org/ 10.1038/ismej.2010.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Caporaso J.G, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. , QIIME allows analysis of high-throughput community sequencing data. Nat Methods, 2010. 7(5): p. 335-6; PMID:20383131; http://dx.doi.org/ 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Haas B.J, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, et al. , Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res, 2011. 21(3): p. 494-504; PMID:21212162; http://dx.doi.org/ 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cole J.R., Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res, 2007. 35(Database issue): p. D169-72; PMID:17090583; http://dx.doi.org/ 10.1093/nar/gkl889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol, 2007. 73(16): p. 5261-7; PMID:17586664; http://dx.doi.org/ 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


